Background: Argonaute proteins associate with siRNAs and miRNAs to repress gene expression.
Results: The N-terminal lobe of Argonaute regulates slicer activity on target mRNAs.
Conclusion: RNA binding interactions with the Argonaute proteins determine whether the target mRNA is regulated through slicer-dependent or -independent pathway.
Significance: The study provides mechanistic understanding of how Argonaute proteins differentially mediate the RNAi and miRNA-mediated repression pathways.
Keywords: MicroRNA, Protein Domains, Ribonuclear Protein (RNP), Ribonuclease, RNA Interference (RNAi), RNA-Protein Interaction, Argonaute, PAZ, siRNA, Slicer
Abstract
Structural studies indicate that binding of both the guide RNA (siRNA and miRNA) and the target mRNA trigger substantial conformational changes in the Argonaute proteins. Here we explore the role of the N-terminal lobe (and its PAZ domain) in these conformational changes using biochemical and cell culture-based approaches. In vitro, whereas deletion (or mutation) of the N-terminal lobe of DmAgo1 and DmAgo2 had no effect on binding affinity to guide RNAs, we observed a loss of protection of the 3′ end of the guide RNA and decreased target RNA binding; consistent with this, in cells, loss of function DmAgo1 PAZ variant proteins (PAZ6 and ΔN-PAZ) still bind RNA, although the RNAs are shorter than normal. We also find that deletion of the N-terminal lobe results in constitutive activation of endogenous PIWI domain-based cleavage activity in vitro, providing insights into how cleavage activity may be regulated in vivo in response to different types of pairing interactions with the target mRNAs.
Introduction
MicroRNAs (miRNAs)3 and siRNAs are 21–23-nt small noncoding RNAs broadly involved in post-transcriptional gene regulation (1, 2). These short RNAs associate with multiple protein components and recognize target mRNAs with partial or perfect complementarities (3). At the core of this class of ribonucleoprotein (RNP) complexes is an Argonaute protein family member that is responsible for directly interacting with the short (guide) RNA, eliciting repressed gene expression for partially complementary target mRNAs (miRNA-like) and cleavage for fully complementary target mRNAs (siRNA-like) (4). Structural studies showed that the Argonautes interact with the small RNAs through conserved domains; the RNA 5′ end binds to the MID domain, the backbone phosphates of the “seed” region traverse, and engage the P-element Induced Wimpy Testis (PIWI) domain, and the 3′ end binds in the Piwi-Argonaute-Zwille (PAZ) domain (5–8).
In the miRNA pathway, biochemical studies have shown that loading of miRNA into the Argonaute is linked to the recruitment of an important downstream factor GW182 (9, 10). In support of this model was the observation that mutations in the MID domain that decreased miRNA binding also abrogated binding to GW182 (11, 12). We note, however, that studies focused on interactions between the miRNA and the PAZ domain reached different conclusions (13, 14); in particular, a set of Argonaute variants known as PAZ4 and PAZ6 (having four and six point mutations, respectively) appear to interact with GW182 in cells but not with miRNA, suggesting that miRNA binding and GW182 recruitment may be uncoupled (14, 15).
Biochemical and structural studies on interactions of the Argonautes with not only miRNA but also with target RNA yielded insight into how the discrepancy might be reconciled (16, 17). The observation that siRNA-dependent target RNA slicing activity (the kcat) was decreased by base mismatches in the 3′ region of guide RNA led to the proposal of a two-state model in which extended small RNA-target interactions (in the form of longer duplex) activate the slicer function of the Argonaute protein (18, 19). Corroborating these ideas, structures of Thermus thermophilus Argonaute that include both 21-nt DNA and target RNA strands with 12, 15, or 19 complementary bases, showed that the PAZ domain interaction with the 3′ terminus is lost when the region of complementarity exceeds 15 bp (17). These observations suggested that formation of the RNA duplex ultimately strains interactions between the 3′ terminus and the PAZ domain (because the helix cannot form with the 3′ end bound to the PAZ domain), resulting in global conformation changes. Consistently, structural studies of eukaryotic Argonaute MID and PIWI domains from Neurospora crassa (12), as well as recent structures of human Ago2 (7, 8), proposed that an adaptable surface loop in the PIWI domain interacts with the PAZ domain only when miRNA is bound.
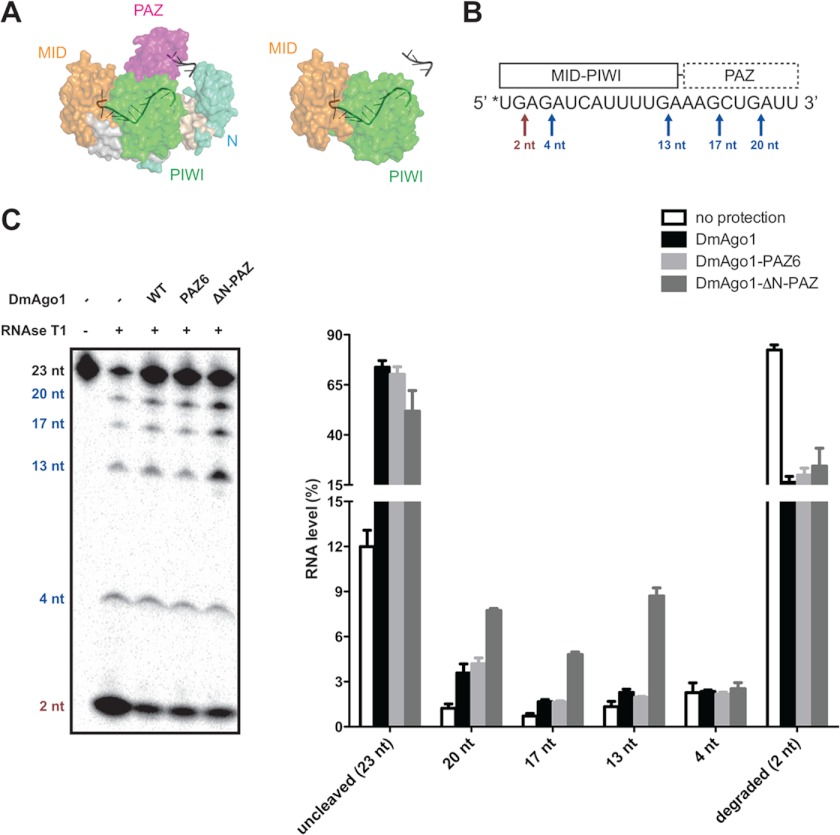
Interestingly, targets with imperfect complementarity also trigger a reorganization of the Argonaute protein; however, this reorganization fails to liberate the 3′ end of the guide RNA from the PAZ domain (6). Structural alignment of Argonautes with miRNA and imperfect or perfect targets shows prominent differences localized within the PAZ domain (see Fig. 1A).
FIGURE 1.
