Abstract
Inactivation of the Staphylococcus aureus tricarboxylic acid (TCA) cycle delays the resolution of cutaneous ulcers in a mouse soft tissue infection model. In this study, it was observed that cutaneous lesions in mice infected with wild-type or isogenic aconitase mutant S. aureus strains contained comparable inflammatory infiltrates, suggesting the delayed resolution was independent of the recruitment of immune cells. These observations led us to hypothesize that staphylococcal metabolism can modulate the host immune response. Using an in vitro model system involving RAW 264.7 cells, the authors observed that cells cultured with S. aureus aconitase mutant strains produced significantly lower amounts of nitric oxide (NO•) and an inducible nitric oxide synthase as compared to those cells exposed to wild-type bacteria. Despite the decrease in NO• synthesis, the expression of antigen-presentation and costimulatory molecules was similar in cells cultured with wild-type and those cultured with aconitase mutant bacteria. The data suggest that staphylococci can evade innate immune responses and potentially enhance their ability to survive in infected hosts by altering their metabolism. This may also explain the occurrence of TCA cycle mutants in clinical S. aureus isolates.
Keywords: Staphylococcus aureus, Aconitase, Nitric oxide, RAW 264.7 cells, Immune evasion
Introduction
Staphylococcus aureus is a gram-positive bacterium capable of surviving extreme environmental conditions and causing severe infections in both immunocompetent and immunodeficient individuals. It is estimated that worldwide S. aureus colonize, both transiently and persistently, two billion people at any given time [1]. During an infection, phagocytes recognize bacteria through toll-like receptor (TLR)-2 and produce bactericidal oxidants such as, reactive oxygen species (ROS) and nitric oxide (NO•), via nicotinamide adenine dinucleotide phosphate-oxidase, myeloperoxidase, and inducible nitric oxide synthase (iNOS) [2–4]. Despite the presence of bactericidal oxidants, staphylococci can survive in macrophages through robust anti-oxidant defense mechanisms such as carotenoid pigments, superoxide dismutases, manganese homeostasis, and catalases [5]. In addition, it was recently shown that S. aureus adapts to nitrosative stress by expressing the NO•-inducible L-lactate dehydrogenase (ldh) as one mechanism to evade the bactericidal effects of NO• [6].
The regulation of virulence in S. aureus is complex, involving the agr quorum-sensing system/riboregulator RNAIII, the SarA family of regulators, and an alternative sigma factor (σB) [7]. In addition to these regulatory elements, the authors have demonstrated a causal relationship between tricarboxylic acid (TCA) cycle activity and virulence factor synthesis [7, 8]. During those studies, the authors created a TCA cycle mutant of S. aureus by inactivating aconitase and demonstrated that mice infected with this TCA cycle mutant strain required a longer time to develop cutaneous ulcers and to resolve the infection as compared to mice infected with wild-type (wt) strain [7]. These observations led us to investigate function of the TCA cycle in the host-pathogen interaction.
Materials and methods
Bacterial strains and growth conditions
Staphylococcus aureus wt strains UAMS-1 and SA564 and the isogenic aconitase mutant strains UAMS-1-acnA and SA564-acnA have been described [7, 9]. Bacteria were grown in tryptic soy broth (TSB; BD Biosciences, San Jose, CA) or on TSB-containing agar (15 g per l) at 37°C with a flask volume-to-medium ratio of 10:1 aerated by shaking at 225 rpm.
Mouse soft-tissue infection model and histology
Immunocompetent, hairless outbred mice (Crl:SKH1-hrBR•SKH1) were procured from the Charles River Laboratories International (Wilmington, MA). The mice were maintained in accordance with the guidelines of the Animal Care and Use Committee at Rocky Mountain Laboratories, Hamilton, MT.
Exponential growth phase bacteria (O.D.600 ≈ 1.0; 25 ml) were harvested by centrifugation, washed two times with ice-cold phosphate buffered saline (PBS), centrifuged, suspended in 20 ml PBS, and frozen at −80°C until use. Prior to use and at the time of inoculation, the colony forming units (CFU) per ml were determined. Mice were inoculated subcutaneously in the dorsal neck region with 5 × 107 CFU and monitored for the development and progression of cutaneous ulcers at the sites of infection [7]. A total of 40 animals were used with five mice per group, each inoculated with either wt or mutant bacteria. At 3, 6, 24, and 48 h postinfection, mice were sacrificed and skin samples were taken from injection sites and fixed by immersion in 10% phosphate buffered formalin. Tissues were sectioned in 5 μM thickness and stained with hematoxylin and eosin [H and E; 10] and evaluated for inflammatory changes such as abscessation, epidermitis, and epidermal and dermal necrosis.
