Regulation of intestinal microbiota by NLR proteins.
Keywords: Crohn’s disease, inflammasome, NLRP6, NOD2, microbiome
Abstract
The human intestine harbors a diverse microbial community consisting of a large number of bacteria and other micro-organisms that have co-evolved with the host intestinal immune system. During this process, microbiota and the host immune system shape one another by various mechanisms to achieve a successful symbiotic relationship. An increasing amount of evidence suggests that dysbiosis—the breakdown of such harmonized colonization—may result in infectious and inflammatory disorders, and recent advances in our studies indicate that receptors such as Toll-like receptors and NLR (nucleotide-binding oligomerization domain-like receptor; or nucleotide-binding domain- and leucine-rich repeat-containing receptor) proteins that detect micro-organisms and their products play a critical role in maintaining intestinal homeostasis. In this review, we summarize the role of NLR proteins in the regulation of intestinal microbiota. NLR proteins belong to a diverse family of cytoplasmic microbial sensors, mutations of which are involved in various disorders, including inflammatory bowel diseases. Understanding of the different roles of NLR family proteins in the intestine is, therefore, an important step towards the development of therapeutics against digestive diseases.
Introduction
The human gastrointestinal tract is colonized by a diverse microbial population, consisting of approximate 100 trillion micro-organisms, including bacteria, fungi and viruses (1). The major population of intestinal microbiota consists of bacteria, comprising 500–1000 different species (1, 2). Having a symbiotic relationship with the host, the microbiota plays a pivotal role in both physiological and pathophysiological conditions. In the healthy human intestine, a constant homeostasis is maintained by the stringent regulation of microbial load and the immune response generated against it. Failure of such a harmonized balance may result in various pathological conditions in the intestines. For instance, disruption of the epithelial barrier by opportunistic infection by resident bacteria or invasion by pathogenic bacteria may result in infectious diseases. In addition, the outgrowth of antibiotic-resistant pathogenic bacterial strains may lead to antibiotic-induced enteritis. The breakdown of homeostasis by either dysbiosis or dysregulation of immune responses may, furthermore, increase susceptibility to inflammatory bowel diseases (3–5). To minimize such pathological conditions, the intestinal tract has evolved to regulate microbiota through various strategies that allow a symbiotic relationship with microbiota and restrict the invasion of micro-organisms through the epithelial barrier. Innate immune sensors belonging to the cytoplasmic NLR [nucleotide-binding oligomerization domain (NOD)-like receptor; or nucleotide-binding domain (NBD)- and leucine-rich repeat (LRR)-containing receptor] family of proteins play an important role in shaping intestinal microbiota. Here, we summarize recent advances in the NLR field, which is critical to understanding infectious and inflammatory diseases in the intestine as well as systemic inflammatory diseases that are also influenced by intestinal microbiota.
The bacterial community in the intestine: a symbiotic relationship
The microbiota in the human intestine consists of a complex and diverse community. The composition and concentration of intestinal microbiota are regulated by a multitude of factors such as genetic background, diet and interactions between commensal and pathogenic bacteria (6). The majority of the intestinal microbiota is in the large intestine and over 99% of it is composed of four major bacterial divisions: Bacteroides, Firmicutes, Proteobacteria and Actinobacteria (4). The small intestine harbors significantly fewer bacteria, having loads of 103 and 107–8 in the proximal and terminal ileum, respectively, compared with 1011–12 in the colon (4).
This intestinal ecosystem has evolved to allow both host and microbiota to benefit from the balanced symbiotic relationship (7). The bacterial flora is assured of a stable habitat, and, in exchange, it aids host metabolic pathways with an array of enzymes and other bacterial products (8). The recent studies on bacterial communities in the intestine and other organs by the Human Microbiome Consortium confirmed that commensal flora diversity varies widely even among healthy individuals (9, 10). In spite of such microbial diversity, the metagenomic analysis of metabolic pathways demonstrated that metagenomic carriage of pathways is surprisingly stable among individuals, suggesting that the whole metabolism by the microbiota community in any person’s intestines remains similar (10).