Binding of guide RNA to Drosophila Argonaute 1 and N-PAZ lobe variants. A, superimposition of T. thermophilus Argonaute structures with imperfect (green, 20-nt target with mismatches at positions 10 and 11; Protein Data Bank code 3F73) or perfect (magenta, 19 base pairs of complementarity; Protein Data Bank code 3DLH) target bound. B, schematic of DmAgo1 PAZ variants. Represented are DmAgo1 variants with N, PAZ, MID, and PIWI domains and linkers (L). DmAgo1-PAZ6 variant contains six point mutations (R352A, K353A, Y354A, T412A, Y413A, and L414A), whereas DmAgo1-ΔN-PAZ lacks the N and PAZ domains. C, competitive double filter binding assays (nitrocellulose and nylon). DmAgo1 was preincubated with radiolabeled 23-nt RNA and unlabeled 23-nt RNA, as homologous inhibitors. Shown is the protein-bound RNA captured on nitrocellulose with different concentrations of competitor. D, apparent Kd values of DmAgo1 variants showing the estimated dissociation constants of the DmAgo1 variants for 23-nt RNA based on competitive filter binding assay. E, competitive binding assay for DmAgo1 variants with radiolabeled 23-nt RNA in the presence of 5-, 15-, or 23-nt RNA competitors.
In this study, we developed in vitro and cell-based assays to study the interactions between DmAgo1 and DmAgo2, miRNA, and various target RNAs to investigate how the PAZ domain regulates interdomain interactions and slicer activity. Our biochemical data indicate that the interactions between the 3′ end of the miRNA and the N-PAZ lobe do not increase the binding affinity of DmAgo1 and DmAgo2 with the miRNA or siRNA but do stabilize binding to the target RNA. Interestingly, using a tethered reporter assay, the PAZ variants of DmAgo1 are still able to induce gene silencing. Moreover, deletion of the N-PAZ domains from either DmAgo1 or DmAgo2 results in constitutive activation of slicer activity in vitro.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Protein expression and purification from Escherichia coli was performed as previously described (11, 20). Drosophila melanogaster Argonaute variants were prepared by PCR from full-length clones of DmAgo1 (gi: 17647145) and DmAgo2 (gi: 24664664) as described (11): DmAgo1 (residues 1–950), DmAgo1-ΔN-PAZ (residues 538–950), DmAgo2 (without the Q-rich N-terminal region, residues 403–214), and DmAgo2-ΔN-PAZ (residues 816–1214). DmAgo1-PAZ6 and DmAgo1-YK were created by using QuikChange (Stratagene) to make the following mutations: DmAgo1-PAZ6 (R352A,K353A,Y354A,T412A,Y413A,L414A) and DmAgo1-YK (Y619L,K623E) (11, 13). Following protein purification, repeated passages over a size exclusion matrix decreased the amount of co-purifying nucleic acid (as assessed by the loss in A260 relative to A280) (21).
RNA Preparation
Bantam miRNA (23 nt) and shorter fragments (5 and 15 nt) were chemically synthesized (Dharmacon). Radiolabeled bantam was prepared using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (PerkinElmer Life Sciences) with the chemically synthesized RNA oligonucleotide. Radioactively labeled RNA was PAGE-purified, eluted with 300 mm NaOAc (pH 5.2), and EtOH-precipitated. For analysis of RNA extracted from immunoprecipitates, we treated the RNA samples with calf intestinal phosphatase and then radiolabeled at the 5′ end with PNK and visualized reaction products on 15% denaturing PAGE. Target RNA sequences were derived from 3′-UTR of HLHmΔ gene encompassing one bantam binding site. The RNAs were transcribed using T7 RNA polymerase and DNA templates with a T7 promoter (IDT DNA) (22). Following transcription, target RNAs were PAGE-purified and 3′ end labeled with T4 RNA ligase (Promega) and [32P]cytidine 3′,5′-bisphosphate, and PAGE-purified again.
RNA Sequences
The Bantam miRNA (dme-bantam-3p, MIMAT0000365) are: 23 nt, UGAGAUCAUUUUGAAAGCUGAUU; 15 nt, UGAGAUCAUUUUGAA; and 5 nt, UGAGA. The RNA sequence from HLHmΔ (natural bantam binding site underlined) was GTATTTAGTTGGTAGTTGATTTGATATGATGTGATGTTCCTGTGATAGTTCTTAAGTTCGTATTTTGTATTTGATCTCCCATAGGCTTACTGATTCATG. The RNA sequence from HLHmΔ (perfect bantam binding site underlined) was GTATTTAGTTGGTAGTTGATTTGATATGATGTGATGTTCCTGTGATAGTTCTTAAGAATCAGCTTTCAAAATGATCTCACATAGGCTTACTGATTCATG.
The 43-nt target RNA sequences were: perfect match, AGTTCTTAAGAATCAGCTTTCAAAATGATCTCACATAGGCTTA; 1–12 match, AGTTCTTAAGTTAGTCGAAACAAAATGATCTCACATAGGCTTA; 1–13 match, AGTTCTTAAGTTAGTCGAATCAAAATGATCTCACATAGGCTTA; 1–14 match, AGTTCTTAAGTTAGTCGATTCAAAATGATCTCACATAGGCTTA; 1–15 match, AGTTCTTAAGTTAGTCGTTTCAAAATGATCTCACATAGGCTTA; 1–16 match, AGTTCTTAAGTTAGTCCTTTCAAAATGATCTCACATAGGCTTA; 1–17 match, AGTTCTTAAGTTAGTGCTTTCAAAATGATCTCACATAGGCTTA; 1–18 match, AGTTCTTAAGTTAGAGCTTTCAAAATGATCTCACATAGGCTTA; and 1–19 match, AGTTCTTAAGTTACAGCTTTCAAAATGATCTCACATAGGCTTA.
The RNA primer for RNase protection analysis of RNA co-purified with DmAgo1 and variants was AGTTCTTAAGAATCAGCTTTCAAAATGATCTCACATAGGCTTA. The primers for quantitative RT-PCR in tethering assay were: firefly luciferase forward, 5′-CCAGGGATTTCAGTCGATGT-3′; firefly luciferase reverse, 5′-AATCTCACGCAGGCAGTTCT-3′; Renilla luciferase forward, 5′-GGTATGGGCAAATCAGGC-3′; and Renilla luciferase forward, 5′-GCACCCCAATCATGGCCG-3′.
Filter Binding Assay
The assay was performed as previously described (11, 23). Briefly, synthetic 23-nt RNA was 5′ end-labeled with polynucleotide kinase and [32P]ATP. Subnanomolar concentrations of radiolabeled 23-nt RNA were premixed with unlabeled competitor RNA and then incubated with purified protein for 10 min. For double filter binding, the samples were added to a dot blot apparatus (Bio-Rad) with nitrocellulose and nylon membranes (top to bottom). Each membrane was dried and exposed to a phosphorimaging screen and quantified. Binding data from titrations with competitor RNA were fit to competition curves by Prism to estimate apparent dissociation constants (Kd values).
In Vitro RNase Protection Assay
5′-Radiolabeled 23-nt RNA was preincubated with Argonaute variant proteins to form ribonucleoprotein complexes in 25 mm HEPES-KOH (pH 7.5), 50 mm KOAc, 5 mm Mg(OAc)2, and 5 mm DTT. RNase T1 (0.2 unit/ml; Fermentas) was then added, and the mix was incubated for 1 min, resulting in a partial digest. The reaction was stopped by adding yeast RNA (final concentration, 0.5 mg/μl) and GMP (final concentration, 10 mm) and was followed by phenol-chloroform extraction. The isolated RNA was resolved by denaturing PAGE, and the gel was exposed to a phosphorimaging screen and quantified.