Intracellular detection of ROS and NO• by flow cytometry
The bacterial suspensions were prepared to a final concentration of 1 × 108 CFU per ml in RPMI supplemented with 50% human serum to opsonize bacteria at 37°C for 30 min. After pelleting, the bacteria were washed with 1× endotoxin-free PBS and suspended in DMEM containing 10% fetal bovine serum (FBS). RAW 264.7 cells (clone, TIB-71) obtained from American Type Culture Collection (Manassas, VA) were maintained in antibiotic-free DMEM supplemented with 10% FBS, hereafter called growth medium. To determine intracellular production of ROS and NO• in RAW 264.7 cells exposed to bacteria, 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester (CM-H2DCFDA) and 4-amino-5-methylamino-2′, 7′-difluorofluorescein (DAF-FM; Invitrogen, Eugene, CA) were used as ROS and NO• indicators, respectively [11–13]. In brief, 100 μl each of RAW 264.7 cells (4 × 106 per ml) and bacteria (2 × 107 per ml) were plated in 96-well plates at a 1:5 ratio, and the plates were incubated at 37°C for 6 h. The oxidation sensitive dyes CM-H2DCFDA and DAF-FM were added at various concentrations (0–1 μM) during the last 20 min of incubation, and the cells were washed twice with 1× PBS. After staining the cells with a cell death marker, 7-aminoactinomycin D (7-AAD; Invitrogen), cells were acquired by flow cytometry (FC; FACScan, BD Bio-sciences), and the fluorescence intensity of live cells (7-AAD−) positive for CM-H2DCFDA and DAF-FM was analyzed by Flow Jo software (Tree Star, Ashland, OR).
Analysis of iNOS mRNA expression
RAW 264.7 cells (6 × 106) and the bacteria (3 × 107) were plated in 6-well plates at a 1:5 ratio in 3 ml of growth medium, and the plates were incubated for 6 h. In addition, lipopolysaccharide (LPS) (100 ng per ml) was used as a positive control [14]. To extract RNA, the medium was removed and the cells were lysed using RLT buffer containing guanidium thiocyanate (Qiagen, RNeasy kit, Valencia, CA), and the samples were treated with RNAse-free DNAse I according to the manufacturer’s recommendations (Qiagen). To make certain that the RNA samples were free of residual DNA, a second round of DNAse digestion was performed using amplification grade DNAse I (Invitrogen), and cDNAs were synthesized utilizing Superscript III reverse transcriptase kit as recommended (Invitrogen). First, it was qualitatively verified the expression of iNOS mRNA by PCR, the levels of which were compared with an endogenous control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA, using a Gradient Thermal cycler (Eppendorf, Hauppauge, NY) as described [15]. The primer sets used were: iNOS-forward, 5′ CCTCCTCCACCCTACCAAGT 3′; iNOS-reverse, 5′ CACCCAAAGTGCTTCAGTCA 3′; GAPDH-forward, 5′ CGGCAAATTCAACGGCACAGTCAA 3′; GAPDH-reverse, 5′ CTTTCCAGAGGGGCCATCCACAG 3′. The PCR products were stained with ethidium bromide and resolved in 1% agarose gel electrophoresis. Second, it was quantitatively analyzed the relative fold induction of iNOS mRNA expression utilizing commercially obtained TaqMan PCR probes and primers (ABI Biosystems, Carlsbad, CA) using an iCycler (Bio-Rad Laboratories, Hercules, CA). The cDNAs derived from three replicates for each treatment group were used for analysis, and the fold induction of iNOS mRNA expression was calculated by normalizing to GAPDH mRNA [15, 16].
Detection of surface molecules by flow cytometry
RAW 264.7 cells and the bacteria were plated as above at a 1:5 ratio, and after 6 h of incubation, the cells were harvested and stained with antibodies for TLR-2, CD80, CD86, major histocompatibility complex (MHC) class II, CD40 (eBioscience, San Diego, CA), and 7-AAD. The clones of the respective antibodies were 6C2, 16-10A1, GL1, M5/114.15.2, and 1C10. The reaction mixtures were incubated for 20 min on ice, and the cells were washed and acquired by FC (FACScan). Percentages of cells positive for each marker were then determined in the live (7-AAD−) subset.
Intracellular cytokine detection
RAW 264.7 cells and the bacteria were plated in 6-well plates at 1:5 ratio in growth medium. After incubating for 2.5 h, the cultures were supplemented with 2 mM monensin [Golgi stop, BD Biosciences; 10, 17] and incubated further for 4 h. It was also used cells in medium alone or cells treated with LPS (1 μg per ml) as negative and positive controls, respectively. At the end of incubation, cells were harvested and washed once with 1× PBS. After fixation and permeabilization, the cells were stained with antibodies for interleukin (IL)-1β (rabbit polyclonal), IL-6 (clone, MPS-20F3), and tumor necrosis factor (TNF)-α (clone, MP6-XT22) and their respective isotype controls (eBioscience) and 7-AAD. Cells were acquired by FC, and frequencies of cytokine-secreting cells were then enumerated in live (7-AAD−) cell populations [10].