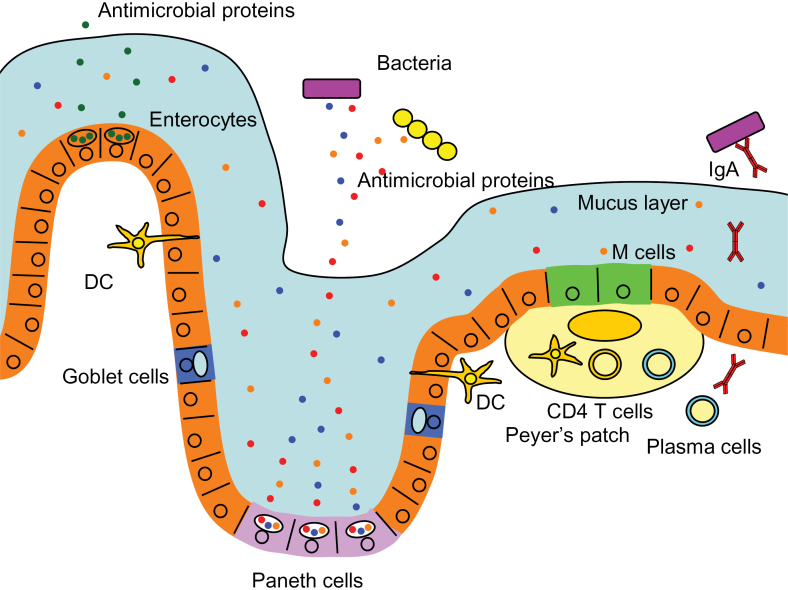
In spite of the host–microbiota symbiotic relationship, a large array of intestinal micro-organisms can become a potential threat to the host. The outgrowth of pathogenic bacteria or opportunistic invasion past the epithelial barrier by resident bacteria should, therefore, be minimized. Consequently, the human intestinal immune system has evolved to protect the host tissue from micro-organisms and simultaneously maintain the symbiotic benefits from the microbial presence (11). In order to accomplish this complex task, the host intestinal tract has evolved to be equipped with multiple layers of security strategies (Fig. 1). First, the intestine minimizes the number of harmful bacteria by shaping the microbiota through a symbiotic relationship. The commensal microbiota competes with pathogenic invaders and thus wards off the latter from colonizing the intestinal tract (7, 12). Second, thick mucus layers composed of mucin glycoproteins secreted from Goblet cells create a physical barrier, which separates bacterial flora and the intestinal epithelial cells. There are two mucus layers in the colon, of which the inner attached layer is resistant to bacterial penetrations (13, 14). The small intestine possess only a single layer of mucus, which minimizes direct bacterial contact with epithelial cells by creating a barrier of anti-microbial peptides, which are secreted from epithelial cells and enriched in the mucus layer (13, 14) (Fig. 1). Third, secreted antibacterial factors from epithelial cells directly regulate microbiota by their bactericidal activity (Fig. 1). Paneth cells at the base of crypts of Lieberkühn in the ileum are a specialized cell type that produce and secrete multiple arrays of such compounds, including α-defensins (cryptdins in mice), C-type lectins such as hepatointestinal pancreatic/pancreatitis-associated protein (HIP/PAP) in humans, and regenerating gene IIIγ (RegIIIγ) and RegIIIβ in mice, lysozyme and phospholipase A2 (15). Both mice lacking functional α-defensins and mice carrying exogenous human α-defensin 5 (HD5) have been shown to display an altered composition within the bacterial community, although the total number of bacteria was not affected (16). Furthermore, secretion of HD5 has been shown to protect the intestine from pathogenic bacterial infection (17). The antibacterial lectin, RegIIIγ, on the other hand, can limit the penetration or invasion of bacteria to epithelial cell layers but does not change the overall composition of luminal microbiota. HD6, secreted from Paneth cells, does not possess bactericidal activity but still inhibits in vitro and in vivo bacterial invasion by a unique mechanism. It has recently been demonstrated that after stochastic binding to bacterial surfaces, HD6 undergoes self-assembly to form fibrils and nanonets that surround and entangle enteric bacteria (18). Fourth, secreted IgA binds to intestinal micro-organisms, preventing their invasion through epithelial cell layers. Sampling of intestinal micro-organism products by dendritic cells leads to antigen presentation to lymphocytes in Peyer’s patches. This results in the development of mature IgA-secreting plasma cells in the lamina propria and their production of IgA, which becomes transcytosed to the luminal surface of the intestine (Fig. 1).
Fig. 1.
Epithelial barriers of the intestine. The human intestine has developed multiple strategies to protect the intestinal epithelial cells from invasion by resident and pathogenic bacteria. Enterocytes, the most abundant cell type in the intestinal epithelium, secrete anti-microbial proteins such as RegIIIγ. Paneth cells, which are specialized epithelial cells in the ileal crypts, produce abundant anti-microbial compounds such as α-defensins to regulate bacterial populations. Goblet cells produce mucin glycoproteins, constituents of the mucus layer, where anti-microbial proteins are enriched and resistance against bacteria is enforced. Sampling of bacterial products by dendritic cells (DCs) through their dendrites or by M cells via transcytosis leads to antigen presentation to the lymphocytes in Peyer’s patches. This results in the development of plasma cells, which produce and secrete IgA into the luminal surface of the intestine.