Cell Culture, Transfection, and Immunoprecipitation
S2 cell cultures were grown in Express Five SFM medium (Invitrogen) with supplements: 100 units/ml penicillin and streptomycin (Cambrex BioScience) and 45 ml/500 ml l-glutamine (200 mm, Invitrogen). Transfections were performed with Effectene (Qiagen) for luciferase and immunoprecipitation assays. For immunoprecipitation, transfection of 4 μg of plamids encoding FLAG-tagged DmAgo1 wild type and variants were performed in 15-cm plates. After 48 h, the cells were harvested and spun in 50-ml conical tubes at 150 × g for 5 min. The cell pellet was washed once with PBS and resuspended in hypotonic lysis buffer (10 mm, Tris-HCl, pH 7.5, 10 mm KOAc, 1 mm DTT, protease inhibitor EDTA-free) for lysis. The lysates were clarified by spinning at 5000 × g for 10 min at 4 °C. Cleared lysate was incubated with prewashed anti-FLAG M2-agarose beads (Sigma) and then rotated in the cold room for 15 min. The beads were washed three times with wash buffer (20 mm HEPES-KOH, pH 7.5, 150 mm KCl, 5 mm DTT) and eluted with 3× FLAG peptide (Sigma) for analysis of protein and RNA content. Western blot analyses of FLAG-tagged proteins were performed using HRP-conjugated antibody against FLAG (Sigma). For GW182, we used rabbit anti-GW182 primary antibody and HRP-conjugated secondary goat anti-rabbit antibody (Promega). GFP-tagged GW182 was detected using a primary antibody against GFP (JL-8; Living Colors, Clonetech; batch no. 632381) and secondary HRP-conjugated goat anti-mouse IgG2a (Jackson ImmunoResearch, catalog no. 115-035-206).
Analysis of RNAs Co-purified with Argonaute Proteins from Cells
For detection of small RNAs that co-purified with Argonaute proteins, we extracted RNAs from the IP, removed the 5′ phosphate with calf intestinal phosphatase, and radiolabeled with polynucleotide kinase and [32P]ATP. To detect partially degraded bantam miRNA species in the co-purified RNAs, we also performed RNase protection assays using the mirVANA miRNA detection kit (Ambion) following the manufacturer's protocol. The extracted RNA was hybridized overnight at 16 °C in hybridization buffer with radioactively body-labeled target RNA (43 nt) containing a perfectly complementary bantam binding site. The RNA was then treated with RNase A/T1, phenol-chloroform-extracted, and ethanol-precipitated. The pellet was resuspended in loading dye, resolved on denaturing PAGE, and exposed to a phosphorimaging screen.
Tethering Assay to Assess Regulation of Reporter by DmAgo1 Variants
The assays were conducted as described (11). S2 cells were co-transfected with DmAgo1 variants or GW182 fused with amino acids 1–22 of bacteriophage lambda N protein, and a luciferase reporter containing five BoxB elements in the 3′-UTR. Transfections were performed in 24-well plates with 5 ng of firefly luciferase reporter plasmid, 5 ng of Renilla luciferase control plasmid and 200 ng of plasmid coding for DmAgo1 variants or GW182 with N-terminal fusion. For normalization, control experiments were conducted by co-expression of a nonfused form of each effector protein. After 48 h, luciferase levels were measured using the dual luciferase luminometer detection reagents (Promega), and mRNA levels were assessed by quantitative RT-PCR.
RNA Cleavage Assays
The assays were conducted as previously described (20). Recombinant DmAgo1 or DmAgo2 variants (500 nm) were incubated with bantam miRNA (1 mm) in cleavage buffer (20 mm HEPES-KOH, pH 7.5, 50 mm KCl, 5 mm DTT, 5 mm Mg(OAc)2) and then added to radiolabeled target RNAs ([32P]cytidine 3′,5′-bisphosphate-labeled at the 3′ end). After cleavage (time course or fixed time of 1 h), the reactions were stopped by adding 2× formamide loading buffer and resolved on denaturing PAGE.
RESULTS
The PAZ Domain Does Not Contribute to Small RNA Binding Affinity of Argonautes in Vitro
Interested in the relationship between RNA binding by the Argonaute PAZ domain and distinct downstream functions, we focused on the Drosophila Argonaute proteins, DmAgo1 and DmAgo2, implicated in miRNA-mediated gene silencing and RNAi, respectively (24). We purified a collection of recombinant MBP-tagged Argonautes expressed in E. coli that included: full-length WT, a mutant in the 3′ RNA binding site (PAZ6), and N-terminal and PAZ domains deleted (ΔN-PAZ) (11, 13) (Fig. 1B and data not shown).
To assess the impact of mutation of the PAZ domain on binding affinity of the Argonaute proteins to small RNA, we carried out filter binding assays with purified Argonuate proteins and radiolabeled bantam miRNA 23 nt in length by standard procedures (23); importantly, DmAgo1-bound miRNAs were not degraded during the time course of the experiment (data not shown). To quantify the strength of the protein-RNA interaction, we conducted competitive binding assays by titration of unlabeled homologous competitor RNA and estimated the binding affinity of DmAgo1 for the bantam miRNA as 2.9 ± 0.3 nm (Fig. 1C). We were surprised to observe that the DmAgo1-PAZ6 and DmAgo1-ΔN-PAZ variants exhibited equivalent binding affinities relative to the wild type (2.3 ± 1.1 and 2.7 ± 1.1 nm, respectively) (Fig. 1D). We also investigated DmAgo2 binding to guide RNA and the effects of an equivalent deletion. DmAgo2 exhibited a somewhat higher dissociation constant (10.7 ± 1.0 nm) in binding guide RNA compared with DmAgo1, but deletion of the N-terminal lobe again had no effect on binding (9.5 ± 1.3 nm) (Fig. 1D). These results indicate that, in vitro, the interactions between the 3′ end of the miRNA and the PAZ domain make no substantial contribution to the overall equilibrium binding constants of Argonautes with their small RNAs.
Because DmAgo1 associates with miRNAs ∼21–23 nt in length in vivo and we previously documented length specific affinity (11), we further examined whether the PAZ domain contributes to binding affinities for certain length substrates. In this case, we compared DmAgo1 wild type and the PAZ variant proteins in competition binding assays with radiolabeled bantam miRNA and competitor (unlabeled) RNAs of length 5, 15, and 23 nt (Fig. 1E). Based on known structural features, we suspected that the smaller RNAs would anchor their 5′ ends in the MID domain of the Argonaute protein but would fail to engage the PAZ domain. Therefore, if the interaction of the PAZ domain with the 3′ end of the guide RNA is important for binding, only the wild type protein might exhibit a preference for the full-length RNA. Contrary to these expectations, the full-length and PAZ variants exhibited binding patterns very similar to one another, indicating that, in vitro, PAZ domain interactions with the 3′ end do not confer length specificity on Argonuate RNA binding (Fig. 1E).