Statistics
Differences in the levels of iNOS mRNA, ROS, NO•, and cytokine induction in RAW 264.7 cells cultured with wt or mutant strains of S. aureus were analyzed by Student’s t test. To determine differences in inflammatory changes in cutaneous tissues at each time point postinfection with wt or mutant bacteria, skin sections were examined for abscessation, epidermitis, and necrosis and the differences between groups were compared using one-tailed Fisher’s exact test. P ≤ 0.05 values were considered significant.
Results and discussion
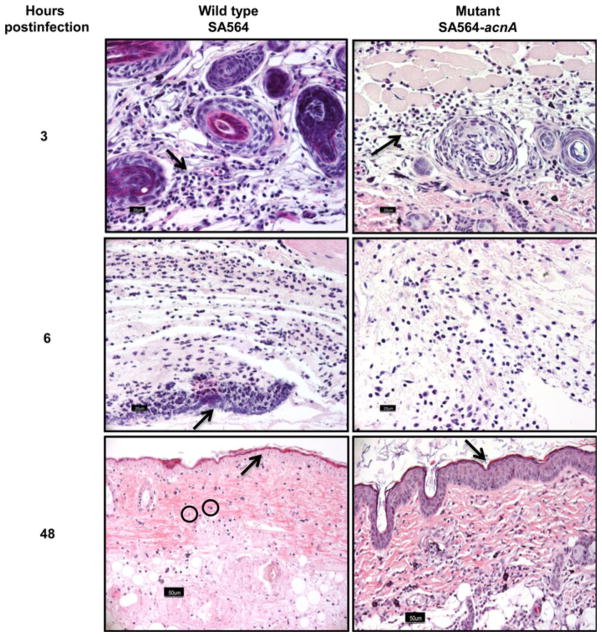
Aconitase inactivation in S. aureus altered the temporal synthesis of secreted virulence and cell-associated adhesion factors [7]. Using a mouse soft tissue infection model, mice infected with an aconitase mutant strain (SA564-acnA) took longer to develop lesions, re-epithelialize the sites of infection, and to regain their pre-inoculation body weight relative to mice infected with an isogenic wild-type strain (SA564) [7], suggesting that disruption of the TCA cycle in S. aureus alters the host’s response to the bacteria. In this report, it was demonstrate that S. aureus metabolism does not affect the recruitment of inflammatory cells but does alter the host immune response, raising the possibility that one mechanism by which S. aureus evades the immune response is by altering its central metabolism. Support for this possibility was recently reported by Richardson et al. [6]. To test this possibility, skin samples were harvested from mice infected with S. aureus wt (SA564) or aconitase mutant (SA654-acnA) strains and assessed by histological evaluation for inflammatory changes. At 3 h postinfection, histological evaluation of skin sections from mice infected with either wt or aconitase mutant strains had identical inflammatory infiltrates, predominantly comprised of neutrophils and a few macrophages (Fig. 1). As the infection progressed, cellular infiltrates and bacteria were less pronounced in tissues sampled at 6, 24, and 48 h from mice infected with strain SA654-acnA as compared to strain SA564. Notably, skin samples obtained from mice infected with wt (10/10) but not mutant bacteria (4/10) consistently showed necrosis of the epidermis and dermis beyond 24 h (P = 0.0054). Similarly, at 48 h there was remarkable epidermitis in mice infected with wt bacteria (5/5) but not in the aconitase mutant (1/5)-infected mice (P = 0.024). These observations indicate the host recognizes TCA cycle inactivated bacteria and recruits phagocytes to the sites of infection; however, it does not address whether differences in the activation state of infiltrates exist. To address this question, an in vitro culture system using the mouse macrophage cell line, RAW 264.7, was established to examine the production of bactericidal oxidants in response to wt and mutant strains of S. aureus at a single cell level by FC. Specifically, CM-H2DCFDA was used as a broad range indicator of ROS, such as hydrogen peroxide, hydroxyl radicals, peroxyl radicals, and peroxynitrite anions, while NO• production was assessed using DAF-FM [11–13]. Figure 2 and Table 1 show that cells cultured with S. aureus aconitase mutants UAMS-1-acnA and SA564-acnA produced significantly lower amounts of NO• as indicated by the lower frequency of DAF-FM-positive cells when compared with the cells exposed to wt bacteria (P<0.05). Similarly, cells cultured with wild-type S. aureus strains produced greater amounts of ROS relative to the aconitase mutant strains when analyzed using CMH2DCFDA-positive cells (Table 1). Taken together, these data suggest that TCA cycle inactivation does not alter the recruitment of phagocytes; however, it does alter the activation of those infected phagocytes.
Fig. 1.
