Misfolded endogenous ER proteins can be presented intact by MHC class II and stimulate B cells.
Keywords: antigen presentation, B cell activation, MHC class II, misfolded protein
Abstract
Nascent MHC class II molecules are associated with the invariant chain and are transported to the endolysosomal pathway, where MHC class II molecules acquire peptide antigens. On the other hand, misfolded endoplasmic reticulum (ER) proteins are generally degraded in the cells and are neither expressed on the cell surface nor secreted. Here, we found that MHC class II molecules associate with some misfolded ER proteins via the peptide-binding groove in competition with invariant chain. The misfolded proteins associated with MHC class II molecules are transported intact to the cell surface without processing to peptides. Furthermore, these complexes efficiently stimulate antigen-specific B cells. These findings reveal that MHC class II molecules function as a chaperone for the cell surface expression of misfolded ER proteins. In addition, we suggest that MHC class II molecules present not only peptides but also intact host-cell-derived proteins on the cell surface. These findings provide new insights into the function of MHC class II molecules.
Introduction
MHC class I and II molecules play a central role in the immune system by presenting peptide antigens to T cells. In general, intracellular proteins are a source of peptides for MHC class I molecules, whereas peptides derived from extracellular proteins are presented by MHC class II molecules. Structural analyses of MHC class I and II molecules have revealed that both ends of the peptide-binding groove of the class I proteins are closed and thus only short peptides are typically presented (1), whereas both ends of the MHC class II peptide-binding groove are open and long peptides can therefore be presented (2–4). Accordingly, peptides of different lengths, both long and short, can be eluted from MHC class II molecules (5, 6). Because of this flexibility of the MHC class II peptide-binding groove, newly synthesized class II proteins associate with the invariant chain (Ii), which contains an endosomal sorting signal, and are then transported to the late endosomal compartment, termed the MHC class II compartment, where MHC class II molecules acquire peptides derived from extracellular antigens (7, 8). However, considering the structure of the MHC class II peptide-binding groove, it is possible that nascent MHC class II products associate with a variety of different proteins other than Ii, as noted previously (9). Here, we demonstrate that both human and mouse MHC class II molecules bind to host-cell-derived misfolded proteins via the peptide-binding groove and transport them to the cell surface.
Methods
Plasmids
First, cDNAs for different HLA-class I and -class II alleles and Ii (accession No. NM_004355.2) were cloned from cDNAs prepared from pooled human PBMCs (3H Biomedical) or human cell lines. Dr P. Parham (Stanford University) kindly provided the HLA-Cw*04:01 (HLA-Cw4) vector. All cDNA sequences for HLA were based on information contained in the IMGT/HLA Database (www.ebi.ac.uk/ipd/imgt/hla/index.html). A cDNA of HLA-Cw4 lacking the transmembrane region was generated by PCR by replacing a glutamine at amino acid residue 303 with a stop codon. cDNAs of HLA-Cw4 and hen egg lysozyme (HEL) lacking signal sequences (amino acid residues from 1 to 24 for HLA-Cw4 and from 1 to 18 for HEL) were also generated by PCR. These mutated cDNAs were cloned into the pME18S expression vector. HLA-DRB1*01:01 containing a covalently attached transferrin receptor peptide (TfRpep-HLA-DRB1*01:01) and the I-Ak β chain containing a covalently attached HEL peptide (HELpep-I-Ak) were generated by adding the peptide sequence RVEYHFLSPYVSPKESP (TfR) or DGSTDYGILQINSR (HEL) and a linker (GSGSGS) between the signal sequence and extracellular domain of these MHC class II cDNAs, as previously described (10). A cDNA for a HEL mutant (mutHEL) in which two cysteine residues at positions 30 and 64 were replaced with alanine was generated by using Quickchange multi-mutagenesis kits. The cDNA for membrane-tethered wild-type HEL (Membrane wtHEL) protein was constructed by inserting the transmembrane and cytoplasmic domains of mouse PILRα (amino acid residues 196–256, accession No. NM_153510.3) just before the stop codon of HEL. Variable regions for HyHEL10 and Gloop4 anti-HEL mAbs were synthesized according to published sequences (accession Nos for HyHEL10 heavy chain: 3D9A_H; for HyHEL10 κ chain: 3D9A_L; Gloop4 heavy chain: X02185; and Gloop4 κ chain: X02180). These variable regions were cloned into pME18S expression vectors containing human secreted IgG1 constant region (accession No. DQ487208.1) or human immunoglobulin κ constant region (accession No. NG_000834.1). The heavy and immunoglobulin κ chains were also cloned into pMx retrovirus vectors containing the mouse membrane IgM constant region (accession No. BC018315.1) and the human immunoglobulin κ constant region.
Antibodies
Dr P. Giacomini kindly provided the L31 mAb, which is specific for misfolded HLA-C (11). Dr J. Neefjes generously provided the HC10 mAb, which is specific for misfolded HLA class I heavy chain (12). HL-40 (EXBIO) mAb was used for detection of HLA-DR by flow cytometry and L243 (American Type Culture Collection) mAb was used for immunoprecipitation of HLA-DR proteins. Anti-HLA class I mAb (W6/32, Sigma), anti-Flag mAb (clone M2, Sigma), anti-DYKDDDDK tag (Flag) mAb (BioLegend), anti-His mAb (Wako), anti-I-Ak mAb (OX-6, BD Biosciences) and anti-human CD74 (Ii) mAb (LN2, BD Biosciences) were used for flow cytometry or immunoprecipitation. HC10 mAb or rabbit anti-HLA-DRα Ab (FL-254, Santa Cruz Biotechnology, Inc.) was used for western blotting of HLA class I or HLA class II proteins, respectively. HyHEL10 and Gloop4 mAbs were produced by co-transfection of their immunoglobulin heavy- and light-chain cDNAs into 293T cells, as described earlier.
cDNA library screening
N-terminal Flag-tagged HLA-Cw*04:01 (Flag-Cw4) was stably transfected into 293T cells by using the pCDNA3.1-IRES-GFP expression vector, and cells stably expressing green fluorescent protein (GFP) were purified using a cell sorter. A cDNA library from 721.221 cells was generated, as previously described (13), by using a Superscript cDNA library construction kit with the pSPORT expression vector (Invitrogen). Complexity of the library was >1×106. The cDNA library was transfected into 293T cells carrying Flag-Cw4 using Lipofectamine-plus reagents (Invitrogen), and Flag-positive cells were isolated 2 days later by using a flow cytometer. Plasmids were purified from the sorted cells and transformed into DH5α bacteria. Plasmids purified from 24 pools containing ~400 DH5α bacterial clones were transfected into 293T cells carrying Flag-Cw4, and a pool that facilitated cell surface expression of Flag-Cw4 was identified. Pools containing ~80 clones derived from the pool of 400 clones that induced cell surface expression of HLA-Cw4 were analyzed, and a pool that induced cell surface Flag expression was identified. The procedure was repeated six times; however, the enrichment was quite low compared with the expected efficiency. The low efficiency of enrichment suggested that HLA-Cw4-inducing cDNA might not be a single molecule. We identified a pool containing ~12 clones that supported cell surface expression of Flag-Cw4. Thereafter, 12 pools containing 8 independent clones were transfected into 293T cells expressing Flag-Cw4 and only 1 pool was identified that caused cell surface expression of Flag-Cw4. Finally, cDNA sequences of these eight clones were determined.