The NLR protein family
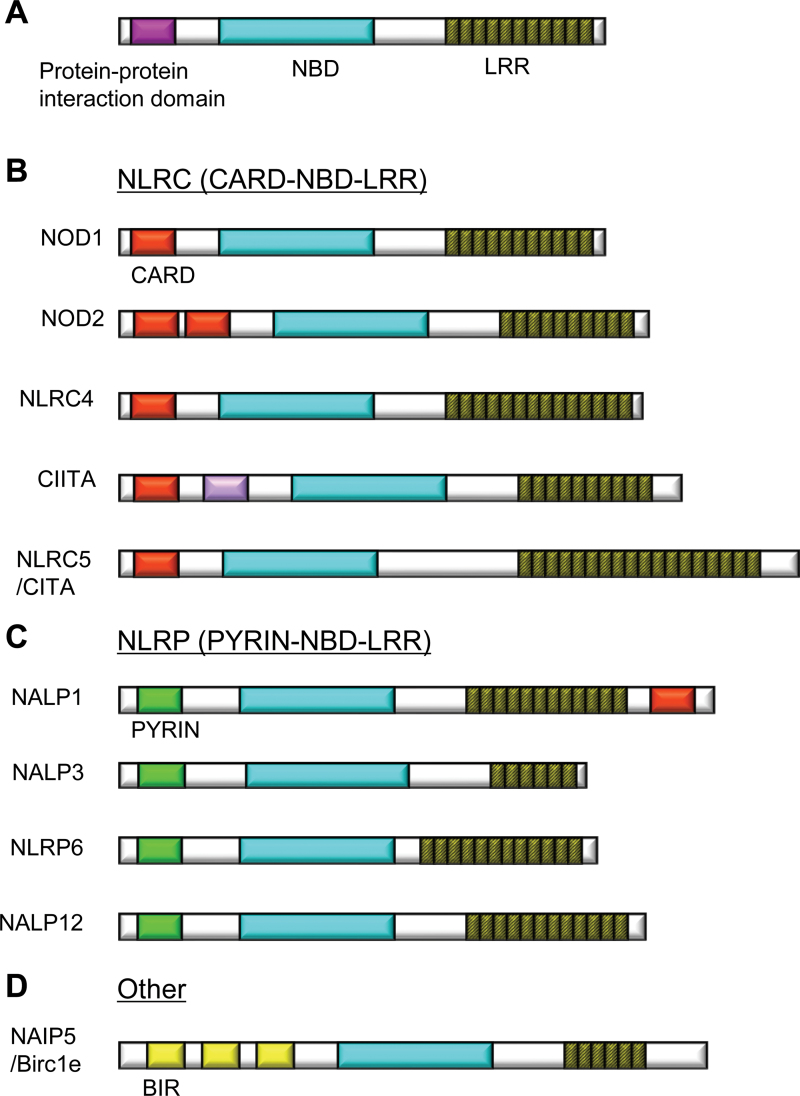
In the intestine and other organs, micro-organisms and their products are detected by pattern-recognition receptors such as Toll-like receptors and NLR proteins to elicit initial host defense responses. NLR proteins in the cytoplasm, consisting of 22 members in humans and about 34 in mice, are prevalent in a wide variety of cells, including immune and epithelial cells (19). NLR proteins play an important role in the recognition and defense against pathogens or extracellular danger signals (20, 21). NLRs consist of three domains characterized by an amino-terminal protein-interaction domain, a central NBD and a carboxy-terminal LRR (20) (Fig. 2A). NLR proteins can be subclassified by their amino-terminal protein-interaction domain into NLRs containing caspase-recruitment domains [(CARDs) NLRCs], NLRs containing PYRIN domains (NLRPs) or other NLR family proteins (19) (Fig. 2B–D). Except for NOD1 and NOD2, which are involved in activation of inflammatory gene expression, several NLRs are involved in the activation of caspase-1-activating complexes called inflammasomes (21, 22). These NLRs, including NLRP1, NLRP3 and the NLRC4, respond to various microbial products or damage-associated molecular patterns and lead to the release of IL-1-family inflammatory cytokines including IL-1β, IL-18 and IL-33 through the formation of the inflammasome (21, 23). NLRP1 senses the Bacillus anthracis lethal toxin, which is delivered into the cytoplasm by receptor-mediated endocytosis (24). NLRP3 senses exogenous and host danger signals such as pore-forming toxins, extracellular ATP and crystals such as uric acid, cholesterol, silica, asbestos or alum (25). NLRC4 senses a component of the bacterial type III secretion system and bacterial flagellin, which is typically translocated into the cytosol by a secretion system (26–28). The inflammasome is a multiprotein complex consisting of NLRs, caspase-1 and the adaptor protein ASC (apoptosis-associated speck-like protein containing a CARD). Caspase-1, also known as IL-1β-converting enzyme, mediates the processing of the pro-form of the cytokines into a mature form, which results in the secretion of bioactive cytokines. Activation of the inflammasome also causes a form of programmed cell death called pyroptosis, which contributes to the elimination of pathogens infecting host cells (29). Various NLR proteins, in both the NLRC and NLRP subfamilies, are involved in the regulation of intestinal microbiota with various mechanisms as discussed in the sections below
Fig. 2.
Structures of NLR protein family. NLR proteins are classified into subfamilies by protein-interaction domains such as CARD or PYRIN. NBDs [NACHT (NAIP CIITA HET-E TP1) domains or NODs] and LRRs are domains common to all NLRs. Two major subfamilies are the CARD- and PYRIN-containing subfamilies. (A) Schema of basic structure of NLR proteins. (B) Representatives of the of the NLRC (NLR family, CARD containing) subfamily proteins. (C) Representatives of the NLRP (NLR family, PYRIN containing) subfamily proteins. (D) An example from the other NLR subfamily, NAIP5. Birc1e, baculovirus inhibitor of apoptosis protein repeat c1e.
NOD2-dependent regulation of microbiota in the ileum
NOD2 was one of the first NLRs reported to function as an intracellular pattern-recognition receptor. NOD2 carries two CARDs at the amino terminus (30–32) and is thus a member of the NLRCs (Fig. 2B). NOD2 is highly expressed in dendritic cells (33), macrophages (32), intestinal, lung and oral epithelial cells (34–36), particularly in Paneth cells (37) and to a lesser extent in T cells (38, 39). Muramyl dipeptide (MDP), found in both Gram-positive and Gram-negative bacterial cell walls (40, 41), is recognized by NOD2, allowing for the detection of a wide variety of bacteria. After recognizing MDP, NOD2 undergoes a conformational change and activates the downstream signaling partner receptor-interacting protein 2 (RIP2) (31, 32). Upon activation, RIP2 switches on the downstream NF-κB and mitogen-activated protein kinase signaling cascades (42, 43) leading to the induction of immune response genes such as those encoding pro-inflammatory cytokines and chemokines (44).