Histological evaluation of skin samples harvested from mice infected with wt and aconitase mutant S. aureus. Skin samples were harvested from mice infected with wt or mutant S. aureus at the indicated time intervals postinfection, and the cutaneous sections were stained with H and E to evaluate inflammatory changes (arrows). Note similar suppurative dermatitis at 3 h postinfection with wt or mutant bacteria. At 6 h, suppuration and bacterial colonization was more extensive in mice infected with wt than with mutant bacteria. Tissues sampled at 48 h from mice infected with wt but not mutant bacteria consistently showed epidermal and dermal necrosis, including pyknosis and karyorrhexis of dermal fibroblasts (shown in circles). Original magnification, ×400 (bar 20 mM)
Fig. 2.
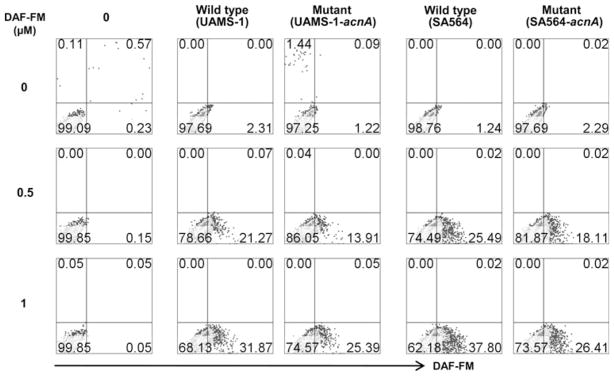
RAW 264.7 cells exposed to aconitase-mutant S. aureus produce less nitric oxide than wt strains. RAW 264.7 cells were cultured with wt or aconitase-inactivated S. aureus mutants at 1:5 ratio for 6 h at 37°C. The cultures were then exposed to DAF-FM during the last 20 min of incubation as an NO indicator. After washing, cells were stained with 7-AAD and acquired by FC. Percentages of DAF-FM+ cells were then determined in the live (7-AAD−) subset. Representative data from four individual experiments are shown
Table 1.
Induction of reactive oxygen species and nitric oxide by wild type and aconitase-mutant S. aureus in RAW 264.7 cells
| Dye (μM) | Medium control | Wild typea (UAMS-1) | Mutanta (UAMS-1-acnA) | Wild typeb (SA-564) | Mutantb (SA-564-acnA) | |
|---|---|---|---|---|---|---|
| ROS | 0 | 0.04 ± 0.04 | 1.41 ± 1.17 | 1.6 ± 1.29 | 0.75 ± 0.66 | 1.63 ± 1.22 |
| 0.5 | 0.19 ± 0.085 | 34.36 ± 6.18 | 23.00 ± 5.43 | 36.18 ± 3.17 | 29.07 ± 7.64 | |
| 1 | 0.39 ± 0.15 | 62.78 ± 10.00 | 50.16 ± 8.44 | 71.48 ± 2.16 | 54.12 ± 5.92 | |
| NO | 0 | 0.21 ± 0.11 | 3.08 ± 0.36 | 2.16 ± 0.51 | 1.20 ± 0.23 | 2.60 ± 0.39 |
| 0.5 | 0.12 ± 0.02 | 23.01 ± 2.13† | 13.54 ± 1.88† | 29.37 ± 4.59‡ | 18.03 ± 1.13‡ | |
| 1 | 0.12 ± 0.05 | 39.41 ± 4.02 | 27.55 ± 5.33 | 50.20 ± 6.28 | 34.43 ± 2.99 |
Numbers are Mean ± SEM (n = 4)
P = 0.01
P = 0.04
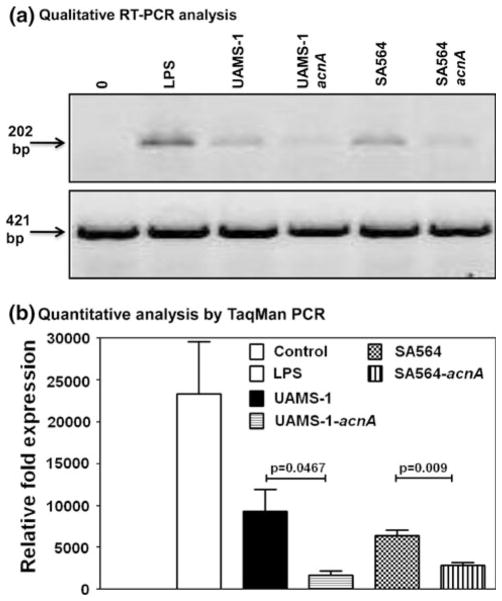
The decreased synthesis of NO• by RAW 264.7 cells exposed to aconitase mutant strains UAMS-1-acnA and SA564-acnA relative to the isogenic wild-type strains (Fig. 2) could occur at the level of transcription or by bacterial interference with the iNOS protein-mediated NO• synthesis [18, 19]. To determine if the decreased production of NO• in cells exposed to aconitase mutant strains was due to a decrease in iNOS transcription, iNOS mRNA expression was analyzed by RT-PCR in cells exposed to wt or mutant bacteria using LPS as a positive control. As shown in Fig. 3, both wt strains (UAMS-1 and SA564) induced the expression of iNOS mRNA in RAW 264.7 cells; however, iNOS mRNA expression was significantly decreased in cells exposed to the bacterial mutants (P ≤ 0.05). Together these data indicate that aconitase inactivation in S. aureus suppresses iNOS synthesis, resulting in decreased NO• production. This has important implications for the host-pathogen interaction because NO• is a critical mediator of bacterial killing [5, 20] and when produced in excess, NO• can contribute to tissue damage during inflammation [21, 22]. These observations and the fact that naturally occurring S. aureus TCA cycle mutants have been reported [23], suggest there is immune system selective pressure on bacterial TCA cycle activity. Any selective pressure that results in decreased TCA cycle activity will create metabolic signals within the bacteria [24] that result in enhanced bacterial survival, decreased virulence factor synthesis, and an alteration of bacterial metabolism [7, 25]; thus, staphylococci may use to evade the host’s defense mechanisms. Finally, these observations may explain the long duration needed to establish and resolve infections in mice inoculated with aconitase mutant bacteria [7].