Transfectants
Next, pME18S expression plasmids containing each cDNA were transiently transfected into 293T or CHO cells with pMx-GFP using Lipofectamine 2000 (Invitrogen) or PEI max (Polyscience). Flag-Cw4 plasmid was transfected into 721.221 cells by using the pMx retrovirus vector, and Flag-positive cells were purified using a cell sorter as previously described (13). The mouse B-cell line A20 expressing the HEL-specific Gloop4 B-cell receptor (BCR) was generated by transfecting Gloop4 mouse IgM heavy chain and Gloop4 human immunoglobulin κ chain into the A20 B-cell line using the pMx retrovirus vector. We used human immunoglobulin κ chain to distinguish transfected κ chain from endogenous κ chain. Transfectants expressing mouse IgM and human κ chain were purified using a cell sorter, and a single-cell clone that expressed CD69 upon stimulation with immobilized (plate-bound) HEL was obtained.
Flow cytometry
Cells were incubated with primary mouse mAbs, rat mAbs or KIR2DL1-human Ig Fc fusion protein, followed by allophycoerythrin (APC)-conjugated anti-mouse IgG, APC-conjugated anti-rat IgG or phycoerythrin-conjugated anti-human IgG Ab (all from Jackson ImmunoResearch), respectively. For intracellular staining, cells were fixed and permeabilized with Fixation/Permeabilization solution (BD Bioscience). Intracellular Ii was detected with anti-human CD74 mAb (BD Bioscience), followed by APC-conjugated anti-mouse IgG antibody. Stained cells were analyzed on a FACSCalibur (Becton Dickinson). PHA-stimulated PBMCs were prepared by culture of freshly isolated PBMCs for 3 days in the presence of PHA (1 µg ml–1, Sigma) and human recombinant IL-2 (200U ml–1, Peprotec). Cells were stained with HC10 (IgG2a), W6/32 (IgG2a) or an isotype-matched control mAb, followed by APC-labeled anti-mouse IgG2a-specific antibody in combination with FITC-conjugated anti-HLA-DR mAb (IgG1).
Structural analysis of HLA-DR
HLA-DR 01:01 structure (PDB code: 3QXA) is illustrated using MacPyMOL software (DeLano Scientific LLC) and the amino acids of HLA-DR 01:03 that are different from those of HLA-DR 01:01 are indicated based on the IMGT/HLA Database (www.ebi.ac.uk/ipd/imgt/hla/index.html).
Immunoprecipitation and immunoblotting
Cells were lysed in lysis buffer (20mM Tris, 150mM NaCl, pH 7.5) containing 1% Brij 98 (Sigma). Lysates were immunoprecipitated with Protein A- or Protein G-Sepharose (GE Healthcare). The immunoprecipitates were eluted by boiling with SDS–PAGE sample buffer, separated on 10–12% polyacrylamide gels and transferred onto polyvinylidene fluoride membranes (Millipore). The membranes were incubated with anti-MHC class I or class II Abs, followed by HRP-conjugated anti-mouse or anti-rabbit IgG Ab (Amersham Biosciences). Peroxidase activity was detected with the SuperSignal reagent (Thermo Fisher Scientific Inc.).
B-cell activation
First, wtHEL and cysteine-mutant HEL (mutHEL) were co-transfected into CHO cells with GFP in the presence or absence of I-Ak. Membrane-tethered wtHEL was transfected with GFP as a positive control. We used CHO cells because these cells adhere to the culture plate more strongly than 293T cells do and their cell surface is larger than that of 293T cells. GFP-positive cells were purified 2 days later by using a cell sorter. After overnight culture, these transfectants were co-cultured with the Gloop4-BCR-transfected A20 B-cell line, designated G4-A20. G4-A20 cells were also stimulated with soluble HEL protein (30 µg ml–1) and immobilized (plate-bound) HEL protein (30 µg ml–1). After 4h, the cells were stained with anti-CD69 and B220 (anti-CD45) mAbs and analyzed by flow cytometry.
Results
MHC class II molecules induce cell surface expression of HLA-Cw4
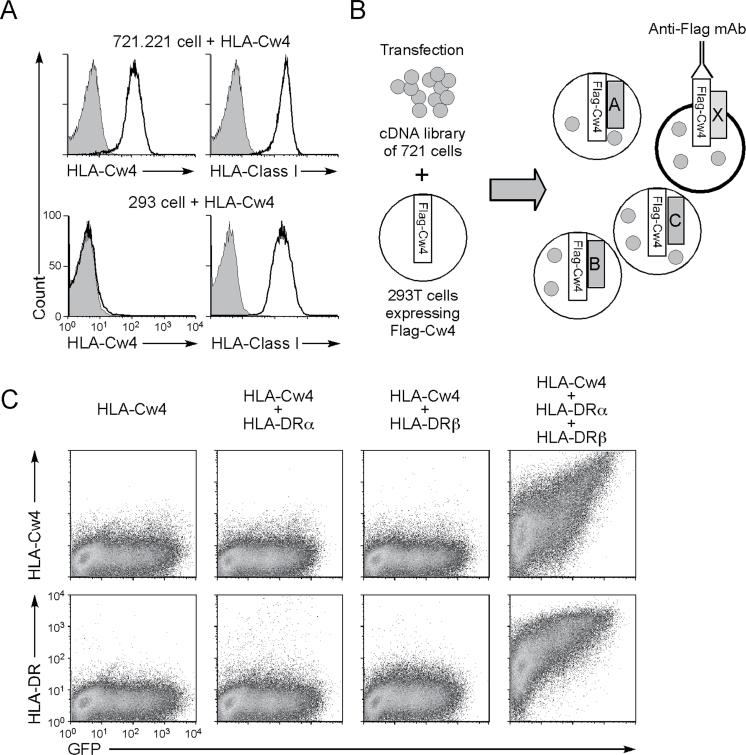
We inadvertently found that the MHC class I molecule, HLA-Cw4, cannot be expressed on the cell surface of human 293T kidney epithelial cells, despite the fact that 293T cells express endogenous MHC class I molecules. On the other hand, when HLA-Cw4 was transfected into the class I-deficient EBV-transformed B-lymphoblastoid cell line 721.221, HLA-Cw4 was readily expressed on the cell surface (Fig. 1A). We hypothesized that 293T cells may lack certain molecular mechanisms required for surface expression of HLA-Cw4. To identify the mechanism, we generated a cDNA library from 721.221 cells and transfected the library into 293T cells stably transfected with a Flag epitope-tagged HLA-Cw4 (Fig. 1B). The HLA-Cw4 was detected on the cell surface after transfecting these 293T cells with the 721.221 cDNA library, although the proportion of HLA-Cw4-expressing cells was low (1.26%; Supplementary Figure 1A, available at International Immunology Online). We then enriched the cDNAs enabling cell surface expression of HLA-Cw4 and finally identified a pool containing eight different clones (Supplementary Figure 1B, available at International Immunology Online). However, none of the cDNAs in the pool individually allowed cell surface expression of HLA-Cw4. Sequencing of the eight clones revealed that they included cDNAs for the HLA-DRα (DRA*01:01) and HLA-DRβ (DRB1*01:01) MHC class II genes. We then co-transfected HLA-Cw4 and GFP with HLA-DRα and/or -DRβ into 293T cells and found that HLA-Cw4 was then expressed on the surface of GFP-expressing cells transfected with both HLA-DRα and HLA-DRβ chains (Fig. 1C). Expression levels of HLA-Cw4 correlated with the amount of HLA-DR on the cell surface. In addition, HLA-Cw4 expression was observed even on GFP-negative cells expressing low levels of HLA-DR, indicating that HLA-Cw4 induction is not a result from over-expression of HLA-DR. These data suggest that HLA-DR permits HLA-Cw4 expression by 721.221 cells.
Fig. 1.