PAZ Domain Protects the 3′ End of the Guide RNA from Degradation in Vitro
Although our data indicated that the N-terminal and PAZ domains fail to make substantial contributions to binding affinity, structural studies have indicated that the guide RNA 3′ end is anchored by the PAZ domain (Fig. 2A) (6). To validate that guide RNAs bind in the predicted conformation and to probe the role of the PAZ domain in interactions with the 3′ end of the guide RNA, we used RNase protection assays to ask whether the PAZ domain protects bound guide RNA from nuclease attack. For these experiments, we preincubated 5′ end-labeled bantam miRNA (23 nt) with DmAgo1 wild type and PAZ variants and treated the complexes with RNase T1 for partial RNA digestion. Because RNase T1 specifically cleaves at the 3′ side of guanosine residues, cleaved bantam yielded five distinct products (Fig. 2B). In this assay, the unprotected digestion reaction (without protein) resulted in the accumulation of completely digested products, whereas protection by DmAgo1 wild type or PAZ mutants resulted in greater amounts of uncleaved or partially cleaved RNA (Fig. 2C). Examining the partial cleavage products of RNase T1, DmAgo1 wild type and the DmAgo1-PAZ6 variant showed equivalent protection of bantam miRNA at all positions, whereas the DmAgo1-ΔN-PAZ variant showed increased levels of cleavage at positions 20, 17, and 13 nt, but not at 4 nt. These data indicate that an Argonaute protein missing the N-terminal lobe can still protect the 5′ end of guide RNA, but not the 3′ end (Fig. 2C). Similar experiments using RNase A yielded equivalent results (data not shown).
FIGURE 2.
Argonaute N-PAZ lobe protects the 3′ end of guide RNA. A, surface representation of T. thermophilus Argonaute structure in complex with 21-nt DNA (Protein Data Bank code 3DLH). Light blue, N; magenta, PAZ; yellow, MID; green, PIWI; gray, bound DNA (only nucleotides 1–11 and 18–21 are traced in structure). Wild type is shown on the left with theoretical deletion of N-PAZ-lobe on the right. B, schematic of RNase T1 protection by DmAgo1 variants: 5′ radioabeled (*) bantam miRNA (23 nt) is depicted with known interactions to the PAZ, MID, and PIWI domains. RNase T1 cleavage sites, which occur 3′ of guanosine residues, are marked with arrows. The red and blue arrows indicate cleavage sites protected by the MID-PIWI lobe and N-PAZ lobe, respectively. C, DmAgo1-miRNA complex subject to RNase T1 protection assay. 5′-Labeled bantam miRNA was incubated with excess amount of purified DmAgo1 variants and was subjected to partial digestion by RNase T1. The amount of each RNA species was normalized with respect to total RNA.
PAZ Domain Variants Bind Short RNAs in Vivo
Given the accumulated in vitro observations regarding the behavior of the PAZ variant Argonautes (that small RNA binding is not compromised in these variants but that the 3′ end of the bound RNA is vulnerable to endonucleolytic cleavage), we hypothesized that in cells, these PAZ variants might be bound to RNA of some sort. More specifically, we speculated either that truncated miRNAs or other nonspecific cellular RNAs might be found in place of full-length canonical miRNA.
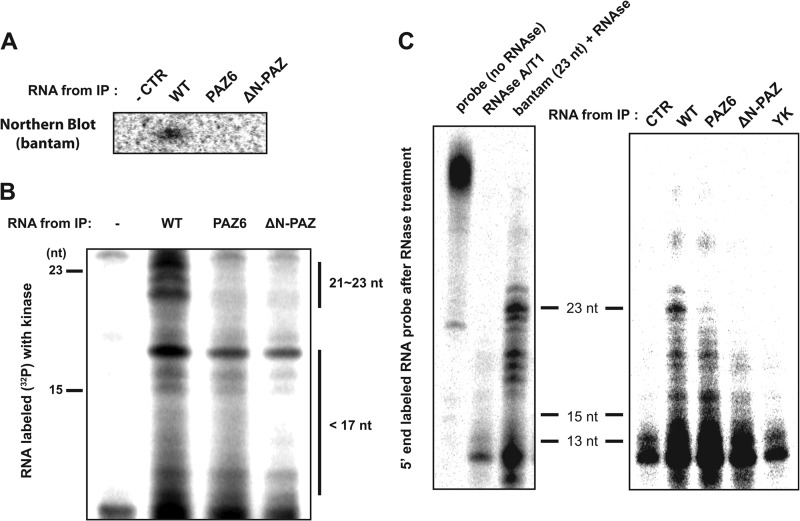
To explore this possibility, we examined the small RNA species associated with DmAgo1 and its variants in cell culture. We expressed FLAG-tagged DmAgo1 variants in S2 cells and directly analyzed the RNA that co-immunoprecipitated (co-IPed) with the tagged proteins. Northern blot analyses of the RNAs with probes to bantam revealed full-length 23-nt bantam associated with DmAgo1 but not with either PAZ domain mutant (Fig. 3A). Moreover, we could not detect any truncated forms of bantam using this assay consistent with earlier studies (13).
FIGURE 3.
Association of Argonaute N-PAZ variants with small RNAs. A, FLAG-tagged DmAgo1 variants (WT, PAZ6, and ΔN-PAZ) were expressed in S2 cells and immunoprecipitated. Co-purification of bantam miRNA was assessed by Northern blot analysis. A mock transfection served as a negative control. B, RNA that co-purified with DmAgo1 variants was radiolabeled with [γ-32P]ATP and T4 polynucleotide kinase and resolved on denaturing PAGE and exposed to phosphoimaging screen. C, detection of small RNA species associated with DmAgo1 in co-IP by RNase protection. RNA associated with DmAgo1 wild type or variants was extracted, hybridized with 43-nt radioactive body-labeled RNA containing a perfect bantam binding site, and digested with RNase specific for single-stranded RNA. The digestion products were then resolved by PAGE and exposed to a phosphorimaging screen. As a control, the probe was digested in the presence of 1 pmol of bantam miRNA (23 nt) or without any RNA. CTR, control.
To detect with higher sensitivity shorter RNAs that are possibly associated with Argonaute proteins in the cell, we performed 5′ end radioactive labeling of RNAs isolated from immunoprecipitated DmAgo1 wild type and PAZ variants and resolved them by denaturing PAGE (Fig. 3B). We observed association of ∼21–23-nucleotide-long RNAs only with DmAgo1 wild type, whereas smaller RNA species (15–17 nt) co-purified with wild type and PAZ variants, albeit to a lesser extent with the variants. Next, to more sensitively probe for bantam miRNA, we performed an RNase protection assay where the same extracted RNA was hybridized directly with anti-bantam body labeled probes (43 nt). The hybridized samples were treated with single-stranded RNA specific RNase, and the remaining regions of probes that hybridized to bantam (full-length or truncated) were resolved by PAGE. We observed that RNAs from the IP of DmAgo1 wild type contained full-length as well as partially degraded bantam miRNA, whereas the PAZ6 and ΔN-PAZ variants co-IPed only with partially degraded bantam miRNA, again to a lesser extent than wild type (Fig. 3C). Importantly, no bantam (either full-length or truncated) could be detected bound to the previously characterized 5′-miRNA binding site mutant (DmAgo1-YK) (11). Together, these results suggest that, in cells, PAZ variant proteins predominantly bind RNA species shorter then ∼21 nt in length.