Fig. 3.
RAW 264.7 cells exposed to aconitase-mutant S. aureus express lower amounts of iNOS mRNA than wt strains. a Qualitative RT-PCR analysis. RAW 264.7 cells were cultured with wt or aconitase-inactivated S. aureus mutants at 1:5 ratio for 6 h at 37°C. After washing, total RNA was extracted, and synthesized cDNAs and iNOS mRNA expression was examined by PCR using sequence specific primers. The ethidium bromide-stained PCR products were resolved in 1% agarose gel electrophoretic analysis, and shown are the expected sizes of iNOS (top panel) and GAPDH (bottom panel) PCR products. b Quantitative analysis by TaqMan PCR. cDNAs were generated from RAW 264.7 cells cultured with wt or aconitase-mutant bacteria as above and the relative fold induction of iNOS mRNA expression was determined by TaqMan PCR analysis by normalizing the expression levels of iNOS mRNA to GAPDH mRNA. Mean ± SEM values are shown (n = 3)
Transcription of iNOS and the synthesis of NO• are regulated by inflammatory cytokines [26–28]. To determine if aconitase mutant strains altered the production of inflammatory cytokines, the authors enumerated by FC the frequencies of cells producing IL-1β, IL-6, and TNF-α at the single cell level. The expression of IL-1β and TNF-α in RAW 264.7 cells infected with wt and those infected with mutant strains was similar; however, the number of IL-6-secreting cells was approximately twofold lower in cells cultured with UAMS-1-acnA but not strain SA564-acnA (Fig. 4). Since both mutant strains carry the same mutations, it was anticipated that cytokine responses would be similar; however, S. aureus strain-dependent variations in host immune responses are common [29, 30]. Traditionally, IL-6 is regarded as one of the prototypical inflammatory cytokines produced in response to S. aureus infection [31], but recent reports indicate that IL-6 can also have an anti-inflammatory function [32, 33]. Although the data provide an association between aconitase inactivation and decreased secretion of IL-6, additional studies are required to investigate whether suppressed IL-6 production favors bacterial survival or their destruction.
Fig. 4.

RAW 264.7 cells exposed to aconitase-mutant S. aureus produce lower amounts of IL-6 than wt strains. RAW 264.7 cells were incubated with wt or aconitase-inactivated S. aureus mutants at 1:5 ratio for 2.5 h in growth medium, and after adding Golgistop, cells were further incubated for 4 h. Cells were then stained with 7-AAD, fixed and permeabilized, followed by staining with anti-IL-6 or isotype control. Cells were acquired by FC, and the percentage of IL-6-secreting cells was enumerated in the live (7-AAD−) subset. Representative data from three individual experiments are shown
Upon entry into a host, S. aureus produce numerous virulence determinants [34, 35] that can induce phagocyte recognition through TLR-2 via pathogen-associated molecular patterns. In addition, during the formation of an abscess, T cells are activated through the CD28-CD80/CD86 costimulation pathway [36], while CD4 helper cells interact with macrophages via the CD40–CD40 ligand pathway to potentiate their bactericidal effects [37]. S. aureus aconitase mutants have been shown to decrease synthesis of virulence determinants such as lipase (geh), enterotoxin C (sec), α-toxin (hla), β-toxin (hlb), δ-toxin (hld), σB (sigB), capsule (cap), and clumping factor (clfA) [7]; thus, it is possible that antigen presentation could be affected by aconitase inactivation. To determine if the decreased synthesis of virulence determinants by the aconitase mutant strains affect adaptive immunity, the authors assayed for the presence of antigen-presentation (MHC class-II) and costimulatory molecules (CD80, CD86, and CD40; Table 2). Both the wt and mutant strains of S. aureus induced CD80, MHC class-II, and CD40 molecules in RAW 264.7 cells to a similar level. Although the expression of CD80 tended to be lower in cells cultured with aconitase mutant strains as compared to those cultured with the wt bacteria, the differences were not significant. These data strongly suggest that phagocyte antigen presentation is independent of S. aureus TCA cycle activity.
