Abstract
Introduction
Serum paraoxonase-1 (PON1) retards the oxidation of low-density lipoprotein and cell membranes and is atheroprotective. Polymorphisms in the promoter region of the PON1 gene have been shown to affect serum PON1 levels and have been related to the presence of coronary heart disease (CHD) in some studies. However, contradictory results have been reported with regard to promoter region polymorphisms and CHD presence; therefore we have re-examined the effects of the C-108T and G-909C promoter polymorphisms on PON1 levels and the presence of CHD in a large case-control study.
Material and methods
Paraoxonase-1 activity, concentration and the C-108T and G-909C polymorphisms were measured in 417 people with CHD and 282 healthy controls, in a case control study.
Results
Paraoxonase-1 activity and concentration were significantly lower in the CHD population compared to controls regardless of their C-108T and G-909C genotype (p < 0.001). Paraoxonase-1 activity was significantly different in the C-108T genotypes in both the control and CHD groups in the order TT < TC < CC (p < 0.01). Paraoxonase-1 concentration was significantly different in the CHD group only in the G-909C genotype in the order GG > GC > CC (p < 0.01). Haplotype analysis revealed no consistent patterns of PON1 activity in the CHD population; however, in the controls PON1 activity differed between haplotypes GGCC > GGTC > GGTT (p < 0.05) and GCCC > GCTC > GCTT (p < 0.02). Neither promoter polymorphism was associated with CHD presence.
Conclusions
Paraoxonase-1 status was significantly lower in people with CHD and was affected by the promoter region polymorphisms.
Keywords: paraoxonase-1, coronary heart disease, genetic polymorphisms, high-density lipid, oxidation
Introduction
The paraoxonase (PON) multi-gene family comprises 3 members, PON1, PON2 and PON3 [1]. The genes for all 3 members of the family are widely expressed in mammalian tissues [2]; however, PON1 and PON3 are predominantly located in the plasma associated with high-density lipoprotein (HDL) while PON2 is not found in the plasma but has a wide cellular distribution [3]. PON1, PON2 and PON3 all retard the proatherogenic oxidative modification of low-density lipoprotein (LDL) and cell membranes and are therefore considered to be anti-atherogenic [4]. Paraoxonase-1 is now considered to be a major factor in the antioxidative activity of HDL [3].
The transgenic expression of human PON1 in various mouse models of atherosclerosis has been shown to retard or reverse atherosclerosis by mechanisms which include a reduction in circulating and aortic oxidised LDL (ox-LDL), a reduction in macrophage oxidative stress and foam cell formation, an increase in reverse cholesterol transport and a normalisation of endothelial function [5–7]. Several prospective studies have shown that low PON1 is an independent risk factor for new coronary events, independent of all other risk factors including HDL [8–12]. Although this finding is not universal [13, 14], low PON1 is a general feature of people who develop CHD.
The PON1 gene contains 2 exonic polymorphisms at positions 55(L/M) and 192 (Q/R), resulting in 4 possible isoenzymes. These isoenzymes hydrolyse many substrates, including lipid peroxides, at different rates [15, 16]. HDL from different individuals varies in how active it is in retarding the oxidation of LDL according to the particular PON1 isoforms it contains [15, 16].
Recently, several polymorphisms have been reported to be present in the promoter region of the PON1 gene [17, 18]. Research has indicated that these promoter polymorphisms are associated with highly significant differences in serum concentrations and activities of PON1 and are important genetic determinants of PON1 levels. It has also been reported that the promoter polymorphisms are also associated with the presence of CHD in non-diabetic and type 2 diabetic people [19, 20]. However, contradictory results have been reported with regard to promoter region polymorphisms and CHD presence; therefore we have re-examined the effects of the C-108T and G-909C promoter polymorphisms on PON1 levels and the presence of CHD in a large case-control study of non-diabetic patients with CHD and controls.
Material and methods
Subjects
Patients included in the study were all outpatients attending the cardiology departments at either Manchester Royal Infirmary, UK or the Royal Victoria Hospital, Blackpool, UK. All patients had angiographically proven CHD, which was visually assessed. All patients had stenosis severe enough to require intervention either by coronary angioplasty or surgery. No patients had sustained a myocardial infarction (MI) within 6 months prior to taking part in the study. Patients with diabetes, renal and hepatic disease were excluded from the study. Four hundred and seventeen sequential patients who fulfilled the inclusion criteria were studied. Fully informed consent was obtained and the Central Manchester Local Research Ethics Committee approved the study. One hundred and four patients were receiving β-adrenoreceptor blocking drugs, 89 were taking calcium channel blockers and 154 were on lipid-lowering medication (151 statins, 3 fibrates). All patients were receiving aspirin 75 or 150 mg daily.