Target RNA Binding Is Negatively Affected by Mutations in the PAZ Domain
We next asked whether the PAZ domain plays a critical role in binding interactions between Argonautes and their mRNA targets. We first preformed miRNA ribonucleoprotein complexes (miRNPs) with bantam miRNA and DmAgo1 proteins (wild type, PAZ6, and ΔN-PAZ proteins) and then performed in vitro filter binding assays with radiolabeled target RNAs (Fig. 4A). For the target RNAs, we prepared radiolabeled RNA 43 nt in length with complementarity to nucleotide positions 1–12 of the bantam miRNA. As a negative control, we prepared miRNP complexes with miR-279, which is not complementary to the chosen target RNAs. Duplex formed between the target RNA and positions 1–12 of the miRNA (containing the “seed” region) is predicted to be short enough to permit binding of the 3′ end of bantam miRNA to the PAZ domain (17). As anticipated, we observed minimal target binding to the control complexes DmAgo1-wild type:miR-279 with substantial binding to DmAgo1-wild type:bantam (Fig. 4B). Moreover, DmAgo1-wild type:bantam showed substantially higher target binding affinity than both the PAZ6 and ΔN-PAZ mutants (Fig. 4B). Because Argonautes can induce cleavage of target RNAs that can in turn affect binding affinities, we performed the assays in magnesium-free buffer in which DmAgo1 and the PAZ variants do not exhibit cleavage activity. Second, to determine whether magnesium alters the binding affinity of the complexes, we conducted filter binding assays with magnesium-supplemented buffers at low temperatures where cleavage was minimized; these experiments indicate that magnesium has a minimal effect on the relative affinities of DmAgo1 wild type and PAZ mutants for their target RNAs (data not shown).
FIGURE 4.
Target RNA binding and silencing are affected by mutation of the N-PAZ lobe. A, schematic of target RNA binding. Drosophila Argonaute variants were preincubated with saturating amounts of guide RNA and radiolabeled target RNA was added to the complex. B, target binding of DmAgo1 with bantam versus miR-279. DmAgo1 bound to either bantam or miR-279 was titrated in filter binding assay with radiolabeled 43-nt target RNA, which base pairs with positions 1–12 of bantam. Binding was performed in the absence of magnesium to prevent cleavage. C, tethered luciferase reporter assay for evaluating silencing activity of Argonaute variants. DmAgo1 variants and GW182 were co-expressed as λN fusion proteins in S2 cells with a luciferase reporter containing box B elements in the 3′-UTR. DmAgo1-YK and GW182 served as negative and positive controls, respectively. D, Western blot analysis of immunoprecipitated DmAgo1 variants and co-purified GW182 protein. FLAG-tagged DmAgo1 variants were expressed, immunoprecipitated, and then probed using anti-FLAG antibody. The asterisk indicates a nonspecific band that served as loading control. Levels of co-purified endogenous GW182 were determined using GW182 antibody. E, GFP-tagged GW182 and FLAG-tagged DmAgo1 variants were co-expressed in S2 cells. Anti-FLAG antibody was used to pull-down the Argonaute variant, and Western blot analysis was performed with anti-GFP antibody to determine the extent of association with GW182. CTR, control.
In Vivo Characterization of Argonaute PAZ Variant Silencing Activity
We next investigated the role of the PAZ domain of DmAgo1 in miRNA-mediated post-transcriptional silencing of target mRNA. Previous studies showed that mutations in the DmAgo1 MID or PIWI domain that abolish miRNA binding also abrogate gene regulatory function and GW182 interactions (9–12). However, other studies suggested that Argonaute binding to miRNAs is independent of GW182 interactions; two DmAgo1 PAZ mutants (PAZ6 and PAZ4) appeared to lose the ability to bind miRNA but retained interactions with GW182 (13, 14). A recent study has proposed a GW182-independent mechanism for miRNA-mediated gene silencing (25) although this view is still subject to discussion (26).
To approach these questions, we next asked whether the DmAgo1 PAZ variant proteins are capable of potentiating the down-regulation of gene expression on a given target mRNA. Based on our results indicating that the PAZ variants will not efficiently bind target RNA (Fig. 4B), we assessed the gene regulatory function of the PAZ variants utilizing a previously described tethered luciferase reporter system (Fig. 4C) (27, 28). In this assay, DmAgo1 (or variants) was expressed as a fusion protein with the λN peptide, and the reporter gene contained BoxB elements in the 3′-UTR that directly recruit the fusion proteins; in this experiment, there are no miRNA binding sites in the 3′-UTR of the reporter gene. As a negative control, we again used the DmAgo1-YK variant, which is defective in small RNA binding and GW182 recruitment and as anticipated observed no repression of luciferase reporter gene expression. As a positive control, we utilized a tethered GW182 construct that elicited robust (∼20-fold) down-regulation of gene expression. Interestingly, we found that all three DmAgo1 proteins (WT, PAZ6, and ΔN-PAZ) exhibited comparable levels of repression (∼5-fold) of luciferase protein expression in the tethered assay (Fig. 4C), with similar in magnitude decreases in mRNA levels (data not shown).
Next, we asked whether DmAgo1 and the PAZ variants (all of which are active in silencing) interact with GW182. We conducted co-IP experiments by expressing and isolating FLAG-tagged DmAgo1 (or variants) in S2 cells and then probing for co-purified endogenous GW182 (Fig. 4D). In this experiment, we observed effective co-IP of GW182 with DmAgo1 and DmAgo1-PAZ6, but not with DmAgo1-ΔN-PAZ or with the DmAgo1-YK mutant. However, under conditions where GW182 is overexpressed (GFP-tagged GW182), the ΔN-PAZ variant could be seen to loosely interact with GW182, although to a considerably lower extent than for the DmAgo1 wild type or PAZ6 variant (Fig. 4E).
The PAZ Domain Regulates the Slicer Activity of the Argonaute
We next investigated the impact of the PAZ domain on the activation of endonuclease activity by the Argonaute proteins using in vitro cleavage assays (11, 20). To confirm that DmAgo1 only cleaves target RNAs with perfect complementarity to guide RNA, we formed catalytically competent RNP complexes with bantam and performed cleavage assays with two target RNAs: one with perfect complementarity to bantam and the other a natural bantam site containing mismatches (Fig. 5A). The assays were performed under single turnover conditions where concentrations of catalytic RNP are in substantial excess (500 nm) relative to target RNA (<1 nm) and relative to the measured binding constants (Fig. 4B). As anticipated, time-dependent accumulation of cleaved product was detected only for the perfectly matched target; cleavage assays of DmAgo2 yielded similar results (Fig. 5A). We next evaluated the cleavage of various targets with 12 nt of complementarity to the miRNA (at various positions along the miRNA) and observed that such targets are poorly cleaved if at all, despite the presence of base pairing at the canonical Argonaute-mediated cleavage site between positions 10 and 11 nt of the miRNA (Fig. 5B).
FIGURE 5.