Table 2.
Comparative analysis of antigen-presentation and costimulatory molecules induced by wild type and aconitase-mutant S. aureus in RAW 264.7 cells
| Medium control | LPS (100 ng/ml) | Wild type (UAMS-1) | Mutant (UAMS-1-acnA) | Wild type (SA-564) | Mutant (SA-564-acnA) | |
|---|---|---|---|---|---|---|
| TLR-2 | 99.96 ± 0.01 | 99.92 ± 0.024 | 97.98 ± 0.77 | 98.11 ± 0.47 | 98.51 ± 0.70 | 98.33 ± 0.32 |
| CD80 | 59.16 ± 8.77 | 70.65 ± 6.13 | 90.52 ± 3.08 | 83.90 ± 8.48 | 88.8 ± 3.62 | 78.22 ± 12.44 |
| CD86 | 73.24 ± 13.38 | 76.58 ± 12.06 | 72.96 ± 12.74 | 64.24 ± 16.23 | 69.84 ± 13.71 | 71.56 ± 13.67 |
| MHC-II | 0.57 ± 0.46 | 2.42 ± 1.94 | 1.49 ± 0.46 | 1.57 ± 0.36 | 1.28 ± 0.22 | 1.047 ± 0.62 |
| CD40 | 6.05 ± 2.18 | 81.35 ± 2.89 | 8.97 ± 1.71 | 7.77 ± 1.00 | 8.51 ± 0.83 | 8.34 ± 0.82 |
Numbers are Mean ± SEM (n = 3)
Staphylococcus aureus have evolved several mechanisms that favor their survival in infected hosts. These include interference with chemotaxis, opsonin-mediated phagocytosis, and detoxification of bactericidal oxidants [5]. Recently it was shown that staphylococci can adapt to nitrosative stress and maintain virulence by promoting the production of lactate through NO•-inducible ldh [6]. Normally, bacteria induce lactate dehydrogenases when the oxidation of NADH through the respiratory chain is inhibited. In the case of the S. aureus NO•-inducible ldh, NO• prevents the respiratory chain from oxidizing NADH and forces carbon through an NO•-inducible ldh to oxidize NADH [6, 38, 39]. Similarly, inactivation of the TCA cycle redirects carbon away from the TCA cycle and into over-flow metabolism pathways, including ldh. In other words, when phagocyte-produced NO• decreases bacterial respiratory activity, NADH accumulates, synthesis of the NO•-inducible ldh is activated, and carbon flow-through the TCA cycle is redirected into ldh, allowing for the oxidation of NADH. Concomitantly, the decrease in TCA cycle activity results in a decrease in phagocyte-produced NO• (Fig. 2 and Table 1) and an increase in S. aureus survival [7]. In total, it is tempting to speculate that mammalian immune responses evolved to recognize bacterial metabolic signatures, and this has resulted in the repurposing of bacterial metabolic pathways to suppress the host’s ability to mount an effective innate immune response.
Acknowledgments
This manuscript is a contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act and by the COBRE Program from the National Center for Research Resources (P20-RR-17675, NIH), Redox Biology Center, University of Nebraska–Lincoln. Additional funding was provided by the National Institutes of Health to GAS (AI087668) and by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Contributor Information
Chandirasegaran Massilamany, School of Veterinary Medicine and Biomedical Sciences, University of Nebraska-Lincoln, Room 202, Bldg VBS, Lincoln, NE 68583, USA.
Arunakumar Gangaplara, School of Veterinary Medicine and Biomedical Sciences, University of Nebraska-Lincoln, Room 202, Bldg VBS, Lincoln, NE 68583, USA.
Donald J. Gardner, Mountain Veterinary Branch, Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA
James M. Musser, Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA
David Steffen, School of Veterinary Medicine and Biomedical Sciences, University of Nebraska-Lincoln, Room 202, Bldg VBS, Lincoln, NE 68583, USA.
Greg A. Somerville, School of Veterinary Medicine and Biomedical Sciences, University of Nebraska-Lincoln, Room 202, Bldg VBS, Lincoln, NE 68583, USA
Jay Reddy, Email: nreddy2@unl.edu, School of Veterinary Medicine and Biomedical Sciences, University of Nebraska-Lincoln, Room 202, Bldg VBS, Lincoln, NE 68583, USA.