MHC class II molecules induce cell surface expression of HLA-Cw4. (A) Expression levels of Flag-Cw4 on 293T cells (293) and 721.221 cells transfected with Flag-HLA-Cw4 were analyzed (continuous lines). 293T cells (293) and 721.221 cells transfected with Flag-HLA-Cw4 were stained with anti-Flag (HLA-Cw4) or anti-HLA class I mAb W6/32 (which recognizes properly folded HLA class I proteins) (continuous lines). Control staining: shaded histograms. (B) A cDNA library generated from 721.221 cells was transfected into 293T cells carrying Flag-HLA-Cw4 (Flag-Cw4) intracellularly, and cDNA pools that induce cell surface Flag expression were enriched for several cycles. (C) 293T cells were transfected with Flag-HLA-Cw4-IRES-GFP and HLA-DRα and/or HLA-DRβ expression vectors. Expression of HLA-Cw4, HLA-DR and GFP is shown.
MHC class II molecules induce misfolded HLA-Cw4 but not normal HLA-Cw4
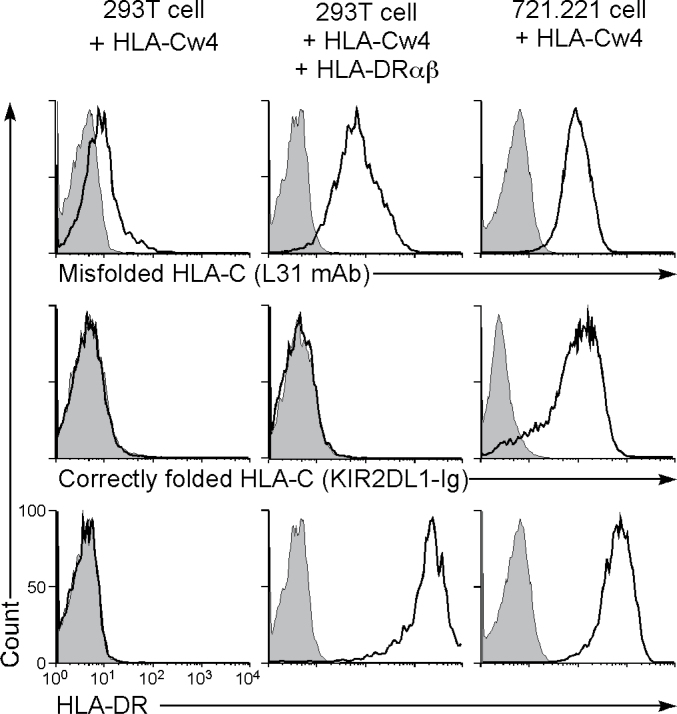
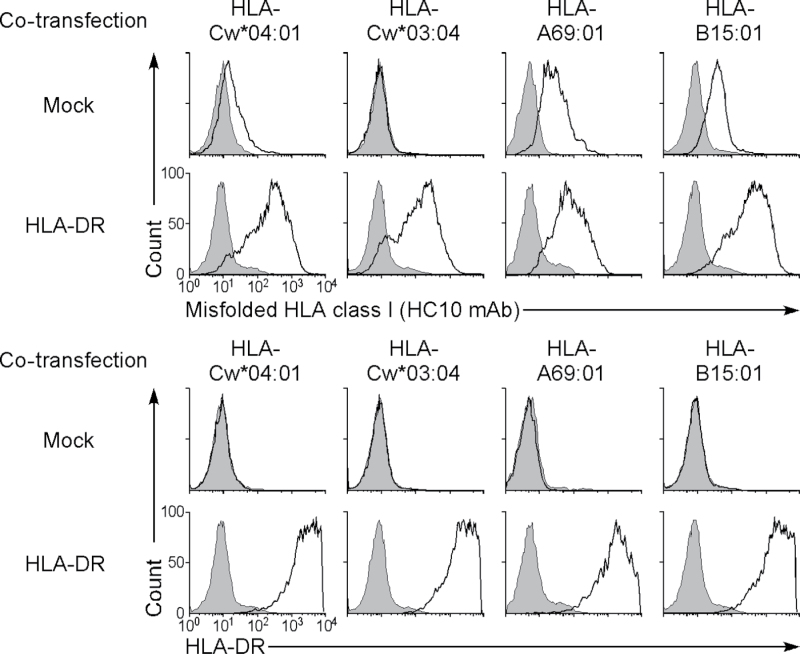
We analyzed the characteristics of the HLA-Cw4 protein expressed by HLA-DRαβ-transfected cells. Because KIR2DL1, an inhibitory NK cell receptor, specifically recognizes correctly folded HLA-Cw4 (14), we used a KIR2DL1-Ig fusion protein to test whether the HLA-Cw4 was properly folded (Fig. 2). Although KIR2DL1-Ig did bind to HLA-Cw4 when it was expressed on 721.221 cells, it did not bind to the HLA-Cw4 induced by co-transfection of HLA-DRαβ into 293T cells. On the other hand, this form of HLA-Cw4 on 293T cells was recognized by the mAb L31, which is specific for misfolded HLA-C heavy chains that lack peptide and β2 microglobulin (11, 15, 16). HLA-Cw4 expressed on 721.221 cells was also recognized by L31 mAb. These data suggest that 293T cells express only structurally altered, or misfolded, HLA-Cw4 in the presence of HLA-DR proteins, whereas 721.221 cells express both correctly folded and misfolded HLA-Cw4 on the cell surface. When wild-type HLA-Cw4 that had no epitope tag sequence was transfected into CHO cells, expression of misfolded HLA-Cw4 in the presence of HLA-DR was also observed on the cell surface (Fig. 3). In addition, expression of misfolded HLA class I caused by HLA-DR co-expression was observed for the other HLA class I isoforms, HLA-A and HLA-B, using the HC10 mAb, which is specific for misfolded HLA class I heavy chains (12, 15, 16), although low levels of misfolded HLA-A and HLA-B expression were detected even in the absence of HLA-DR (Fig. 3). These data suggest that HLA-DR molecules are involved in the expression of misfolded HLA class I molecules; however, the results do not shed light on the reasons why fully processed HLA-Cw4 is not expressed on 293T cells, which express endogenous HLA class I proteins. Because association with appropriate peptides is required for MHC class I to be correctly folded, 293T cells may lack certain molecular mechanisms to generate such peptides for HLA-Cw4.
Fig. 2.
MHC class II molecules induce cell surface expression of misfolded HLA-Cw4. Here, 293T cells transiently transfected with Flag-HLA-Cw4-IRES-GFP and HLA-DRαβ and Flag-HLA-Cw4 stably transfected 721.221 cells were stained with L31 and anti-HLA-DRαβ mAb or KIR2DL1-Ig (specific for misfolded HLA-C, HLA-DR and correctly folded HLA-C, respectively; continuous lines) and their fluorescence intensities on GFP-positive cells or total cells are shown. Control staining: shaded histograms. Representative data of three independent experiments are shown.
Fig. 3.
Expression of misfolded HLA class I on CHO cells induced by HLA-DR. Wild-type HLA-Cw*04:01, HLA-Cw*03:04, HLA-A69:01 and HLA-B15:01 plasmids were transiently transfected into CHO cells with GFP plasmids in the presence (HLA-DR) or absence (Mock) of HLA-DRαβ (01:01) plasmids. Expression of misfolded HLA class I (HC10 mAb, thick lines, upper panel) and HLA-DR (thick lines, lower panel) on GFP-positive cells is shown. Control staining: shaded histograms. Data are representative of three independent experiments.
