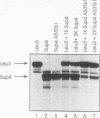
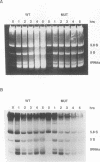
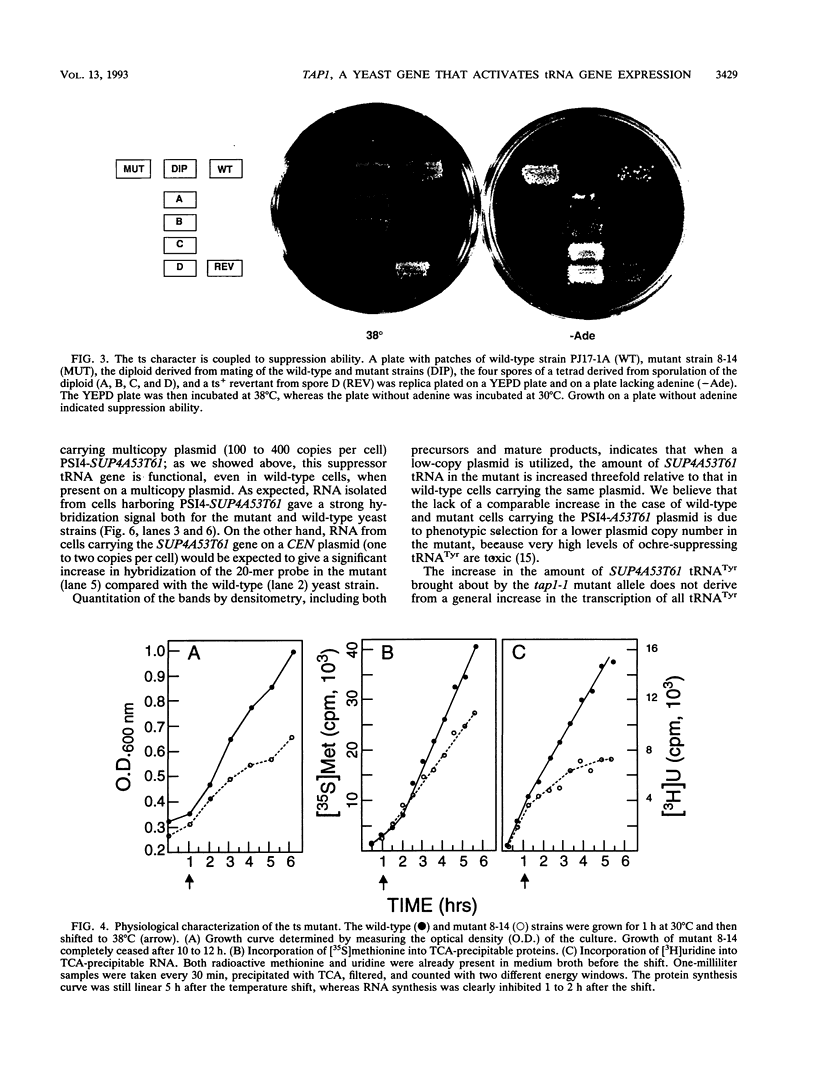
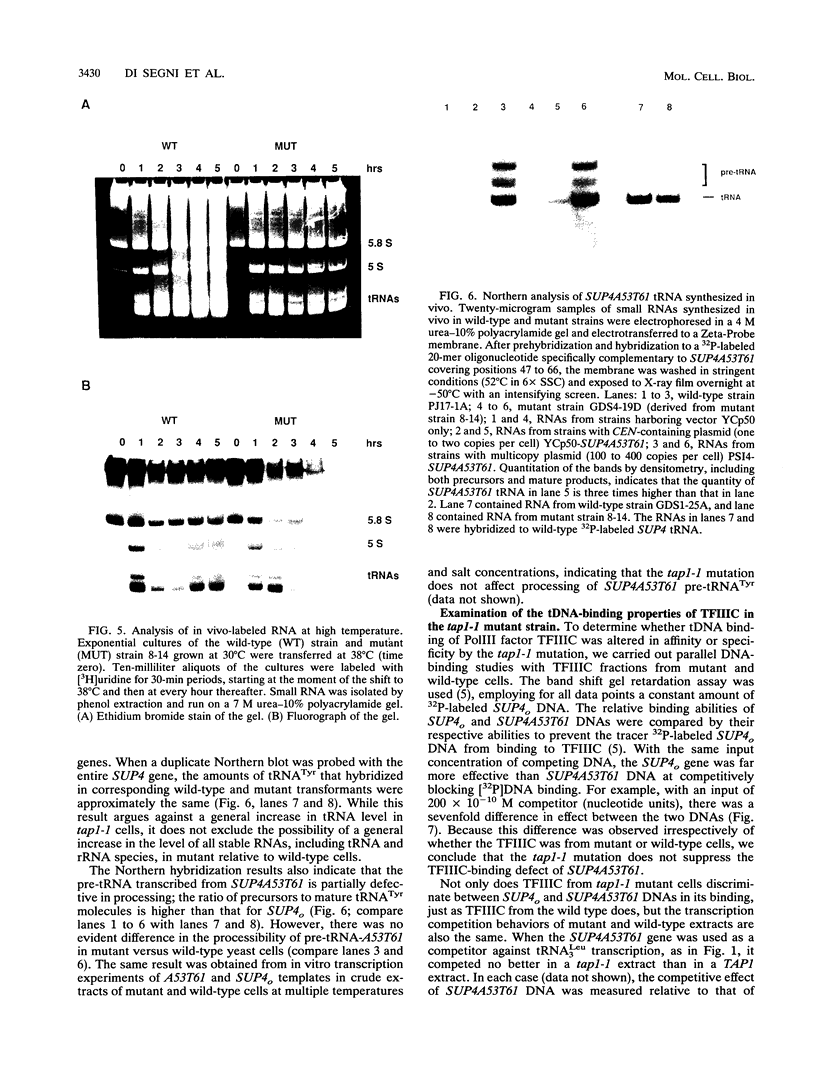
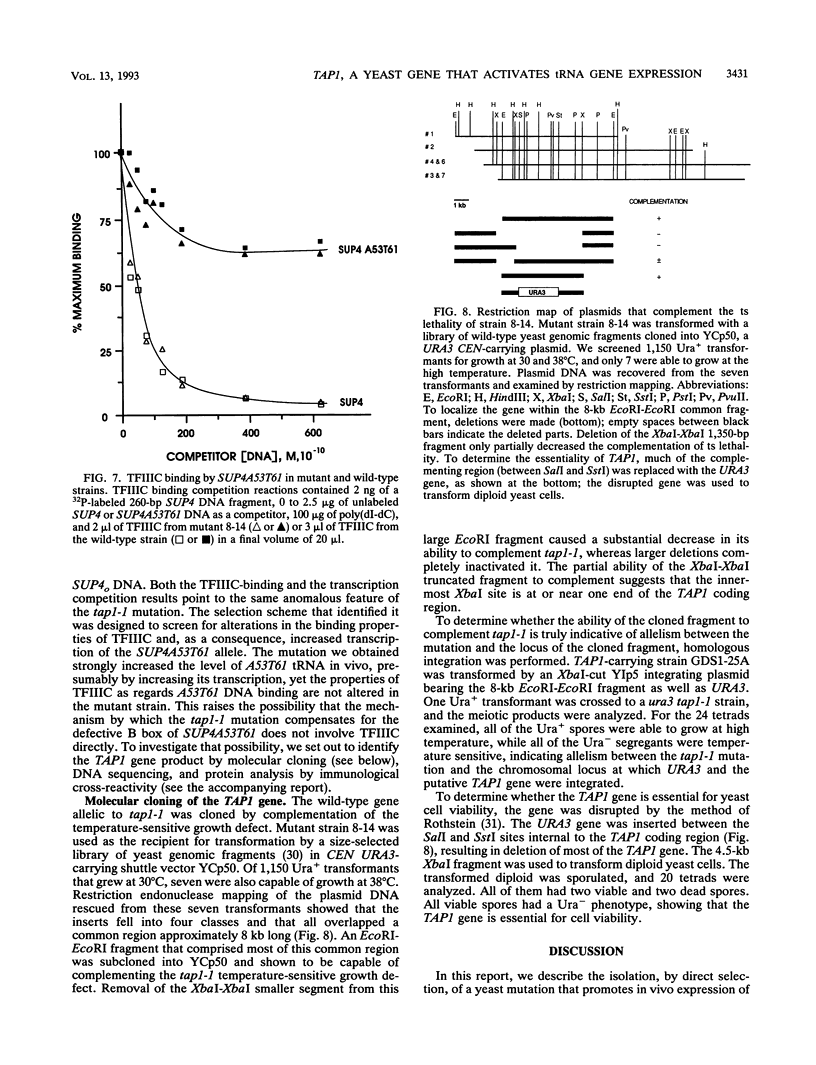
Abstract
We developed a genetic selection system based on nonsense suppression in Saccharomyces cerevisiae to identify mutations in proteins involved in transcription initiation by RNA polymerase III. A SUP4 tRNA(Tyr) internal promoter mutation (A53T61) that was unable to suppress ochre mutations in vivo and was incapable of binding TFIIIC in vitro was used as the target for selection of trans-acting compensatory mutations. We identified two such mutations in the same gene, which we named TAP1 (for transcription activation protein). The level of the SUP4A53T61 transcript was threefold higher in the tap1-1 mutant than in the wild type. The tap1-1 mutant strain was also temperature sensitive for growth. The thermosensitive character cosegregated with the restorer of suppression activity, as shown by meiotic linkage analysis and coreversion of the two traits. At 1 to 2 h after a shift to the restrictive temperature, RNA synthesis was strongly inhibited in the tap1-1 mutant, preceding any effect upon protein synthesis or growth. A marked decrease in tRNA and 5S rRNA synthesis was seen, and shortly after that, rRNA synthesis was inhibited. By complementation of the ts- growth defect, we cloned the wild-type TAP1 gene. It is essential for yeast growth. We show in the accompanying report (T. L. Aldrich, G. Di Segni, B. L. McConaughy, N. J. Keen, S. Whelen, and B. D. Hall, Mol. Cell. Biol. 13:3434-3444, 1993) that TAP1 is identical to RAT1, a yeast gene implicated in poly(A)+ RNA export and that the TAP1/RAT1 gene product has extensive sequence similarity to the protein encoded by another yeast gene (variously named DST2, KEM1, RAR5, SEP1, or XRN1) having exonuclease and DNA strand transfer activity (reviewed by Kearsey and Kipling [Trends Cell Biol. 1:110-112, 1991]).