References
- 1.Enwemeka CS, Williams D, Hollosi S, Yens D, Enwemeka SK. Visible 405 nm SLD light photo-destroys methicillin-resistant Staphylococcus aureus (MRSA) in vitro. Lasers Surg Med. 2008;40:734–737. doi: 10.1002/lsm.20724. [DOI] [PubMed] [Google Scholar]
- 2.Kengatharan KM, De Kimpe S, Robson C, Foster SJ, Thiemermann C. Mechanism of gram-positive shock: identification of peptidoglycan and lipoteichoic acid moieties essential in the induction of nitric oxide synthase, shock, and multiple organ failure. J Exp Med. 1998;188:305–315. doi: 10.1084/jem.188.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothfork JM, Timmins GS, Harris MN, Chen X, Lusis AJ, Otto M, Cheung AL, Gresham HD. Inactivation of a bacterial virulence pheromone by phagocyte-derived oxidants: new role for the NADPH oxidase in host defense. Proc Natl Acad Sci USA. 2004;101:13867–13872. doi: 10.1073/pnas.0402996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, Golenbock D. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 5.Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- 6.Richardson AR, Libby SJ, Fang FC. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science. 2008;319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- 7.Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun. 2002;70:6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somerville GA, Cockayne A, Durr M, Peschel A, Otto M, Musser JM. Synthesis and deformylation of Staphylococcus aureus delta-toxin are linked to tricarboxylic acid cycle activity. J Bacteriol. 2003;185:6686–6694. doi: 10.1128/JB.185.22.6686-6694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadykov MR, Mattes TA, Luong TT, Zhu Y, Day SR, Sifri CD, Lee CY, Somerville GA. Tricarboxylic acid cycle-dependent synthesis of Staphylococcus aureus Type 5 and 8 capsular polysaccharides. J Bacteriol. 2010;192:1459–1462. doi: 10.1128/JB.01377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massilamany C, Steffen D, Reddy J. An epitope from Acanthamoeba castellanii that cross-react with proteolipid protein 139–151-reactive T cells induces autoimmune encephalomyelitis in SJL mice. J Neuroimmunol. 2010;219:17–24. doi: 10.1016/j.jneuroim.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Devadas S, Zaritskaya L, Rhee SG, Oberley L, Williams MS. Discrete generation of superoxide and hydrogen peroxide by T cell receptor stimulation: selective regulation of mitogen-activated protein kinase activation and fas ligand expression. J Exp Med. 2002;195:59–70. doi: 10.1084/jem.20010659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsue H, Edelbaum D, Shalhevet D, Mizumoto N, Yang C, Mummert ME, Oeda J, Masayasu H, Takashima A. Generation and function of reactive oxygen species in dendritic cells during antigen presentation. J Immunol. 2003;171:3010–3018. doi: 10.4049/jimmunol.171.6.3010. [DOI] [PubMed] [Google Scholar]
- 13.Cohen O, Kfir-Erenfeld S, Spokoini R, Zilberman Y, Yefenof E, Sionov RV. Nitric oxide cooperates with glucocorticoids in thymic epithelial cell-mediated apoptosis of double positive thymocytes. Int Immunol. 2009;21:1113–1123. doi: 10.1093/intimm/dxp079. [DOI] [PubMed] [Google Scholar]
- 14.Ambrozova G, Pekarova M, Lojek A. Effect of polyunsaturated fatty acids on the reactive oxygen and nitrogen species production by raw 264.7 macrophages. Eur J Nutr. 2010;49:133–139. doi: 10.1007/s00394-009-0057-3. [DOI] [PubMed] [Google Scholar]
- 15.Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005;21:389–395. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- 16.Bustin SA. Absolute quantification of mRNA using realtime reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 17.Reddy J, Illes Z, Zhang X, Encinas J, Pyrdol J, Nicholson L, Sobel RA, Wucherpfennig KW, Kuchroo VK. Myelin proteolipid protein-specific CD4 + CD25 + regulatory cells mediate genetic resistance to experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2004;101:15434–15439. doi: 10.1073/pnas.0404444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 19.Chaturvedi R, Asim M, Lewis ND, Algood HM, Cover TL, Kim PY, Wilson KT. L-arginine availability regulates inducible nitric oxide synthase-dependent host defense against Helicobacter pylori. Infect Immun. 2007;75:4305–4315. doi: 10.1128/IAI.00578-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X, Liu Y, Liu S, Diao F, Xu R, Ni X. Lipopolysaccharide primes macrophages to increase nitric oxide production in response to Staphylococcus aureus. Immunol Lett. 2007;112:75–81. doi: 10.1016/j.imlet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Grisham MB, Jourd’Heuil D, Wink DA. Nitric oxide. I. Physiological chemistry of nitric oxide and its metabolites: implications in inflammation. Am J Physiol. 1999;276:G315–G321. doi: 10.1152/ajpgi.1999.276.2.G315. [DOI] [PubMed] [Google Scholar]