Slicer activity is activated on release of guide RNA from the Argonaute N-PAZ lobe. A, cleavage assay with DmAgo1 and DmAgo2. Argonaute protein is preincubated with bantam and added to radiolabeled target RNA that contains either a complete (perfect) or a partial (natural) complementary bantam binding site. Time-dependent accumulation of cleaved product was observed only for target with perfect match. B, cleavage assay with DmAgo1 conducted on targets with 12 consecutive base pairs across the bantam miRNA: 2–13 to 9–20 bp. A target with no base pairing served as a negative control (CTR, sense to bantam). A target RNA with perfect bantam site (1–23 bp, antisense to bantam) served as a positive control. C, RNA slicing activity of DmAgo1 was assessed for targets having different extents of complementarity: from 1–12 to 1–19 bp, and, finally, a complete match (1–23 bp). Cleavage products were observed when base pairing exceeded 15 nt, and cleavage activity was fully activated when pairing reached 17 nt. D, DmAgo1 variants in complex with bantam were added to target RNAs containing 1–13 through 1–16 pairings with bantam. DmAgo1-ΔN-PAZ showed enhanced slicer activity on 1–16 target, which was marginally cleaved by DmAgo1 or DmAgo1-PAZ6. E, potential base pairing between bantam and natural targets. F, extent of cleavage of natural targets with DmAgo1 and variants.
The absence of cleavage of target RNA with 1–12 nt matched to bantam suggested that cleavage of target RNA may be inhibited when the duplex is not long enough to induce disengagement of guide RNA from the PAZ domain. This idea was tested by increasing the extent of base pairing that extends from the 5′ seed region of the miRNA (Fig. 5C). Although virtually no cleavage was observed for targets with pairing in the range from positions 1–12 to 1–15, extension of base pairing beyond position 15 resulted in a sharp increase in the efficiency of target cleavage (Fig. 5C). We next compared the cleavage activity of DmAgo1, PAZ6, and ΔN-PAZ variants on targets with base pairing from positions 1–13, 1–14, 1–15, and 1–16 (Fig. 5D). Notably, at equivalent concentrations, the DmAgo1-ΔN-PAZ variant showed substantially increased cleavage on multiple targets when compared with DmAgo1, although no increases in cleavage were observed for DmAgo1-PAZ6. We observed similar up-regulation of DmAgo2 slicer activity when its N-terminal lobe containing the PAZ domain was deleted (data not shown).
We next investigated the regulation of DmAgo1-WT, PAZ6, and ΔN-PAZ slicer activities on known target sites for bantam (Fig. 5E). We prepared three targets: a fully complementary target as a positive control, a computationally predicted bantam target site found in the 3′-UTR of mRNA of atypical protein kinase C (aPKC) (with abundant mismatches and modest seed complementarity), and an experimentally verified bantam site in the 3′-UTR of Head involution defect (Hid) mRNA with nearly perfect complimentarity and a single mismatch at the site of cleavage (position 11) (29–31). Interestingly, target RNA with a natural Hid bantam site was cleaved with similar efficiency and at the same position, as the perfectly matched target by DmAgo1, and even more efficiently by DmAgo1-ΔN-PAZ (Fig. 5F). More importantly, in support of a model for the PAZ domain regulating cleavage activity, the DmAgo1-ΔN-PAZ variant cleaved all three targets more efficiently than DmAgo1. Together, the results are consistent with the idea that in the absence of the PAZ domain, the Argonaute protein is more easily activated.
DISCUSSION
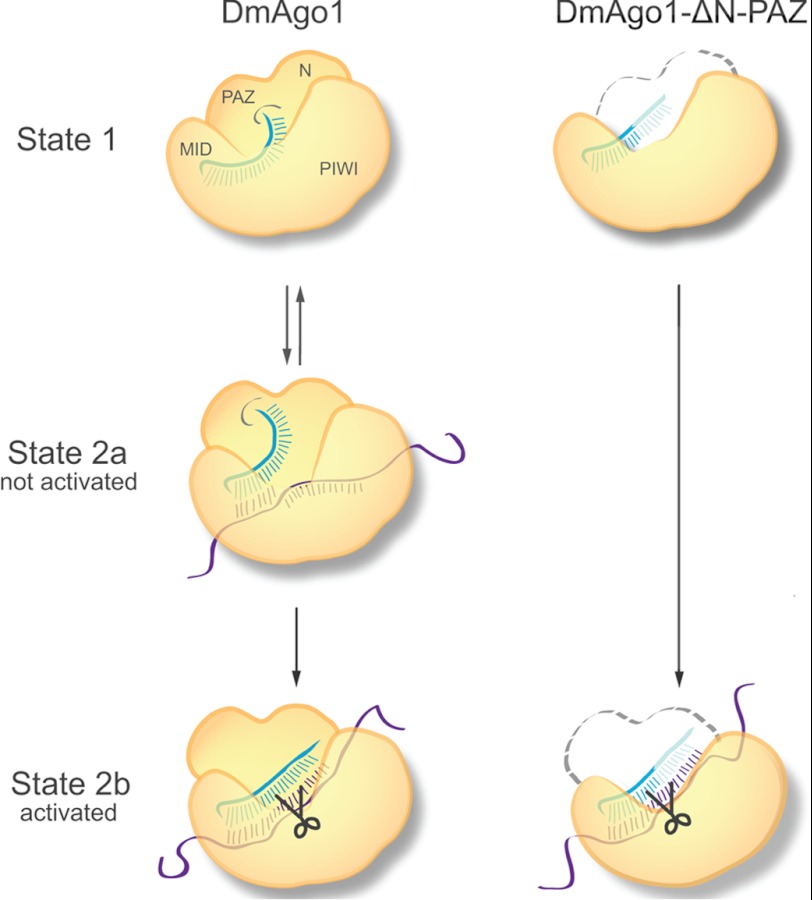
The results presented here provide insights into the role of the N-terminal lobe (N-PAZ) in Argonaute protein function. Although we focus principally on the likely contributions of the PAZ domain (given its clear role in guide RNA 3′ end binding (6, 13)), our conclusions cannot distinguish between a contributory role played by the N domain itself. Our in vitro assays showed that although the N-PAZ lobe does not contribute to small RNA binding affinity (Fig. 1D), the lobe does play a critical role in the maintenance of small RNA integrity (Fig. 2C) and target RNA binding (Fig. 4B). In cells, the PAZ mutants were deficient in full-length bantam miRNA binding but showed some association with <20 nt RNAs (Fig. 3, B and C). In a tethered reporter assay, we observed that N-PAZ mutants are capable of silencing tethered mRNAs (Fig. 4C). We also find that the N-PAZ lobe plays an important role in regulating the catalytic activity (Fig. 5, D and F). These results are broadly consistent with the two-state model for activation of Argonaute slicer activity (18); loss of contact between the miRNA 3′ end and the PAZ domain (as a result of extended duplex formation on an siRNA-like target or by mutation of the PAZ domain) leads to activation of the slicer function of the Argonaute protein.