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldrich T. L., Di Segni G., McConaughy B. L., Keen N. J., Whelen S., Hall B. D. Structure of the yeast TAP1 protein: dependence of transcription activation on the DNA context of the target gene. Mol Cell Biol. 1993 Jun;13(6):3434–3444. doi: 10.1128/mcb.13.6.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison D. S., Goh S. H., Hall B. D. The promoter sequence of a yeast tRNAtyr gene. Cell. 1983 Sep;34(2):655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
- Allison D. S., Hall B. D. Effects of alterations in the 3' flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985 Oct;4(10):2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D. C., Goldstein A. L., Cole C. N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992 Jul;6(7):1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Camier S., Sentenac A., Hall B. D. Gene size differentially affects the binding of yeast transcription factor tau to two intragenic regions. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8768–8772. doi: 10.1073/pnas.84.24.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Gabrielsen O., Hall B. D. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem. 1986 Apr 25;261(12):5275–5282. [PubMed] [Google Scholar]
- Baker R. E., Hall B. D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984 Dec 1;3(12):2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Broach J. R. Construction of high copy yeast vectors using 2-microns circle sequences. Methods Enzymol. 1983;101:307–325. doi: 10.1016/0076-6879(83)01024-1. [DOI] [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi M. S., Melton D. A., Cortese R. Site-directed mutagenesis of a tRNA gene: base alterations in the coding region affect transcription. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1388–1392. doi: 10.1073/pnas.79.5.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby D., Leboy P. S., Guthrie C. Yeast tRNA precursor mutated at a splice junction is correctly processed in vivo. Proc Natl Acad Sci U S A. 1981 Jan;78(1):415–419. doi: 10.1073/pnas.78.