MHC class II molecules present misfolded HLA-Cw4 protein via the peptide-binding groove
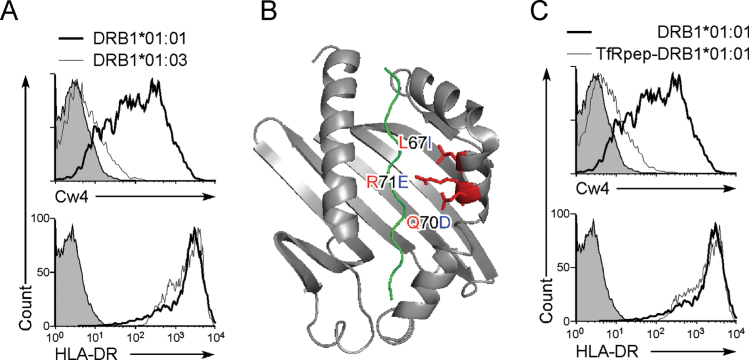
To elucidate how MHC class II molecules support the expression of misfolded HLA-Cw4, we analyzed HLA-Cw4 transport by different HLA-DR β chain alleles because HLA-DR α chain is homogeneous. We found that the HLA-DRA*01:01/HLA-DRB1*01:03 complex did not enable cell surface expression of HLA-Cw4, whereas HLA-DRA*01:01/DRB1*01:01, which we cloned from the 721.221 cDNA library, did so efficiently (Fig. 4A). Three amino acids are different between DRB1*01:01 and DRB1*01:03 at residues 67, 70 and 71. These are the residues that are involved in the formation of pockets 6 and 7 of the HLA-DR peptide-binding groove (Fig. 4B) (2, 3), suggesting that misfolded HLA-Cw4 binds to the peptide-binding groove and is thus transported to the cell surface. To investigate this possibility, a peptide derived from amino acids 680–696 of the transferrin receptor (TfR) and a linker peptide were inserted between the signal sequence and the N-terminus of mature HLA-DRB1*01:01 (TfRpep-HLA-DRB1*01:01), because the TfR peptide is bound by HLA-DRB1*01:01 at relatively high affinity (5) and peptides attached to the N-terminus of mature MHC class II β chain are known to be efficiently presented (10, 17). The HLA-DRA*01:01 / TfRpep-HLA-DRB1*01:01 complex did not allow surface HLA-Cw4 expression, although the TfR peptide did not affect the expression of HLA-DR itself (Fig. 4C). Similarly, HLA-Cw4 expression was also induced by some HLA-DQ and DP alleles (data not shown). Therefore, the HLA-Cw4 heavy chain seems to associate with the peptide-binding groove of certain HLA class II molecules.
Fig. 4.
Misfolded HLA-Cw4 is transported to the cell surface by associating with the peptide-binding groove of MHC class II molecules. (A) HLA-DRB1*01:01, but not HLA-DRB1*01:03, allows cell surface HLA-Cw4 expression. HLA-DRB1*01:01 (thick lines), HLA-DRB1*01:03 (thin lines) or control plasmids (shaded histograms) were co-transfected with HLA-DRA*01:01 and DsRed plasmids into 293T cells stably transfected with Flag-Cw4-IRES-GFP. Expression of Flag-Cw4 and HLA-DR on DsRed- and GFP-positive cells is shown. (B) Amino acid differences between DRB1*01:01 (in red) and DRB1*01:03 (in blue). Structure of DRB1*01:01 with peptide (green) is illustrated. (C) Inhibition of HLA-Cw4 surface expression by a MHC class II-binding peptide. HLA-DRB1*01:01 (thick lines), TfRpep-HLA-DRB1*01:01 (thin lines) or control plasmid (shaded histograms) was co-transfected with HLA-DRA*01:01 and DsRed plasmids into 293T cells stably transfected with Flag-Cw4-IRES-GFP. Expression of Flag-Cw4 and HLA-DR on DsRed- and GFP-positive cells is shown. Data are representative of at least three independent experiments.
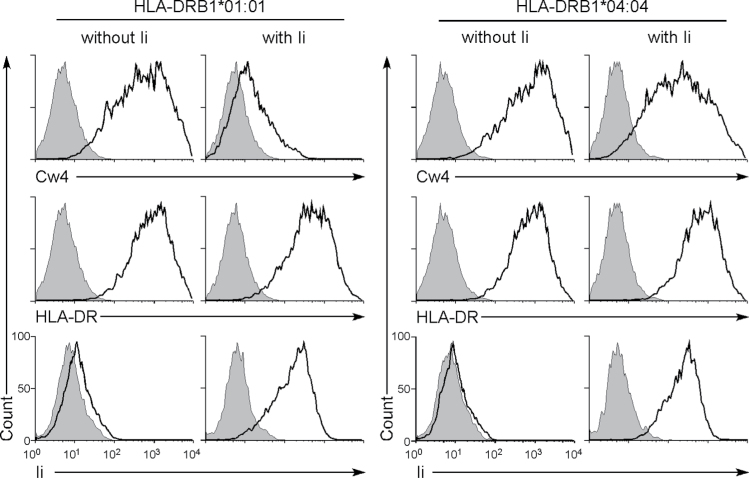
The Ii subunit associates with newly synthesized MHC class II molecules and blocks the peptide-binding site while the complex is transported to the endosomal compartment (7, 8, 18). Because 293T cells do not express Ii, it is possible that newly synthesized HLA-DR proteins do not associate with HLA-Cw4 in the presence of Ii. Accordingly, co-transfection of Ii together with the HLA-DRA*01:01 and HLA-DRB1*01:01 cDNA resulted in a significant decrease of cell surface HLA-Cw4 expression. However, HLA-Cw4 transported by the HLA-DRA*01:01/HLA-DRB1*04:04 complex was only slightly affected by Ii (Fig. 5), suggesting that the binding affinity of Ii to the HLA-DRA*01:01/HLA-DRB1*04:04 complex is weaker than that of HLA-Cw4. Therefore, the efficiency of transportation of misfolded HLA-Cw4 to the cell surface by HLA-DR molecules is affected by a balance between the strength of association of HLA-Cw4 and Ii to HLA-DR and/or the relative amounts of HLA-DR and Ii proteins present.
Fig. 5.
Effect of Ii on induction of cell surface expression of HLA-Cw4 induced by HLA-DR. Plasmids containing Flag-HLA-Cw4 and HLA-DRB1*01:01 or HLA-DRB1*04:04 were co-transfected into 293T cells together with HLA-DRA*01:01, GFP and Ii plasmids or were mock transfected. Expressions of Flag-HLA-Cw4 (HLA-Cw4), HLA-DR or Ii (thick lines) on GFP-positive cells are shown. Expression of Ii was analyzed by intracellular staining. Control staining: shaded histograms. Data are representative of three independent experiments.