In our in vitro binding assays with Argonaute and guide RNA, the observation that the N-PAZ lobe makes no energetic contribution to binding affinity (Fig. 1D) (defined by differences in ΔG) was initially surprising given that the 3′ end of the miRNA is known to directly engage this region (6). The possibility of miRNA binding to Argonaute in a site different from that shown in the structure was first ruled out by showing in in vitro RNase protection assays that deletion of the PAZ domain increases the accessibility of this region to nucleolytic attack (Fig. 2C). The surprising binding results were rationalized by arguing that the loss of the N-PAZ lobe increases the overall entropy of the RNA-protein complex, balancing the losses in enthalpy from deletion of the N-PAZ-lobe-small RNA interaction. Importantly, target RNA binding by Argonaute-guide RNA complexes was substantially impacted by the loss of the N-PAZ lobe (Fig. 4B). These results can similarly be reconciled by entropic and structural considerations (17, 32); in the ternary complex where the guide-target duplex has 12 base pairs, the guide-target duplex traverses between the N-PAZ and MID-PIWI lobes of the Argonaute protein where backbone phosphates at positions 8–10 of the guide strand form intermolecular interactions with side chains of the N-PAZ lobe. Initial binding of guide RNA to the PAZ domain preorganizes the bi-lobal channel (with an associated loss of entropy) to facilitate nucleation in the seed region and duplex propagation toward the 3′ end. Mutation or deletion of the N-PAZ lobe disrupts this fixed conformation, thus reducing duplex stabilization and, hence, target RNA binding. These data support much experimental and bioinformatic data indicating that “seed” pairing plays a critical role in Argonaute function (32–34).
Because our in vitro binding assays showed that the deletion of the N-PAZ lobe had no significant impact on binding affinity of DmAgo1 to small RNA (Fig. 1D), it seemed likely that the N-PAZ mutants might be associated with some RNAs in the cell. It did seem likely, however, that DmAgo1-PAZ6 or the ΔN-PAZ variants might fail to be efficiently loaded with miRNAs in the canonical pathway because the PAZ domain was proposed to be critical in Argonaute loading (35). Despite such predicted poor loading (13, 14), we wondered whether the N-PAZ variants may nevertheless be bound to miRNAs, their fragments, or other short RNAs in the cell via their strong interactions with the MID and PIWI domains. Such bound RNA might be truncated from the 3′ end (Fig. 2C) by nucleases present in the cell. As such, either class of RNA (miRNA or nonspecific RNA) might be difficult to detect by Northern analysis. These ideas were corroborated by the finding that although RNA does co-purify with DmAgo1-PAZ6 and DmAgo1-ΔN-PAZ in the cell, their abundance is lower, and their length is truncated (Fig. 3, B and C). Consistent with these ideas, in Schizosaccharomyces pombe, when Dicer is absent, Argonaute 1 was shown to associate with diverse RNAs representing the degradation products of abundant transcripts (36). Similarly, recent structural studies found that recombinant human Ago2 purified with endogenous cellular RNAs (7, 8).
The deficiencies of the Argonaute PAZ variants in target mRNA binding (Fig. 4B) and in GW182 binding (Fig. 4D) that we report here are consistent with previous findings that the DmAgo1 PAZ6 variant cannot rescue miRNA-mediated gene silencing in a complementation assay (13). Interestingly, however, when we bound the various Argonaute proteins to mRNAs in a tethered reporter assay, we observed that DmAgo1 wild type, PAZ6, and ΔN-PAZ variants are all able to silence gene expression (Fig. 4C). On the other hand, the YK mutant that showed no association with small RNA did not induce repression (Fig. 4C). Together, these data suggest that binding of RNA (even RNA shorter than ∼23 nt) is critical for the activation of gene silencing by DmAgo1, whereas engagement of miRNA with the N-PAZ lobe per se may be dispensable for this activation (Fig. 4C). These results provide a possible explanation for why certain PAZ domain variants showed diminished binding of full-length miRNAs and a loss of silencing activity (in a nontethered assay) but still could bind to GW182 and localize to P-bodies (13, 14). The failure of the N-PAZ variants to fully complement an Argonaute deletion in vivo likely results from poor loading of the Argonaute proteins with the guide RNA, as well as from diminished binding to the mRNA targets; in a tethered reporter assay, these issues are largely eliminated. These ideas are consistent with a recent report establishing an important role for the PAZ domain in the loading of the Argonaute proteins with duplex RNA from the Dicer processing step (37).
Interestingly, despite the robust silencing that we observe in the tethering assay, we find which whereas the PAZ6 variant binds GW182 at a level similar to wild type, only very weak binding is observed for the ΔN-PAZ variant (Fig. 4, D and E). Given the documented importance of GW182 in the miRNA-mediated repression pathway (10, 13), how does DmAgo1-ΔN-PAZ exhibit robust repression in our assay? One possible explanation is that even weak interactions between the MID-PIWI domains of DmAgo1 and GW182 may be sufficient to confer robust silencing in the tethering system (13); the PAZ domain would provide additional, but nonessential, stabilization to the Argonaute-GW182 complex. Alternatively, silencing may be mediated through a pathway that is independent of GW182 protein as recently reported (25). It is also possible that the tethering assay fails to recapitulate key aspects of authentic miRNA-mediated silencing.
Although DmAgo1 is principally involved in miRNA-mediated silencing (in which targets typically do not undergo Argonaute-mediated cleavage), DmAgo1 is known to be capable of cleaving target RNA in vitro given extensive pairing interactions with the guide RNA (Fig. 5F and Ref. 24). Although structural data made clear predictions about whether or not the PAZ domain would be engaged by the 3′ end of the miRNA on different targets (17), the role of the N-PAZ lobe in the activation of the slicer function was not fully understood. Here we provide biochemical data to show that slicer activity is activated when duplex length reaches 15–16 bp (Fig. 5C) or when the PAZ domain is deleted (Fig. 5, D and F), consistent with previous findings (17, 19). We also note smaller cleavage products for target RNAs with 1–14 and 1–15 bp pairs of complementarity with the guide RNA; the appearance of these products suggests that for these target RNAs, there is an imperfect juxtaposition of catalytic residues with the normal cleavage site, perhaps because these duplex RNAs are not long enough to effectively engage the PIWI domain. The observed increase in target cleavage on deletion of the N-PAZ lobe likely results from faster catalysis (kcat) because binding to the target RNA is known to be diminished (Fig. 4B).
Small RNA binding to the Argonaute proteins is central to overall gene silencing. Different conformations of the Argonaute protein are induced by binding of the small RNA to discrete semi-mobile MID, PIWI, and PAZ domains. Our results provide biochemical evidence to support the two-state model proposed by Tomari and Zamore (18) and provide mechanistic insight into how the Argonaute proteins differentiate their behavior in the miRNA and siRNA pathways. Upon binding of the target RNA, the characteristics of the RNA duplex (miRNA-like or siRNA-like) and their interactions with the Argonaute protein modulate further changes in the protein conformation. Our results provide support for the idea that interactions between guide and target RNAs are not simply a substrate selection mechanism but are also an active component of Argonaute protein function in deciding whether the target mRNA is repressed by RNA cleavage (siRNA-like) or by a slicer-independent pathway (miRNA-like) (Fig. 6). In addition, because the Argonaute proteins are differentially activated by distinct guide-target RNA interactions, distinct proteins might be recruited to facilitate downstream events. It also seems likely that the PAZ domain-mediated regulation of slicer activity plays other roles in the life cycle of the miRNA; for example, the slicing-competent conformation of DmAgo1 could be utilized in the cell for maturation, degradation, or protection of the miRNAs (38–41).