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W., Stutz F., Rosbash M., Wickens M. Defects in mRNA 3'-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992 Oct;6(10):1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- Gudenus R., Mariotte S., Moenne A., Ruet A., Memet S., Buhler J. M., Sentenac A., Thuriaux P. Conditional mutants of RPC160, the gene encoding the largest subunit of RNA polymerase C in Saccharomyces cerevisiae. Genetics. 1988 Jul;119(3):517–526. doi: 10.1093/genetics/119.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D. C., Leupold U. Suppressors in yeast. Curr Top Microbiol Immunol. 1974;64(0):1–47. doi: 10.1007/978-3-642-65848-8_1. [DOI] [PubMed] [Google Scholar]
- Heyer W. D., Munz P., Amstutz H., Aebi R., Gysler C., Schuchert P., Szankasi P., Leupold U., Kohli J., Gamulin V. Inactivation of nonsense suppressor transfer RNA genes in Schizosaccharomyces pombe. Intergenic conversion and hot spots of mutation. J Mol Biol. 1986 Apr 5;188(3):343–353. doi: 10.1016/0022-2836(86)90159-2. [DOI] [PubMed] [Google Scholar]
- James P., Hall B. D. ret1-1, a yeast mutant affecting transcription termination by RNA polymerase III. Genetics. 1990 Jun;125(2):293–303. doi: 10.1093/genetics/125.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Kearsey S., Kipling D. Recombination and RNA processing: a common strand? Trends Cell Biol. 1991 Nov;1(5):110–112. doi: 10.1016/0962-8924(91)90101-e. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D., Gillam S., Smith M. Mutations at the yeast SUP4 tRNATyr locus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980 Jul;20(3):701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Lefebvre O., Carles C., Conesa C., Swanson R. N., Bouet F., Riva M., Sentenac A. TFC3: gene encoding the B-block binding subunit of the yeast transcription factor IIIC. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10512–10516. doi: 10.1073/pnas.89.21.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzouki N., Camier S., Ruet A., Moenne A., Sentenac A. Selective proteolysis defines two DNA binding domains in yeast transcription factor tau. Nature. 1986 Sep 11;323(6084):176–178. doi: 10.1038/323176a0. [DOI] [PubMed] [Google Scholar]