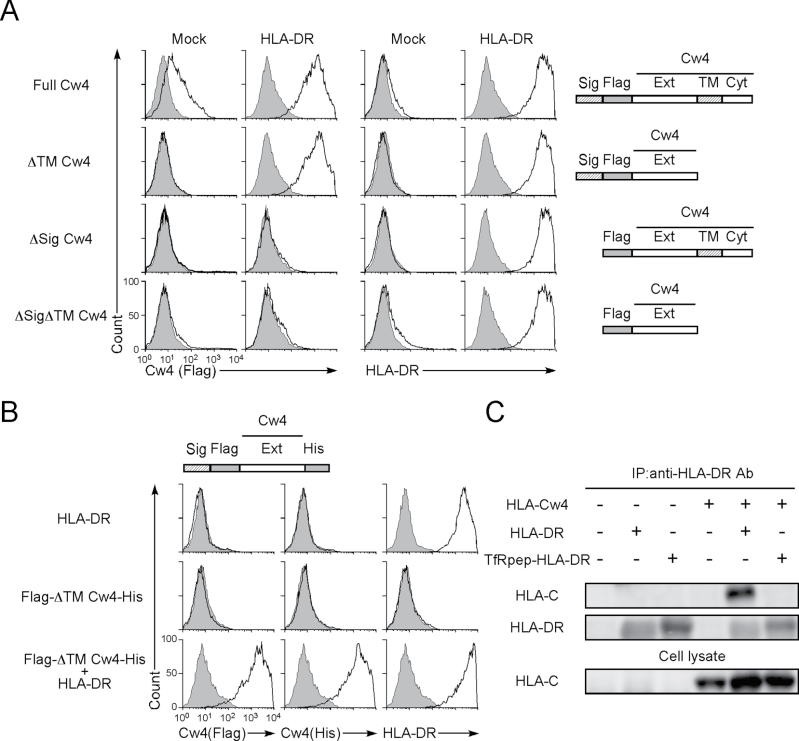
Considering that HLA-Cw4 appears to be transported to the cell surface via the peptide-binding groove, the transmembrane domain of HLA-Cw4 should not be required for its cell surface expression. When the extracellular domain of HLA-Cw4, lacking the transmembrane region, was co-transfected with HLA-DRA*01:01 and HLA-DRB1*01:01, it was detected on the cell surface in the presence of HLA-DR but not in the absence of HLA-DR (Fig. 6A). Similar results were obtained using the extracellular domain of HLA-Cw4 with a His-tag at the C-terminus and a Flag-tag at the N-terminus (Fig. 6B). In contrast, HLA-Cw4 lacking the signal sequence, which is required for nascent proteins to be transported to the endoplasmic reticulum (ER), was not transported to the cell surface by HLA-DR, suggesting that HLA-Cw4 associates with HLA-DR in the ER (Fig. 6A). When HLA-DR protein was immunoprecipitated from lysates of HLA-DR and HLA-Cw4 co-transfectants, a band of the size predicted for HLA-Cw4 was co-precipitated from HLA-DRA*01:01 and HLA-DRB1*01:01 transfectants but not from cells transfected with HLA-DRA*01:01 and TfRpep-HLA-DRB1*01:01 (Fig. 6C).
Fig. 6.
Signal sequence and not the transmembrane region of HLA-Cw4 is required for transport by HLA-DR. (A) Plasmids containing Flag-tagged full-length HLA-Cw*04:01 (Full Cw4), transmembrane and cytoplasmic domain region-deleted HLA-Cw*04:01 (ΔTM Cw4), signal sequence-deleted HLA-Cw*04:01 (ΔSig Cw4) and the signal sequence-, transmembrane- and cytoplasmic region-deleted HLA-Cw*04:01 (ΔTMΔSig Cw4) were co-transfected into 293T cells with HLA-DRαβ (01:01) and GFP plasmids. Expression of HLA-Cw4 (Flag, thick lines, left panel) and HLA-DR (thick lines, right panel) on GFP-expressing cells is shown. Control staining: shaded histograms. Constructs of HLA-Cw*04:01 cDNA (Cw4) are shown on the right-hand side of the figure (Sig: signal sequence; Flag: Flag-tag; Ext: extracellular domain; TM: transmembrane region; and Cyt: cytoplasmic domain). (B) Plasmids containing N-terminal Flag-tagged and C-terminal His-tagged extracellular domain of HLA-Cw*04:01 (Flag-ΔTM Cw4-His) were transfected into 293T cells together with HLA-DRαβ (01:01) and GFP plasmids. The transfectants were stained with anti-Flag mAb (thick lines, left panel), anti-His mAb (thick lines, center panel), anti-HLA-DR mAb (thick lines, right panel) or control mAb (shaded histograms) and their expressions on GFP-positive cells are shown. A Flag-ΔTM HLA-Cw4-His cDNA construct is shown at the top of the figure (Sig: signal sequence; Flag: Flag-tag; Ext: extracellular domain; and His: His-tag). (C) Association of HLA-Cw4 with HLA-DR. Plasmids containing HLA-DRB1*01:01 or TfRpep-HLA-DRB1*01:01, HLA-DRA*01:01 and the extracellular domain of HLA-Cw4 were transfected into 293T cells. HLA-DR was immunoprecipitated from cell lysates, and HLA-Cw4 and HLA-DR in precipitates and whole cell lysates were immunoblotted. All data are representative of at least three independent experiments.
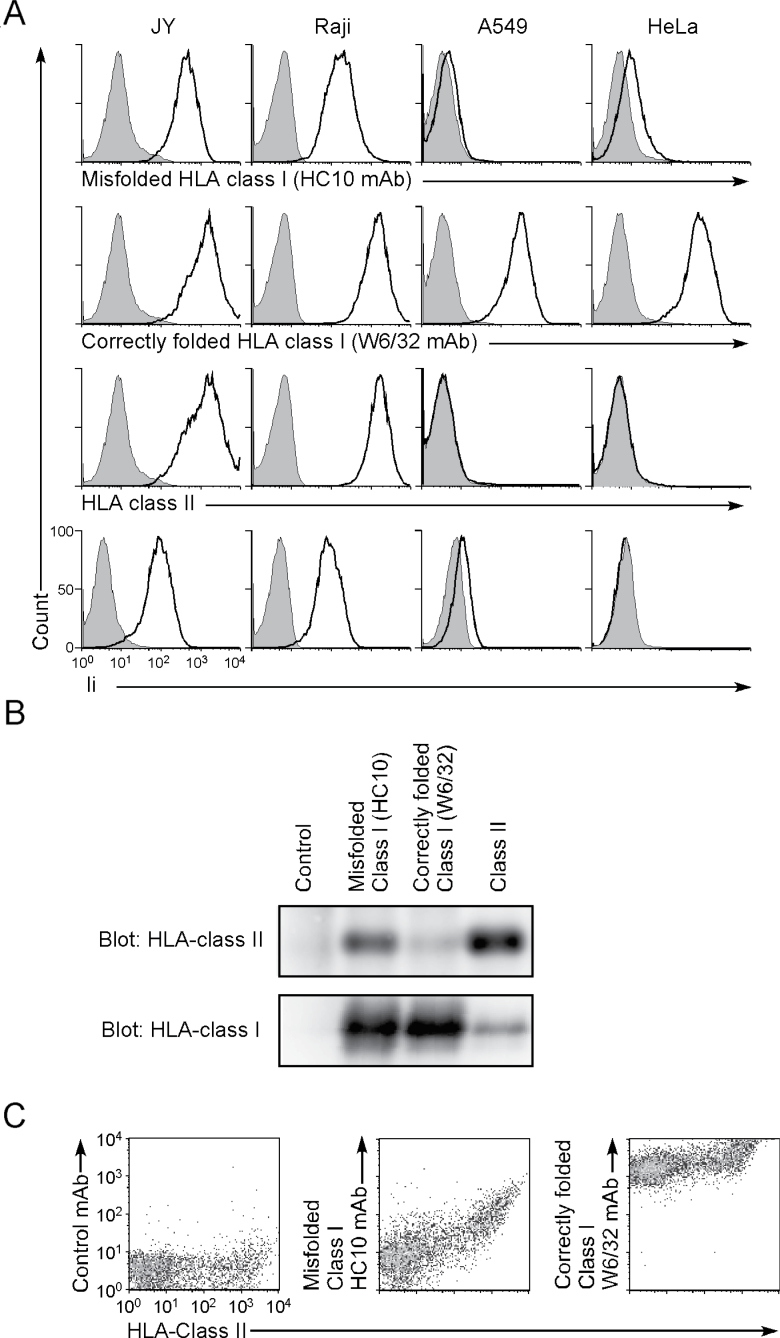
The JY and Raji B-cell lines express both HLA class II and Ii, and they were stained well by the HC10 mAb, which is specific for misfolded HLA class I (Fig. 7A). In contrast, the HeLa and A549 cell lines do not express HLA class II, and the HLA class I proteins on their cell surface were recognized by W6/32, which is specific for correctly folded HLA class I, but not by HC10 mAb, indicating that only correctly folded HLA class I is expressed on these HLA class II-negative cell lines (Fig. 7A). HLA class I was immunoprecipitated from JY and Raji B cells using HC10 or W6/32 mAb and analyzed by western blotting with an anti-HLA-DR mAb. HLA-DR proteins were detected in the HC10 mAb immunoprecipitates, but few HLA-DR proteins were detected in the W6/32 mAb immunoprecipitates (Fig. 7B). These data indicated that misfolded HLA class I, but not correctly folded HLA class I, is associated with HLA class II in non-transfected cell lines. Similarly, expression of misfolded HLA class I on HLA class II-expressing cells was also detected on the surface of PHA-stimulated PBMCs (Fig. 7C). Expression levels of misfolded HLA class I on the PHA-stimulated PBMCs correlated with the amount of HLA class II on the cell surface. Taken together, we conclude that it is the whole misfolded HLA class I protein, and not degraded fragments thereof, which associates in the ER with the HLA-DR peptide-binding groove and is then transported to the cell surface.