The control population consisted of 282 healthy subjects, who attended a routine health check at a general practice. Absence of CHD in the control population was assessed by use of a health questionnaire. None had a history suggestive of CHD. Subjects with diabetes, renal and hepatic disease were excluded. The demographic details of the patients and controls as well as their lipid and lipoprotein concentrations have been reported previously [21].
Methods
Venous blood was collected from all subjects between 9.00 and 11.00 am following fasting from 10.00 pm the previous day. Serum and plasma were isolated by low speed centrifugation. White cells were removed from the buffy coat of the plasma tube. HDL was isolated immediately by precipitating lower density lipoproteins with heparin/Mn2+. Other samples were stored at –20°C for no more than 2 months prior to analysis.
Analysis of PON1 activity
Paraoxonase-1 activity was measured by adding serum to Tris buffer (100 mmol/l, pH 8.0) containing 2 mmol/l CaCl2 and 5.5 mmol/l paraoxon (O,O-die thyl-O-p-nitrophenylphosphate (Sigma Chemical Co.)). The rate of generation of p-nitrophenol was determined at 405 nm, 25°C, with the use of a continuously recording spectrophotometer (Beckman DU-68) as described previously [21].
Determination of PON1 concentration
Paraoxonase-1 concentration was determined using our in-house competitive ELISA employing rabbit anti-human PON1 monospecific antibodies as described previously [21].
Determination of PON1 genotype
DNA was extracted from the white cells and PON1 genotype for the 192 and 55 polymorphisms determined by PCR amplification and restriction enzyme digestion. The Q192R polymorphism was detected by Alw I digestion, and the L55M polymorphism was detected by Nla III digestion, as described [21].
The –108 polymorphism was determined by PCR amplification with primers GACCGCAAGCCACGCC-TTCTGTGCACC and TATATTTAATTGCAGCCGCAG-CCCTGCTGGGGCAGCG-CCGATTGGCCCGCCGC. These primers create a Bst U I site (New England Biolabs) when a C is present at −108. The products were analysed after digestion by a 3% agarose gel (Sigma). Presence of a −108C allele results in a digested band of 52 bp and 67 bp (instead of an undigested band of 119 bp). The −909G/C polymorphism was amplified by PCR with primers AACATGTCACTGTGGCA-TA-TATAATGCTC and TATTATAATATATTATATCATTCACAG-TAACAGCAGACAGCAGAGAAAAGA. The primers remove a second Bsm AI site when a G is present at −909, resulting in digested bands of 50 bp and 206 bp.
Statistical analysis
The Wilcoxon sign rank test was used to test for differences in variables with a non-Gaussian frequency distribution, namely PON1 activity towards paraoxon and PON1 concentration. Probabilities of < 0.05 were considered statistically significant. Differences in gene frequency were sought by the χ2 test. The χ2 test was also used to test whether genotype frequencies deviated from Hardy-Weinberg equilibrium and to test for linkage disequilibrium between pairs of polymorphisms. ANOVA was used to test for differences in parameters between genotypes.
Results
The PON1 parameters of the study populations are given in Table I. The PON1 activity and concentration were significantly lower in the CHD group as we have previously reported [21]. The gene frequencies of the PON1 exonic polymorphisms were as follows: for Q192R, Q = 0.74, R = 0.26 in the control subjects, Q = 0.71, R = 0.29 in the CHD population; and for L55M, L = 0.64, M = 0.36 in controls and L = 0.65, M = 0.35 in those with CHD. There was no statistically significant difference between the frequencies of these polymorphisms in patients and controls.
Table I.