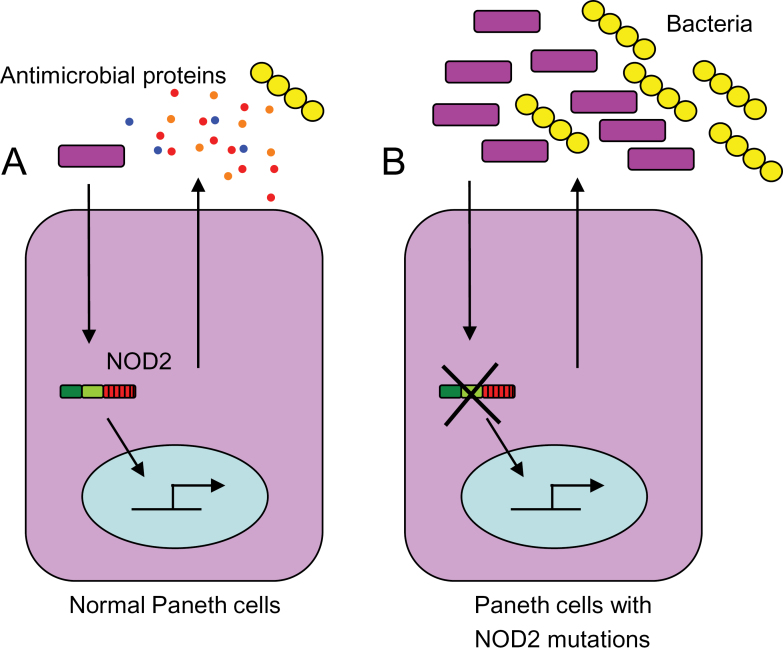
NOD2 has recently been shown to play a crucial role in regulating host–microbe interactions and maintaining intestinal homeostasis (45–47). The function of NOD2 in the Paneth cells of ileal crypts is critical in keeping the bacterial load in the ileum under check (Fig. 3). It was recently demonstrated that NOD2 is an important regulator of crypt anti-microbial function (45). Stimulation of ileal crypts by active forms of MDP can induce secretion by isolated crypts of antibacterial compounds, which can kill both Gram-positive and Gram-negative bacteria effectively (45), but the inactive chiral isomer of MDP (MDPDD or MDPLL) does not induce any such bacterial killing activity. This indicates that Paneth cells induce bacterial killing activity upon sensing the NOD2 ligand MDP. Interestingly, Nod2-deficient crypts are also unable to kill bacteria efficiently upon stimulation by a non-specific secretion inducer, carbamylcholine. Nod2 is, therefore, important not only for MDP sensing in Paneth cells but also for a general effect on secretion and/or composition of antibacterial factors. Ileal crypts lacking Rip2, similarly display an impaired bacterial killing ability, supporting the idea that the Nod2–Rip2 signaling pathway is involved in the regulation of Paneth cell function.
Fig. 3.
NOD2-mediated regulation of bacteria by Paneth cells. (A) With functional NOD2, Paneth cells sense bacteria or bacterial antigens and release anti-microbial peptides, which keep the intestinal flora under control. (B) Deletion of NOD2 or the presence of a NOD2 ‘loss of function’ mutation renders Paneth cells non-functional. Lack of Paneth cell-derived antibacterial compounds leads to dysregulated bacterial colonization of the intestinal mucosa and breakdown of homeostasis.
In agreement with the critical function of Nod2 in Paneth cells, it was discovered that Nod2 is one of the critical factors that effectively regulates the bacterial concentration or load in the intestine. Nod2-deficient mice with the congenic C57BL/6 genetic background from the same parents, kept in the same cages and fed the same diet have a significantly higher amount of Bacteroides, Firmicutes and Bacillus (a genus within phylum Firmicutes) in the terminal ileum compared with their wild-type littermates (45). This difference is less prominent in the feces probably because the expression of Nod2 is specific to Paneth cells, which are mainly localized in the terminal ileum (48, 49). Similarly, another study found that Nod2-deficient mice have an increased load of Bacteroides in the ileum and to a less significant extent in the feces (46). In addition to commensal bacteria, Nod2 also controls the load of pathogenic bacteria in the terminal ileum (45). It was found that Nod2-deficient mice cannot control the colonization of the opportunistic pathogen Helicobacter hepaticus and showed poor bacterial clearing capacity compared with wild-type controls (45). Interestingly, this regulation of ileal bacterial flora by Nod2 is not unidirectional, as it was found that the bacterial flora could also regulate the expression of Nod2 and Rip2. Germ-free mice have poor expression of Nod2 and Rip2 in the intestine in comparison with conventional specific pathogen-free mice with a congenic background. Additionally, it was found that mono-colonization of germ-free mice with a single probiotic bacterial strain, such as Lactobacillus plantarum or Escherichia coli strain Nissle 1917, increased the expression of Nod2 and Rip2. There is, therefore, a feedback loop in which bacteria-mediated up-regulation of Nod2 counteracts the bacterial flora to keep it under control in the ileum. Breakdown in this balanced relationship can lead to dysbiosis, which is known to underlie the pathogenesis of Crohn’s disease (CD) (4).