- 22.Laroux FS, Pavlick KP, Hines IN, Kawachi S, Harada H, Bharwani S, Hoffman JM, Grisham MB. Role of nitric oxide in inflammation. Acta Physiol Scand. 2001;173:113–118. doi: 10.1046/j.1365-201X.2001.00891.x. [DOI] [PubMed] [Google Scholar]
- 23.Somerville GA, Said-Salim B, Wickman JM, Raffel SJ, Kreiswirth BN, Musser JM. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect Immun. 2003;71:4724–4732. doi: 10.1128/IAI.71.8.4724-4732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadykov MR, Zhang B, Halouska S, Nelson JL, Kreimer LW, Zhu Y, Powers R, Somerville GA. Using NMR metabolomics to investigate tricarboxylic acid cycle-dependent signal transduction in Staphylococcus epidermidis. J Biol Chem. 2010;285:36616–36624. doi: 10.1074/jbc.M110.152843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaupp R, Schlag S, Liebeke M, Lalk M, Gotz F. Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms. J Bacteriol. 2010;192:2385–2394. doi: 10.1128/JB.01472-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jana M, Anderson JA, Saha RN, Liu X, Pahan K. Regulation of inducible nitric oxide synthase in proinflammatory cytokine-stimulated human primary astrocytes. Free Radic Biol Med. 2005;38:655–664. doi: 10.1016/j.freeradbiomed.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 27.Taylor BS, de Vera ME, Ganster RW, Wang Q, Shapiro RA, Morris SM, Jr, Billiar TR, Geller DA. Multiple NF-kappaB enhancer elements regulate cytokine induction of the human inducible nitric oxide synthase gene. J Biol Chem. 1998;273:15148–15156. doi: 10.1074/jbc.273.24.15148. [DOI] [PubMed] [Google Scholar]
- 28.Vila-del Sol V, Diaz-Munoz MD, Fresno M. Requirement of tumor necrosis factor alpha and nuclear factor-kappaB in the induction by IFN-gamma of inducible nitric oxide synthase in macrophages. J Leukoc Biol. 2007;81:272–283. doi: 10.1189/jlb.0905529. [DOI] [PubMed] [Google Scholar]
- 29.Graves SF, Kobayashi SD, DeLeo FR. Community-associated methicillin-resistant Staphylococcus aureus immune evasion and virulence. J Mol Med. 2010;88:109–114. doi: 10.1007/s00109-009-0573-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Cheung GY, Hu J, Wang D, Joo HS, Deleo FR, Otto M. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis. 2010;202:1866–1876. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleming SD, Iandolo JJ, Chapes SK. Murine macrophage activation by Staphylococcal exotoxins. Infect Immun. 1991;59:4049–4055. doi: 10.1128/iai.59.11.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, Hirano T, Chien KR, Yoshimura A. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol. 2003;4:551–556. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- 33.Capparelli R, Nocerino N, Lanzetta R, Silipo A, Amoresano A, Giangrande C, Becker K, Blaiotta G, Evidente A, Cimmino A, Iannaccone M, Parlato M, Medaglia C, Roperto S, Roperto F, Ramunno L, Iannelli D. Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS One. 2010;5:e11720. doi: 10.1371/journal.pone.0011720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knuefermann P, Sakata Y, Baker JS, Huang CH, Sekiguchi K, Hardarson HS, Takeuchi O, Akira S, Vallejo JG. Toll-like receptor 2 mediates Staphylococcus aureus-induced myocardial dysfunction and cytokine production in the heart. Circulation. 2004;110:3693–3698. doi: 10.1161/01.CIR.0000143081.13042.04. [DOI] [PubMed] [Google Scholar]
- 35.Vidlak D, Mariani MM, Aldrich A, Liu S, Kielian T. Roles of Toll-like receptor 2 (TLR2) and superantigens on adaptive immune responses during CNS staphylococcal infection. Brain Behav Immun. 2010;28(2):230–234. doi: 10.1016/j.bbi.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzianabos AO, Chandraker A, Kalka-Moll W, Stingele F, Dong VM, Finberg RW, Peach R, Sayegh MH. Bacterial pathogens induce abscess formation by CD4(+) T-cell activation via the CD28–B7-2 costimulatory pathway. Infect Immun. 2000;68:6650–6655. doi: 10.1128/iai.68.12.6650-6655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schrum LW, Bost KL, Hudson MC, Marriott I. Bacterial infection induces expression of functional MHC class II molecules in murine and human osteoblasts. Bone. 2003;33:812–821. doi: 10.1016/s8756-3282(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 38.Brown GC, Borutaite V. Nitric oxide, cytochrome c and mitochondria. Biochem Soc Symp. 1999;66:17–25. doi: 10.1042/bss0660017. [DOI] [PubMed] [Google Scholar]
- 39.McCollister BD, Hoffman M, Husain M, Vazquez-Torres A. Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob Agents Chemother. 2011 doi: 10.1128/AAC.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]