Fig. 7.
Expression of misfolded HLA class I on HLA class II-expressing cells. (A) JY, Raji, A549 and HeLa cells were stained with HC10 (specific for misfolded HLA class I), W6/32 (specific for correctly folded HLA class I), anti-HLA-DR or anti-Ii mAb (thick lines), respectively. Expression of Ii was analyzed by intracellular staining. Control staining: shaded histograms. Data are representative of three independent experiments. (B) Cell lysates of JY cells were immunoprecipitated with HC10, W6/32, anti-HLA-DR or control mAb. Immunoprecipitated proteins were separated by SDS–PAGE and immunoblotted with anti-class I (HC10) or rabbit anti-HLA-DRα antibodies. (C) PHA-stimulated PBMCs were stained with HC10 or W6/32 and anti-HLA-DR mAbs and their correlated expression is shown. Data are representative of three independent experiments.
MHC class II molecules present misfolded HEL protein via the peptide-binding groove
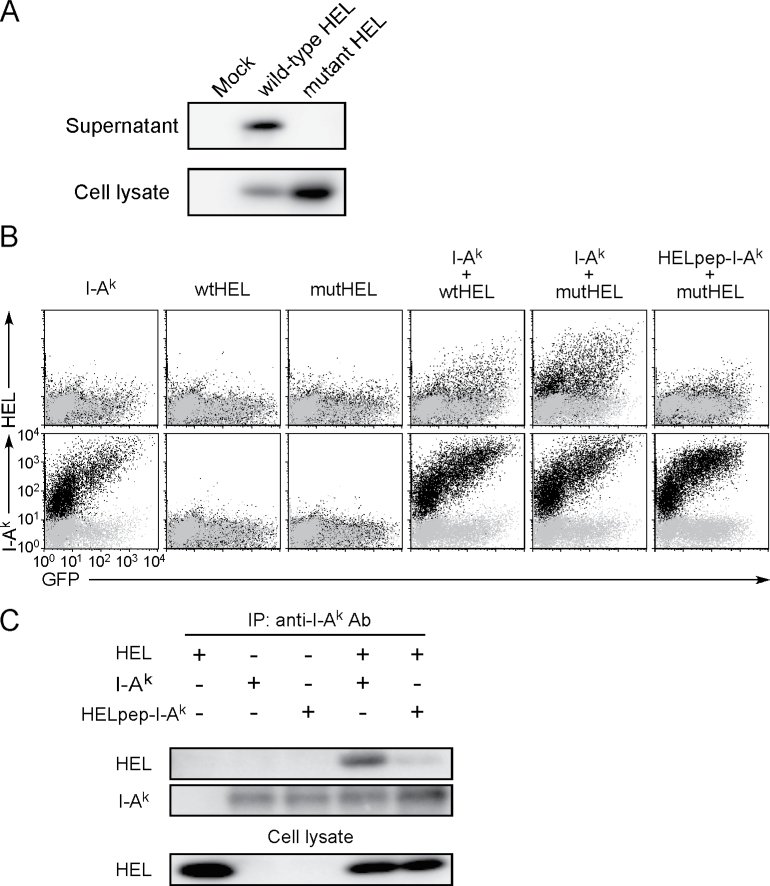
We addressed whether other misfolded proteins could also be presented by MHC class II molecules. HEL is a structurally well-characterized secreted soluble protein the immunological response to which has been well investigated. HEL has several disulfide bonds that play a crucial role in its correct folding (19) and misfolded proteins are usually not secreted (20). When wtHEL and HEL with mutations at two cysteine residues, Cys30 and Cys64, were transfected, wtHEL but not mutant HEL protein was secreted into the culture supernatant, although both were detected in cell lysates (Fig. 8A). This indicated that the cysteine mutations induced misfolding of HEL. We analyzed whether wtHEL or cysteine-mutant HEL is transported to the cell surface by mouse MHC class II, I-Ak. Thus, wtHEL transfected together with I-Ak was weakly detected on the cell surface but was not seen in the absence of I-Ak (Fig. 8B). Cysteine-mutant HEL co-transfected with I-Ak was efficiently presented on I-Ak relative to wtHEL. When we analyzed an I-Ak that was engineered to express a covalently bound HEL peptide (amino acids 48–61; HELpep-I-Ak), which binds to I-Ak with high affinity (21), HELpep-I-Ak failed to present cysteine-mutant HEL, whereas cell surface expression of the HELpep-I-Ak was equivalent to that of wild-type I-Ak. The Ii also blocked HEL presentation by I-Ak, similar to Cw4 presentation by HLA-DRB1*01:01 (data not shown). These data suggest that misfolded intact mutant HEL protein also associates with the peptide-binding groove of MHC class II molecule in the ER and is transported to the cell surface.
Fig. 8.
Presentation of misfolded HEL protein by MHC class II molecules. (A) Western blot of soluble HEL in supernatants and lysates of 293T cells transfected with plasmids containing Flag-tagged wild-type or cysteine-mutant HEL. Flag-tagged HEL was detected by using anti-Flag mAb. (B) Plasmids containing Flag-tagged wild-type HEL (wtHEL) or cysteine-mutant HEL (mutHEL) were transfected into 293T cells together with GFP and I-Ak or HELpep-I-Ak plasmids. Expression of HEL, I-Ak and GFP is shown (black dots). Control staining was overlaid (gray dots). (C) Association of HEL with I-Ak. Plasmids containing Flag-tagged mutant HEL were co-transfected with C-terminus His-tagged I-Akα and I-Akβ or HELpep-I-Akβ, into 293T cells, and I-Ak was precipitated from lysates. HEL and I-Ak in the precipitates and whole cell lysates were blotted with anti-Flag or anti-His mAb, respectively. All data are representative of at least three independent experiments.