The role of NOD2 in the pathogenesis of ileal CD
The regulatory function of NOD2 on the microbiota is particularly important for human health because NOD2 is associated with the pathogenesis of CD, a multifactorial inflammatory disorder of the gastrointestinal tract. Various genetic or environmental factors can lead to the breakdown of intestinal homeostasis and can initiate chronic inflammation, leading to CD (50). NOD2 was found to be strongly associated with ileal CD susceptibility in North American and European populations (51, 52). Three main variants, or polymorphisms, in the NOD2 gene were identified as risk factors for CD: (i) a frameshift mutation at position 1007 (1007 fs); (ii) a glycine-to-arginine conversion at amino acid residue 908 (G908R) and (iii) an arginine-to-tryptophan conversion at amino acid residue 702 (R702W) (51–53). Although the exact mechanism by which NOD2 mutations contribute to CD pathogenesis is still unclear, loss-of-function mutations in NOD2 have been suggested to alter host–microbe interactions through various mechanisms, one of them being altered anti-microbial activity of Paneth cells in the terminal ileum (54). Dysregulated host–microbe interactions due to NOD2 mutations may increase the susceptibility to abnormal gut inflammation and, in combination with other genetic or environmental factors, may result in the development of active CD (55). In humans, patients with ileal CD showed reduced levels of the Paneth cell-derived HD5 and HD6 (56–58). Interestingly, the production of α-defensins is drastically reduced in CD patients with non-functional NOD2 mutations (56). Moreover, it was also found that Nod2-deficient mice have reduced mRNA expression of Paneth cell-derived α-defensins and are more susceptible to oral infection with Listeria monocytogenes (44).
The concept of microbial dysregulation and breakdown of the host–microbe balance behind the development of CD is very tempting, but an appropriate animal model for pre-clinical studies had been lacking until recently (59). It has recently been shown that inoculation of H. hepaticus into Nod2-deficient mice leads to the development of granulomatous inflammation of the ileum, providing a new ileal CD model that has many similarities with CD in human patients. Moreover, Nod2-deficient mice were protected from H. hepaticus-induced ileal inflammation after the restoration of Paneth cell bacterial killing activity, supporting the idea that Nod2 function in Paneth cells is critical in preventing intestinal inflammation. Nod2 is, therefore, necessary for normal Paneth cell function, which, in turn, plays a vital role in maintaining the normal microbial population and gut homeostasis. The significance of Paneth cells for ileal CD has been underscored recently by the observations that a number of CD-associated genes including ATG16L1 and XBP1 are involved in the regulation of Paneth cell anti-microbial function (60, 61). The functional association between ATG16L1 and NOD2 has also been shown (62–65). Moreover, it was found that polymorphism of the Wnt signaling pathway transcription factor, TCF4, which orchestrates Paneth cell differentiation, is genetically associated with ileal CD (66). Therefore, Paneth cell-mediated ileal microbiota regulation plays a vital physiological role against the development of pathological conditions like CD.
NLRP6-dependent regulation of microbiota
NLRP6 carries a PYRIN domain at its amino terminus and thus belongs to the PYRIN-containing NLR protein family (Fig. 2C) (19). NLRP6 is preferentially expressed in the epithelial cells of various organs such as the urinary bladder, kidney, liver, lung, duodenum, jejunum, ileum, cecum and colon (67, 68). In the intestinal tract, NLRP6 is highly expressed in myofibroblasts and epithelial cells but not in CD45+ hematopoietic cells (68, 69). NLRP6 can associate with caspase-1 via the adapter ASC, activating caspase-1, which can cleave pro-IL-1β and pro-IL-18 to produce their mature forms.
It was recently demonstrated that Nlrp6 is critical for regulating the microbiota in the colon (68). Metagenomic microbiota analysis of feces (i.e. study of overall bacterial genetic material recovered directly from feces) using multiplex pyrosequencing revealed that Nlrp6-deficient mice carry altered microbiota. In particular, nine genera belonging to four phyla (Firmicutes, Bacteroidetes, Proteobacteria and TM7) were significantly altered. An unnamed genus in Prevotellaceae (within Bacteroidetes) was most significantly increased in Nlrp6-deficient mice, followed by the phylum TM7 and the named genus Prevotella within Prevotellaceae. On the other hand, members of the genus Lactobacillus in the Firmicutes phylum were reduced. Moreover, electron microscopic studies disclosed aberrant colonization of the crypts of Lieberkühn by bacteria with morphologic features of Prevotellaceae. Interestingly, the altered microbiota is associated with the colitis-prone phenotype of Nlrp6-deficient mice. At steady state, Nlrp6-deficient mice and mice deficient for the downstream adaptor Asc exhibit colonic crypt hyperplasia, changes in crypt-to-villus ratios in the terminal ileum and enlargement of Peyer’s patches with the formation of germinal centers. Furthermore, Nlrp6-deficient mice are highly susceptible to colitis induced by dextran sodium sulfate (DSS) (67–69). Mice deficient in molecules downstream of Nlrp6, either Asc or caspase-1, exhibited a similar phenotype of susceptibility to DSS, indicating that the Nlrp6–Asc–caspase-1 pathway is involved in the regulation of colitogenic microbiota. This colitis-prone phenotype is transmissible to wild-type mice via co-housing, indicating that altered microbiota in Nlrp6- or Asc-deficient mice can be transferred to other strains. Although caspase-1 can activate both IL-1β and IL-18, it was determined that IL-18 plays a critical role in the regulation of microbiota downstream of Nlrp6 because IL-18- but not IL-1β-deficient mice could transfer a colits-prone phenotype to wild-type mice via co-housing. It appears that the regulatory role of microbiota is dominated by Nlrp6, as mice deficient for Nlrp3, Nlrp4, Nlrp10, Nlrp12 or Aim2 (absent in melanoma 2; also involved in the inflammasome formation) did not transfer microbiota with increased colitogenic properties to wild-type mice via co-housing. The altered microbiota in Nlrp6-, Asc- or IL-18-deficient mice resulted in the increased production of chemokine (C-C motif) ligand 5 (Ccl5) in intestinal epithelial cells, which accounts for the colitis-prone phenotype. Indeed, Ccl5-deficient mice do not exhibit severe DSS-induced colitis even upon co-housing with Nlrp6-deficient mice.