- Mosrin C., Riva M., Beltrame M., Cassar E., Sentenac A., Thuriaux P. The RPC31 gene of Saccharomyces cerevisiae encodes a subunit of RNA polymerase C (III) with an acidic tail. Mol Cell Biol. 1990 Sep;10(9):4737–4743. doi: 10.1128/mcb.10.9.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M. C., Weil P. A. Cloning of TFC1, the Saccharomyces cerevisiae gene encoding the 95-kDa subunit of transcription factor TFIIIC. J Biol Chem. 1992 Feb 15;267(5):2894–2901. [PubMed] [Google Scholar]
- Pearson D., Willis I., Hottinger H., Bell J., Kumar A., Leupold U., Söll D. Mutations preventing expression of sup3 tRNASer nonsense suppressors of Schizosaccharomyces pombe. Mol Cell Biol. 1985 Apr;5(4):808–815. doi: 10.1128/mcb.5.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Caspers P., Geiduschek E. P. Effects of temperature and single-stranded DNA on the interaction of an RNA polymerase III transcription factor with a tRNA gene. Cell. 1985 Feb;40(2):311–317. doi: 10.1016/0092-8674(85)90145-x. [DOI] [PubMed] [Google Scholar]
- Swanson R. N., Conesa C., Lefebvre O., Carles C., Ruet A., Quemeneur E., Gagnon J., Sentenac A. Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor tau (TFIIIC). Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4887–4891. doi: 10.1073/pnas.88.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I., Oksman A., López-De-León A. The PCF1-1 mutation increases the activity of the transcription factor (TF) IIIB fraction from Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Jul 25;20(14):3725–3730. doi: 10.1093/nar/20.14.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I., Schmidt P., Söll D. A selection for mutants of the RNA polymerase III transcription apparatus: PCF1 stimulates transcription of tRNA and 5S RNA genes. EMBO J. 1989 Dec 20;8(13):4281–4288. doi: 10.1002/j.1460-2075.1989.tb08614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormington W. M., Bogenhagen D. F., Jordan E., Brown D. D. A quantitative assay for Xenopus 5S RNA gene transcription in vitro. Cell. 1981 Jun;24(3):809–817. doi: 10.1016/0092-8674(81)90106-9. [DOI] [PubMed] [Google Scholar]