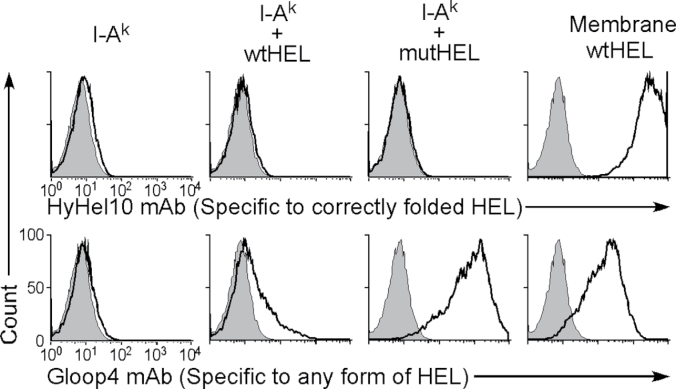
We confirmed that misfolded HEL protein is presented by MHC class II in co-immunoprecipitation experiments. As shown in (Fig. 8C), a protein of the predicted molecular weight of mutant HEL was co-precipitated with I-Ak, but this was not seen using HELpep-I-Ak. This indicates that the whole HEL protein is present at the peptide-binding site of I-Ak. We investigated whether HEL protein presented on I-Ak is misfolded by using the two anti-HEL mAbs, HyHEL10 (22) and Gloop4 (23). HyHEL10 is specific for correctly folded HEL protein, whereas Gloop4 mAb recognizes HEL protein independently of its structure. Both mAbs recognized wtHEL that was expressed on the cell surface by creating a chimeric protein linking HEL to the transmembrane and cytoplasmic domains of the PILRα protein (membrane-tethered HEL; Fig. 9) (24). Mutant HEL protein presented on I-Ak was recognized by Gloop4 mAb but not by HyHEL10 mAb. Notably, the secreted wild-type HEL protein, weakly presented on I-Ak, was also recognized by Gloop4 but not by HyHEL10 mAb, indicating that misfolded HEL protein was specifically presented on I-Ak even when wtHEL protein was also present. These data support the notion that MHC class II molecules associate with misfolded proteins and transport them to the cell surface.
Fig. 9.
Only misfolded HEL is presented on I-Ak. Plasmids containing wild-type and mutant HEL were transfected into 293T cells with I-Ak and GFP. Plasmids containing membrane-tethered wild-type HEL (Membrane wtHEL) and cysteine-mutant HEL (mutHEL) were transfected with GFP plasmids. Cell surface HEL on GFP-positive cells was analyzed using HyHEL10 and Gloop4 mAbs. All data are representative of three independent experiments.
B-cell activation by misfolded proteins presented by MHC class II molecules
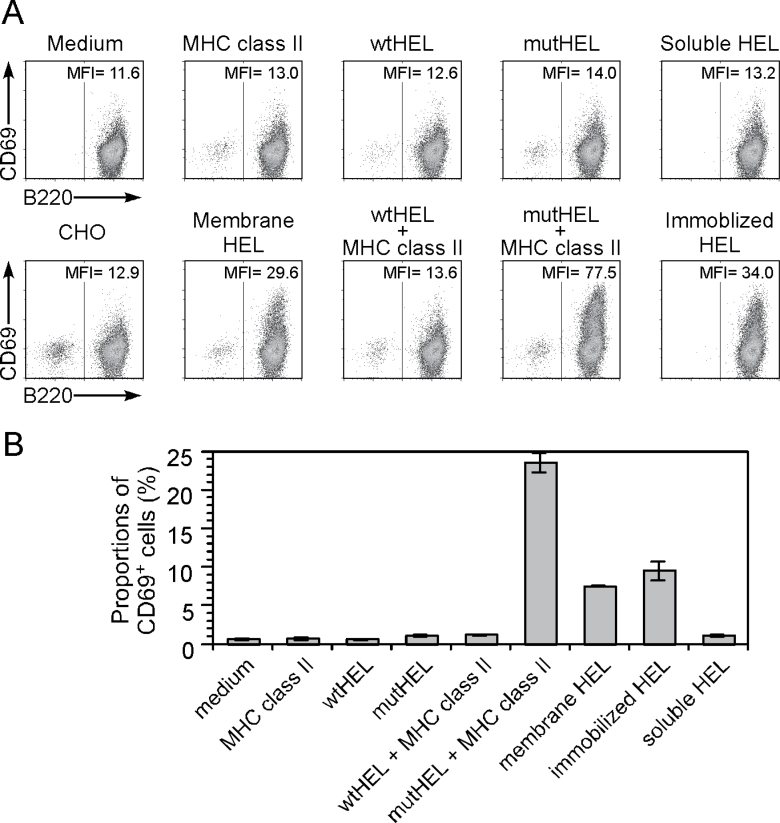
Because it is known that membrane-associated antigens elicit B-cell activation more efficiently than soluble antigens do (25, 26), we analyzed whether misfolded proteins presented on MHC class II molecules induce an antigen- specific B-cell response. We generated a mouse A20 B-cell line expressing the HEL-specific Gloop4 IgM BCR (G4-A20) and analyzed cell activation by monitoring expression of CD69, an early activation marker (Fig. 10A and B). Wild-type HEL or cysteine-mutant HEL transfectants and soluble HEL protein did not stimulate G4-A20 cells. On the other hand, cysteine-mutant HEL protein presented on MHC class II molecules stimulated G4-A20 cells more efficiently than plate-bound or membrane-tethered HEL protein did. These results suggest that misfolded proteins presented on MHC class II molecules can efficiently activate antigen-specific B cells.
Fig. 10.
Activation of HEL-specific B cells by misfolded HEL protein presented on MHC class II molecules. (A) Plasmids containing wild-type HEL (wtHEL) and cysteine-mutant HEL (mutHEL) were co-transfected into CHO cells with GFP plasmid in the presence or absence of I-Ak. Plasmids containing membrane-tethered wild-type HEL (membrane HEL) were also co-transfected with GFP plasmids into CHO cells. GFP-positive cells were sorted 2 days later and co-cultured with the Gloop4-BCR-transfected A20 B cell line, G4-A20. G4-A20 was also stimulated with soluble or immobilized (plate-bound) HEL protein. Cells stimulated for 4h were stained with anti-CD69 and B220 (anti-CD45) mAbs. (B) G4-A20 was stimulated as in (A) and proportions of cells expressing CD69 within B220-positive cells are shown. Data are means ± SD of triplicate cultures, representative of three independent experiments.
Discussion
Although the major function of MHC class II is to present exogenous peptide antigens derived from endosomal compartments of antigen-presenting cells to CD4+ T cells, our data indicate that MHC class II molecules also present endogenous intact misfolded proteins. Many studies have identified peptide antigens presented on MHC class II molecules by eluting them from MHC class II molecules (27). Structural analyses have confirmed the nature of these MHC class II–small peptide complexes (2–4). However, because ultrafiltration is generally used to prepare samples for analyses of peptides eluted from MHC class II molecules to remove other proteins, intact proteins presented on MHC class II might have been overlooked. Structural analyses of MHC class II proteins have revealed that both ends of the MHC class II peptide-binding groove are open. Therefore, unlike MHC class I, it is possible that MHC class II molecules might bind linear epitopes in large intact proteins. Indeed, several studies have suggested that MHC class II molecules have the capacity to associate with large proteins (9, 28, 29).
Ii binds newly synthesized MHC class II molecules and prevents associations with other proteins (8, 18). However, the affinity of Ii-derived peptide (CLIP peptide) binding to MHC class II proteins is not always higher than that of other peptide antigens (30). Therefore, linear epitopes exposed in misfolded proteins might associate with MHC class II molecules in the ER in place of Ii if they have a high affinity or high abundance. Indeed, in our study, the efficiency of the blockade of the binding of misfolded proteins to MHC class II molecules by Ii varied according to the allele examined. The MHC class II molecules that associate with misfolded proteins other than li will not be transported to the endosomal compartments because unlike li most proteins do not have an endosomal sorting signal. Therefore, misfolded proteins associated with MHC class II molecules might be directly transported to the cell surface in an intact form by MHC class II molecules. Taken together, our findings that MHC class II molecules associate with misfolded proteins and transport them to the cells surface are compatible with the known structural characteristics of MHC class II molecules.