These studies demonstrate the striking property of Nlrp6-mediated regulation of colitogenic microbiota by the Nlrp6–Asc–caspase-1–IL-18 axis. This is underscored by the fact that the genes for Nlrp6, Asc and IL-18 are similarly up-regulated during development in the intestine (70). The most intriguing, yet unanswered, questions are the molecular mechanism of Nlrp6 activation in the intestinal epithelium and what its ligand is. Interestingly, the altered microbiota is also associated with another disease entity—non-alcohol fatty liver disease (NAFLD). Mice that are co-housed with Nlrp6-deficient mice (or mice deficient for Asc, capase-1, IL-18 or Nlrp3) and are fed with a methionine–choline-deficient diet (MCDD; this exacerbates NAFLD) developed a more severe NAFLD-like pathology than wild-type mice on an MCDD, including hepatic steatosis, inflammatory cell infiltration and fibrosis (71), providing further supportive evidence that changes in intestinal microbiota may affect susceptibility to systemic disorders.
NOD1-mediated regulation of microbiota
It was recently shown that commensal bacteria in the ileum are regulated by Nod1, which detects the diaminopimeric acid-containing moiety of bacterial peptidoglycan (72). In Nod1-deficient mice, the whole bacterial kingdoms detected by quantitative PCR on bacterial 16S ribosomal DNA increased approximately 100-fold. Specifically, Clostiridiales and Bacteroides and the smaller group of Enterobacteriaceae expanded 100-fold. A small expansion of segmented filamentous bacteria, which are potent inducers of Peyer’s patch function, was also observed in Nod1-deficient mice. Interestingly, the development of isolated lymphoid follicles (ILFs) and crypt patches, both of which are major lymphoid organs in the intestine in addition to Peyer’s patches, is affected significantly in Nod1-deficient mice and mildly in Nod2-deficient mice and Myd88-deficient mice (Myd88 signals downstream of Toll-like receptors and IL-1/IL-18 receptors). This reduction is probably through the chemokine receptor CCR6, which is required for ILF formation because one CCR6 ligand, murine β-defensin 3, is induced by Nod1 activation and another CCR6 ligand, CCL20, is expressed in a Nod1-dependent manner in crypt patches. An impaired crypt-patch–ILF system may, therefore, account for the altered microbiota in Nod1-deficient mice. Interestingly, lymphotoxin βR–Ig-treated mice, which lack ILFs, showed a milder phenotype compared with Nod1-deficient mice, suggesting that there may be alternative mechanisms as to how Nod1 deficiency affects microbiota (73). Comparing the microbiota and intestinal homeostasis among mice of different genetic backgrounds can greatly affect experimental outcomes (73). The aforementioned phenotype of Nod1-deficient mice, therefore, needs to be verified in a more controlled experimental setting, ideally using littermate control mice on a congenic background.
Conclusions
Recent advancements in the NLR field have revealed the critical function of NLR proteins in the intestine. Interestingly, the mechanisms whereby NLR protein activation regulates microbiota varies, despite similar molecular structures of NLRs. Although NOD2 plays a critical role in ileal Paneth cell function, NLRP6 regulates colitogenic bacteria via IL-18 secretion. Because NLRs are such a diverse protein family, it is tempting to speculate that other NLR proteins may also play a role in the regulation of intestinal microbiota. Although recent National Institutes of Health funded, multicentered human mirobiome studies have shown that personal habits and genetic makeup are major factors that affect variations of the bacterial community among individuals, we are still a long way from understanding how a particular microbiome is established under the influence of certain foods, host genetic factors and immune systems. Further studies on the function of NLR proteins and other elements of the innate immune system in the intestine will be pivotal for our understanding of human intestinal microbiota and of intestinal inflammatory and infectious diseases.
Funding
National Institutes of Health (R01DK074738 to K.S.K.); the Broad Medical Research Program of the Eli and Edythe L. Broad Foundations (IBD-0328 to K.S.K.). K.S.K. is a recipient of the Investigator Award from the Cancer Research Institute and the Claudia Adams Barr Award. A.B. is a recipient of fellowship from the Crohn’s and Colitis Foundation of America.
Acknowledgements
The authors thank Yuen-Joyce Liu for proofreading this manuscript.
Conflict of interest: The authors have no conflicting financial interests.