FIGURE 6.
Model describing the impact of RNA binding on functionally relevant conformational changes in Argonaute. State 1, Argonaute-guide RNA complex with 5′ end bound to MID domain and 3′ end bound to the PAZ domain. State 2a, Argonaute-guide RNA bound to miRNA-like target. Argonaute undergoes conformational change but is not activated for cleavage as guide RNA 3′ end is still engaged with the PAZ domain. State 2b, Argonaute-guide RNA bound to siRNA-like target. Duplex region is extended and the 3′ end of guide RNA dissociates from the N-PAZ lobe. Conformational changes occur that activate slicer activity. In the case of DmAgo1-ΔN-PAZ, the 3′ end of the guide RNA is exposed and susceptible to truncation (shown as translucent), and the slicer activity is constitutively on.
Acknowledgments
We are grateful to A. Nahvi for early contributions in the design of the project, E. Izaurralde (Max Planck Institute) for luciferase reporter plasmids, and Julie Brunelle for preparation of GW182 antibody. We also thank Karen Wehner for helpful comments on the manuscript.
This work was supported by funding from the Howard Hughes Medical Institute (to R. G.).
- miRNA
- microRNA
- RNP
- ribonucleoprotein
- PIWI
- P-element Induced Wimpy Testis
- PAZ
- PAZ-Argonaute-Zwille
- nt
- nucleotide(s)
- IP
- immunoprecipitation.
REFERENCES
- 1. Bartel D. P. (2009) MicroRNAs. Target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Djuranovic S., Nahvi A., Green R. (2011) A parsimonious model for gene regulation by miRNAs. Science 331, 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fabian M. R., Sonenberg N., Filipowicz W. (2010) Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 [DOI] [PubMed] [Google Scholar]
- 4. Hutvagner G., Simard M. J. (2008) Argonaute proteins. Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32 [DOI] [PubMed] [Google Scholar]
- 5. Song J. J., Smith S. K., Hannon G. J., Joshua-Tor L. (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437 [DOI] [PubMed] [Google Scholar]
- 6. Wang Y., Sheng G., Juranek S., Tuschl T., Patel D. J. (2008) Structure of the guide-strand-containing argonaute silencing complex. Nature 456, 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elkayam E., Kuhn C. D., Tocilj A., Haase A. D., Greene E. M., Hannon G. J., Joshua-Tor L. (2012) The structure of human argonaute-2 in complex with miR-20a. Cell 150, 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schirle N. T., MacRae I. J. (2012) The crystal structure of human Argonaute2. Science 336, 1037–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M. W., Ladurner A. G. (2007) A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 14, 897–903 [DOI] [PubMed] [Google Scholar]
- 10. Eulalio A., Huntzinger E., Izaurralde E. (2008) GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 15, 346–353 [DOI] [PubMed] [Google Scholar]
- 11. Djuranovic S., Zinchenko M. K., Hur J. K., Nahvi A., Brunelle J. L., Rogers E. J., Green R. (2010) Allosteric regulation of Argonaute proteins by miRNAs. Nat. Struct. Mol. Biol. 17, 144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boland A., Huntzinger E., Schmidt S., Izaurralde E., Weichenrieder O. (2011) Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc. Natl. Acad. Sci. U.S.A. 108, 10466–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eulalio A., Helms S., Fritzsch C., Fauser M., Izaurralde E. (2009) A C-terminal silencing domain in GW182 is essential for miRNA function. RNA 15, 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miyoshi K., Okada T. N., Siomi H., Siomi M. C. (2009) Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA 15, 1282–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E. (2009) Deadenylation is a widespread effect of miRNA regulation. RNA 15, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D. J. (2008) Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456, 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y., Juranek S., Li H., Sheng G., Wardle G. S., Tuschl T., Patel D. J. (2009) Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461, 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tomari Y., Zamore P. D. (2005) Perspective. Machines for RNAi. Genes Dev. 19, 517–529 [DOI] [PubMed] [Google Scholar]
- 19. Haley B., Zamore P. D. (2004) Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 11, 599–606 [DOI] [PubMed] [Google Scholar]
- 20. Miyoshi K., Uejima H., Nagami-Okada T., Siomi H., Siomi M. C. (2008) In vitro RNA cleavage assay for Argonaute-family proteins. Methods Mol. Biol. 442, 29–43 [DOI] [PubMed] [Google Scholar]
- 21. Wang B., Li S., Qi H. H., Chowdhury D., Shi Y., Novina C. D. (2009) Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat. Struct. Mol. Biol. 16, 1259–1266 [DOI] [PubMed] [Google Scholar]
- 22. Milligan J. F., Uhlenbeck O. C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180, 51–62 [DOI] [PubMed] [Google Scholar]
- 23. Wong I., Lohman T. M. (1993) A double-filter method for nitrocellulose-filter binding. Application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. U.S.A. 90, 5428–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Förstemann K., Horwich M. D., Wee L., Tomari Y., Zamore P. D. (2007) Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 130, 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fukaya T., Tomari Y. (2012) MicroRNAs mediate gene silencing via multiple different pathways in Drosophila. Mol. cell 48, 825–836 [DOI] [PubMed] [Google Scholar]
- 26. Huntzinger E., Kuzuoglu-Öztürk D., Braun J. E., Eulalio A., Wohlbold L., Izaurralde E. (2013) The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 41, 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20, 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pillai R. S., Artus C. G., Filipowicz W. (2004) Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10, 1518–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M. (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 [DOI] [PubMed] [Google Scholar]
- 30. Enright A. J., John B., Gaul U., Tuschl T., Sander C., Marks D. S. (2003) MicroRNA targets in Drosophila. Genome Biol. 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nahvi A., Shoemaker C. J., Green R. (2009) An expanded seed sequence definition accounts for full regulation of the hid 3′ UTR by bantam miRNA. RNA 15, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parker J. S., Parizotto E. A., Wang M., Roe S. M., Barford D. (2009) Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell 33, 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 34. Doench J. G., Sharp P. A. (2004) Specificity of microRNA target selection in translational repression. Genes Dev. 18, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang H. W., Noland C., Siridechadilok B., Taylor D. W., Ma E., Felderer K., Doudna J. A., Nogales E. (2009) Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 16, 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Halic M., Moazed D. (2010) Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140, 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gu S., Jin L., Huang Y., Zhang F., Kay M. A. (2012) Slicing-independent RISC activation requires the argonaute PAZ domain. Curr. Biol. 22, 1536–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ameres S. L., Horwich M. D., Hung J. H., Xu J., Ghildiyal M., Weng Z., Zamore P. D. (2010) Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lund E., Sheets M. D., Imboden S. B., Dahlberg J. E. (2011) Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev. 25, 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cifuentes D., Xue H., Taylor D. W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G. J., Lawson N. D., Wolfe S. A., Giraldez A. J. (2010) A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328, 1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Diederichs S., Haber D. A. (2007) Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131, 1097–1108 [DOI] [PubMed] [Google Scholar]