Folding of nascent proteins is an imperfect process and misfolded proteins are constitutively produced in cells under normal conditions (20). However, the misfolded proteins are generally degraded in cells by the process of ER-associated degradation and are not transported outside the cells (20). Therefore, potentially autoreactive B cells may not have been exposed to such misfolded proteins during their development and thus may not be tolerant to them. In situations in which MHC class II expression is increased or induced, misfolded proteins might be transported to the cell surface by the MHC class II molecules. Because misfolded proteins presented on MHC class II molecules stimulated a B-cell line expressing an antigen-specific BCR, misfolded proteins aberrantly transported by MHC class II molecules might induce autoreactive B-cell activation, which could potentially result in auto-antibody production. There are no transgenic mice expressing a BCR reactive to misfolded proteins. Therefore, generation of BCR-transgenic mice expressing a Gloop4 BCR will enable us to examine the function of MHC class II-misfolded protein complexes on B-cell activation under physiological conditions. Further analyses of misfolded protein presentation by MHC class II products might elucidate as yet unknown functions of MHC class II molecules in immune responses.
Supplementary data
Supplementary data are available at International Immunology Online.
Funding
Scientific Research from the Ministry of Education, Science and Culture, Japan (M.K., K.H., T.S. and H.A.); National Institutes of Health (AI068129) to L.L.L.
Supplementary Material
Acknowledgements
We thank Drs B. Diamond and S. Sakaguchi for critical reading of the manuscript, H. Kikutani, S. Yamasaki and N. Sorimachi for helpful comments and Ms R. Hirohata and Ms A. Sakamoto for technical assistance. The authors declare no competing financial interests. L.L.L. is an American Cancer Society Research Professor.
References
- 1. Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. 1987. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329: 506 [DOI] [PubMed] [Google Scholar]
- 2. Stern L. J., Brown J. H., Jardetzky T. S, et al. 1994. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 368: 215 [DOI] [PubMed] [Google Scholar]
- 3. Ghosh P., Amaya M., Mellins E., Wiley D. C. 1995. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature 378: 457 [DOI] [PubMed] [Google Scholar]
- 4. Fremont D. H., Hendrickson W. A., Marrack P., Kappler J. 1996. Structures of an MHC class II molecule with covalently bound single peptides. Science 272: 1001 [DOI] [PubMed] [Google Scholar]
- 5. Chicz R. M., Urban R. G., Lane W. S, et al. 1992. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature 358: 764 [DOI] [PubMed] [Google Scholar]
- 6. Rudensky A. Y., Preston-Hurlburt P., Hong S. C., Barlow A., Janeway C. A., Jr 1991. Sequence analysis of peptides bound to MHC class II molecules. Nature 353: 622 [DOI] [PubMed] [Google Scholar]
- 7. Honey K., Rudensky A. Y. 2003. Lysosomal cysteine proteases regulate antigen presentation. Nat. Rev. Immunol. 3: 472 [DOI] [PubMed] [Google Scholar]
- 8. Neefjes J., Jongsma M. L., Paul P., Bakke O. 2011. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11: 823 [DOI] [PubMed] [Google Scholar]
- 9. Busch R., Cloutier I., Sékaly R. P., Hämmerling G. J. 1996. Invariant chain protects class II histocompatibility antigens from binding intact polypeptides in the endoplasmic reticulum. EMBO J. 15: 418 [PMC free article] [PubMed] [Google Scholar]
- 10. Scott C. A., Peterson P. A., Teyton L., Wilson I. A. 1998. Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues. Immunity 8: 319 [DOI] [PubMed] [Google Scholar]
- 11. Giacomini P., Beretta A., Nicotra M. R, et al. 1997. HLA-C heavy chains free of β2-microglobulin: distribution in normal tissues and neoplastic lesions of non-lymphoid origin and interferon-gamma responsiveness. Tissue Antigens 50: 555 [DOI] [PubMed] [Google Scholar]
- 12. Stam N. J., Spits H., Ploegh H. L. 1986. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 137: 2299 [PubMed] [Google Scholar]
- 13. Shiratori I., Ogasawara K., Saito T., Lanier L. L., Arase H. 2004. Activation of natural killer cells and dendritic cells upon recognition of a novel CD99-like ligand by paired immunoglobulin-like type 2 receptor. J. Exp. Med. 199: 525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan Q. R., Garboczi D. N., Winter C. C., Wagtmann N., Long E. O., Wiley D. C. 1996. Direct binding of a soluble natural killer cell inhibitory receptor to a soluble human leukocyte antigen-Cw4 class I major histocompatibility complex molecule. Proc. Natl. Acad. Sci. U.S.A. 93: 7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sibilio L., Martayan A., Setini A, et al. 2005. Impaired assembly results in the accumulation of multiple HLA-C heavy chain folding intermediates. J. Immunol. 175: 6651 [DOI] [PubMed] [Google Scholar]
- 16. Arosa F. A., Santos S. G., Powis S. J. 2007. Open conformers: the hidden face of MHC-I molecules. Trends Immunol. 28: 115 [DOI] [PubMed] [Google Scholar]
- 17. Kozono H., White J., Clements J., Marrack P., Kappler J. 1994. Production of soluble MHC class II proteins with covalently bound single peptides. Nature 369: 151 [DOI] [PubMed] [Google Scholar]
- 18. Cresswell P. 1994. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 12: 259 [DOI] [PubMed] [Google Scholar]
- 19. Ohkuri T., Ueda T., Tsurumaru M., Imoto T. 2001. Evidence for an initiation site for hen lysozyme folding from the reduced form using its dissected peptide fragments. Protein Eng. 14: 829 [DOI] [PubMed] [Google Scholar]
- 20. Meusser B., Hirsch C., Jarosch E., Sommer T. 2005. ERAD: the long road to destruction. Nat. Cell Biol. 7: 766 [DOI] [PubMed] [Google Scholar]
- 21. Nelson C. A., Viner N. J., Young S. P., Petzold S. J., Unanue E. R. 1996. A negatively charged anchor residue promotes high affinity binding to the MHC class II molecule I-Ak . J. Immunol. 157: 755 [PubMed] [Google Scholar]
- 22. Smith-Gill S. J., Lavoie T. B., Mainhart C. R. 1984. Antigenic regions defined by monoclonal antibodies correspond to structural domains of avian lysozyme. J. Immunol. 133: 384 [PubMed] [Google Scholar]
- 23. Darsley M. J., Rees A. R. 1985. Three distinct epitopes within the loop region of hen egg lysozyme defined with monoclonal antibodies. EMBO J. 4: 383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Satoh T., Arii J., Suenaga T, et al. 2008. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132: 935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qi H., Egen J. G., Huang A. Y., Germain R. N. 2006. Extrafollicular activation of lymph node B cells by antigen-bearing dendritic cells. Science 312: 1672 [DOI] [PubMed] [Google Scholar]
- 26. Batista F. D., Harwood N. E. 2009. The who, how and where of antigen presentation to B cells. Nat. Rev. Immunol. 9: 15 [DOI] [PubMed] [Google Scholar]
- 27. Rammensee H., Bachmann J., Emmerich N. P., Bachor O. A., Stevanović S. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50: 213 [DOI] [PubMed] [Google Scholar]
- 28. Castellino F., Zappacosta F., Coligan J. E., Germain R. N. 1998. Large protein fragments as substrates for endocytic antigen capture by MHC class II molecules. J. Immunol. 161: 4048 [PubMed] [Google Scholar]
- 29. Sercarz E. E., Maverakis E. 2003. MHC-guided processing: binding of large antigen fragments. Nat. Rev. Immunol. 3: 621 [DOI] [PubMed] [Google Scholar]
- 30. Davenport M. P., Quinn C. L., Chicz R. M, et al. 1995. Naturally processed peptides from two disease-resistance-associated HLA-DR13 alleles show related sequence motifs and the effects of the dimorphism at position 86 of the HLA-DR β-chain. Proc. Natl. Acad. Sci. U.S.A. 92: 6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.