References
- 1. Hooper L. V., Gordon J. I. 2001. Commensal host-bacterial relationships in the gut. Science 292: 1115 [DOI] [PubMed] [Google Scholar]
- 2. Guarner F., Malagelada J. R. 2003. Gut flora in health and disease. Lancet 361: 512 [DOI] [PubMed] [Google Scholar]
- 3. Xavier R. J., Podolsky D. K. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448: 427 [DOI] [PubMed] [Google Scholar]
- 4. Sartor R. B. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology 134: 577 [DOI] [PubMed] [Google Scholar]
- 5. Abraham C., Cho J. H. 2009. Inflammatory bowel disease. N. Engl. J. Med. 361: 2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Begue B., Dumant C., Bambou J. C, et al. 2006. Microbial induction of CARD15 expression in intestinal epithelial cells via toll-like receptor 5 triggers an antibacterial response loop. J. Cell. Physiol. 209: 241 [DOI] [PubMed] [Google Scholar]
- 7. Hooper L. V., Macpherson A. J. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10: 159 [DOI] [PubMed] [Google Scholar]
- 8. Nicholson J. K., Holmes E., Kinross J, et al. 2012. Host-gut microbiota metabolic interactions. Science 336: 1262 [DOI] [PubMed] [Google Scholar]
- 9. The Human Microbiome Project Consortium 2012. A framework for human microbiome research. Nature 486: 215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. The Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486: 207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hooper L. V., Littman D. R., Macpherson A. J. 2012. Interactions between the microbiota and the immune system. Science 336: 1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macpherson A. J., Uhr T. 2004. Compartmentalization of the mucosal immune responses to commensal intestinal bacteria. Ann. N. Y. Acad. Sci. 1029: 36 [DOI] [PubMed] [Google Scholar]
- 13. Johansson M. E., Larsson J. M., Hansson G. C. 2011. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1): 4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johansson M. E., Hansson G. C. 2011. Microbiology. Keeping bacteria at a distance. Science 334: 182 [DOI] [PubMed] [Google Scholar]
- 15. Gallo R. L., Hooper L. V. 2012. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12: 503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salzman N. H., Hung K., Haribhai D, et al. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11: 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salzman N. H., Ghosh D., Huttner K. M., Paterson Y., Bevins C. L. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422: 522 [DOI] [PubMed] [Google Scholar]
- 18. Chu H., Pazgier M., Jung G, et al. 2012. Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337: 477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ting J. P., Lovering R. C., Alnemri E. S, et al. 2008. The NLR gene family: a standard nomenclature. Immunity 28: 285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilmanski J. M., Petnicki-Ocwieja T., Kobayashi K. S. 2008. NLR proteins: integral members of innate immunity and mediators of inflammatory diseases. J. Leukoc. Biol. 83: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elinav E., Strowig T., Henao-Mejia J., Flavell R. A. 2011. Regulation of the antimicrobial response by NLR proteins. Immunity 34: 665 [DOI] [PubMed] [Google Scholar]
- 22. Martinon F., Burns K., Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10: 417 [DOI] [PubMed] [Google Scholar]
- 23. Lamkanfi M., Dixit V. M. 2009. Inflammasomes: guardians of cytosolic sanctity. Immunol. Rev. 227: 95 [DOI] [PubMed] [Google Scholar]
- 24. Boyden E. D., Dietrich W. F. 2006. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38: 240 [DOI] [PubMed] [Google Scholar]
- 25. Cassel S. L., Joly S., Sutterwala F. S. 2009. The NLRP3 inflammasome: a sensor of immune danger signals. Semin. Immunol. 21: 194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miao E. A., Alpuche-Aranda C. M., Dors M, et al. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7: 569 [DOI] [PubMed] [Google Scholar]
- 27. Franchi L., Amer A., Body-Malapel M, et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7: 576 [DOI] [PubMed] [Google Scholar]
- 28. Miao E. A., Mao D. P., Yudkovsky N, et al. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 107: 3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miao E. A., Leaf I. A., Treuting P. M, et al. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11: 1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bertin J., Nir W. J., Fischer C. M, et al. 1999. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J. Biol. Chem. 274: 12955 [DOI] [PubMed] [Google Scholar]
- 31. Inohara N., Koseki T., del Peso L, et al. 1999. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J. Biol. Chem. 274: 14560 [DOI] [PubMed] [Google Scholar]
- 32. Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. 2001. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J. Biol. Chem. 276: 4812 [DOI] [PubMed] [Google Scholar]
- 33. Tada H., Aiba S., Shibata K., Ohteki T., Takada H. 2005. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect. Immun. 73: 7967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hisamatsu T., Suzuki M., Reinecker H. C., Nadeau W. J., McCormick B. A., Podolsky D. K. 2003. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology 124: 993 [DOI] [PubMed] [Google Scholar]
- 35. Uehara A., Fujimoto Y., Fukase K., Takada H. 2007. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol. Immunol. 44: 3100 [DOI] [PubMed] [Google Scholar]
- 36. Uehara A., Sugawara Y., Kurata S, et al. 2005. Chemically synthesized pathogen-associated molecular patterns increase the expression of peptidoglycan recognition proteins via toll-like receptors, NOD1 and NOD2 in human oral epithelial cells. Cell. Microbiol. 7: 675 [DOI] [PubMed] [Google Scholar]
- 37. Voss E., Wehkamp J., Wehkamp K., Stange E. F., Schröder J. M., Harder J. 2006. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J. Biol. Chem. 281: 2005 [DOI] [PubMed] [Google Scholar]
- 38. Gutierrez O., Pipaon C., Inohara N, et al. 2002. Induction of Nod2 in myelomonocytic and intestinal epithelial cells via nuclear factor-kappa B activation. J. Biol. Chem. 277: 41701 [DOI] [PubMed] [Google Scholar]
- 39. Caetano B. C., Biswas A., Lima D. S., Jr, et al. 2011. Intrinsic expression of Nod2 in CD4+ T lymphocytes is not necessary for the development of cell-mediated immunity and host resistance to Toxoplasma gondii . Eur. J. Immunol. 41: 3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Girardin S. E., Boneca I. G., Viala J, et al. 2003. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278: 8869 [DOI] [PubMed] [Google Scholar]
- 41. Inohara N., Ogura Y., Fontalba A, et al. 2003. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J. Biol. Chem. 278: 5509 [DOI] [PubMed] [Google Scholar]
- 42. Inohara N., Koseki T., Lin J, et al. 2000. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 275: 27823 [DOI] [PubMed] [Google Scholar]
- 43. Abbott D. W., Wilkins A., Asara J. M., Cantley L. C. 2004. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr. Biol. 14: 2217 [DOI] [PubMed] [Google Scholar]
- 44. Kobayashi K. S., Chamaillard M., Ogura Y, et al. 2005. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307: 731 [DOI] [PubMed] [Google Scholar]
- 45. Petnicki-Ocwieja T., Hrncir T., Liu Y. J, et al. 2009. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. U.S.A. 106: 15813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rehman A., Sina C., Gavrilova O, et al. 2011. Nod2 is essential for temporal development of intestinal microbial communities. Gut 60: 1354 [DOI] [PubMed] [Google Scholar]
- 47. Mondot S., Barreau F., Al Nabhani Z, et al. 2012. Altered gut microbiota composition in immune-impaired Nod2(-/-) mice. Gut 61: 634 [DOI] [PubMed] [Google Scholar]
- 48. Ogura Y., Lala S., Xin W, et al. 2003. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut 52: 1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lala S., Ogura Y., Osborne C, et al. 2003. Crohn’s disease and the NOD2 gene: a role for Paneth cells. Gastroenterology 125: 47 [DOI] [PubMed] [Google Scholar]
- 50. Podolsky D. K. 2002. Inflammatory bowel disease. N. Engl. J. Med. 347: 417 [DOI] [PubMed] [Google Scholar]
- 51. Hugot J. P., Chamaillard M., Zouali H, et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411: 599 [DOI] [PubMed] [Google Scholar]
- 52. Ogura Y., Bonen D. K., Inohara N, et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411: 603 [DOI] [PubMed] [Google Scholar]
- 53. Lesage S., Zouali H., Cézard J. P, et al. 2002. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am. J. Hum. Genet. 70: 845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cho J. H. 2008. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 8: 458 [DOI] [PubMed] [Google Scholar]
- 55. Biswas A., Petnicki-Ocwieja T., Kobayashi K. S. 2012. Nod2: a key regulator linking microbiota to intestinal mucosal immunity. J. Mol. Med. 90: 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wehkamp J., Salzman N. H., Porter E, et al. 2005. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. U.S.A. 102: 18129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simms L. A., Doecke J. D., Walsh M. D., Huang N., Fowler E. V., Radford-Smith G. L. 2008. Reduced alpha-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn’s disease. Gut 57: 903 [DOI] [PubMed] [Google Scholar]
- 58. Perminow G., Beisner J., Koslowski M, et al. 2010. Defective Paneth cell-mediated host defense in pediatric ileal Crohn’s disease. Am. J. Gastroenterol. 105: 452 [DOI] [PubMed] [Google Scholar]
- 59. Biswas A., Liu Y. J., Hao L, et al. 2010. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc. Natl. Acad. Sci. U.S.A. 107: 14739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cadwell K., Liu J. Y., Brown S. L, et al. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456: 259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kaser A., Lee A. H., Franke A, et al. 2008. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134: 743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cooney R., Baker J., Brain O, et al. 2010. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16: 90 [DOI] [PubMed] [Google Scholar]
- 63. Homer C. R., Richmond A. L., Rebert N. A., Achkar J. P., McDonald C. 2010. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology 139: 1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Travassos L. H., Carneiro L. A., Ramjeet M, et al. 2010. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11: 55 [DOI] [PubMed] [Google Scholar]
- 65. Plantinga T. S., Crisan T. O., Oosting M, et al. 2011. Crohn’s disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut 60: 1229 [DOI] [PubMed] [Google Scholar]
- 66. Koslowski M. J., Kübler I., Chamaillard M, et al. 2009. Genetic variants of Wnt transcription factor TCF-4 (TCF7L2) putative promoter region are associated with small intestinal Crohn’s disease. PLoS ONE 4: e4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen G. Y., Liu M., Wang F., Bertin J., Núñez G. 2011. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 186: 7187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Elinav E., Strowig T., Kau A. L, et al. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145: 745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Normand S., Delanoye-Crespin A., Bressenot A, et al. 2011. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc. Natl. Acad. Sci. U.S.A. 108: 9601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kempster S. L., Belteki G., Forhead A. J, et al. 2011. Developmental control of the Nlrp6 inflammasome and a substrate, IL-18, in mammalian intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 300: G253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Henao-Mejia J., Elinav E., Jin C, et al. 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482: 179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bouskra D., Brézillon C., Bérard M, et al. 2008. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456: 507 [DOI] [PubMed] [Google Scholar]
- 73. Chen G. Y., Núñez G. 2009. Gut immunity: a NOD to the commensals. Curr. Biol. 19: R171 [DOI] [PubMed] [Google Scholar]