PON1 parameters in the populations studied
| Parameter | Healthy control | CHD |
|---|---|---|
| N | 282 | 417 |
| PON1 activity [nmol/min/ml serum] | 214.6 (26.3-620.8) | 122.8* (3.3-802.8) |
| PON1 concentration [µg/ml] | 89.1 (16.8-527.4) | 71.6* (11.4-489.3) |
Data are median (range). Significantly different from control
p < 0.001
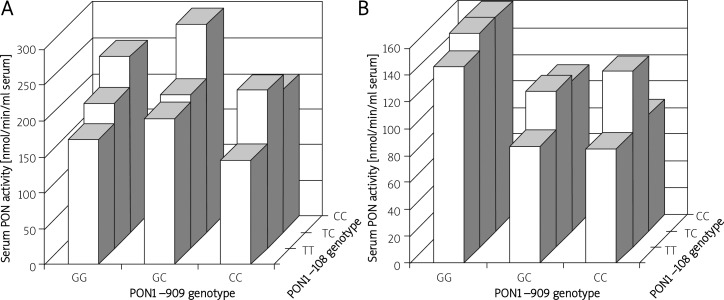
Paraoxonase-1 activity was significantly different in the C-108T genotypes (Table II) in both the control and CHD groups in the order TT < TC < CC. Paraoxonase-1 concentration was not different between genotypes in the control population. However, there were significant differences in the CHD population in the order GG > GC > CC of the −909 genotype (Table II). Both PON1 activity and concentration were significantly lower in the CHD group in all –108 and –909 genotypes. When the populations were divided by –108 and –909 haplotype (Figure 1), there were no consistent patterns of PON1 activity in the CHD population. However, in the controls PON1 activity differed between haplotypes in the order GGCC > GGTC > GGTT (p < 0.05) and GCCC > GCTC > GCTT (p < 0.02). This pattern was not repeated within the –909CC haplotypes (Figure 1). Neither the –108 nor the –909 genotypes influenced serum PON1 concentration.
Table II.
PON1 activity and concentration in the control and CHD populations according to the PON1 promoter polymorphisms
| PON1 polymorphism | PON1 activity [mmol/min/ml] | PON1 concentration [µg/ml] | |||
|---|---|---|---|---|---|
| Controls | CHD | Controls | CHD | ||
| −108 | TT | 159.8 (26.3-518.7) | 85.93 (24.6-494.1) | 86.4 (25.1-527.4) | 71.53 (14.7-267.1) |
| TC | 211.8 51.8-592.5) | 131.92,3 (8.3-802.8) | 91.3 (16.8-509.5) | 73.63 (12.5-433.7) | |
| CC | 245.31 (78.6-620.8) | 145.62,3 (41.7-653.9) | 90.4 (27.3-464.7) | 65.33 (11.4-489.3) | |
| −909 | GG | 211.8 (89.9-574.9) | 156.63 (56.0-663.4) | 86.9 (27.3-464.7) | 68.43 (16.4-342.7) |
| GC | 227.0 (51.8-620.8) | 113.63 (8.3-802.8) | 88.5 (16.8-490.5) | 73.43 (17.7-489.3) | |
| CC | 191.4 (26.3-592.5) | 101.03,4 (30.3-653.9) | 91.4 (25.1-527.4) | 72.83 (11.4-433.7) | |
Figures are median (range).
Figures are median (range). Significantly different from TT p < 0.001
significantly different from TT p < 0.01
significantly different from control p < 0.001
significantly different from GG p < 0.01
Figure 1.
PON1 activity according to the –108, –909 polymorphism haplotypes: A – controls, B – CHD
There were no differences in allele frequencies between the controls and patients with CHD for either the –108 or the –909 polymorphism (Table III). However, all the polymorphisms studied, including the 55 and 192 coding region polymorphisms, were in linkage disequilibrium with each other (a minimum of p < 0.01). In a further analysis of the relationship between the PON1, –108 and –909 promoter polymorphisms and the presence of CHD, we calculated the odds of having CHD according to promoter polymorphism haplotype (Table IV). Again, we could find no significant association between the promoter polymorphisms and the presence of CHD.
Table III.
Allele frequencies for the PON1 promoter polymorphisms in the control and CHD populations
| PON1 polymorphism | Control | CHD | |
|---|---|---|---|
| −108 | T | 0.47 | 0.49 |
| C | 0.53 | 0.51 | |
| −909 | G | 0.44 | 0.45 |
| C | 0.56 | 0.55 |
Table IV.
The odds of having CHD by promoter polymorphism haplotype
| −909 Polymorphism | ||||||
|---|---|---|---|---|---|---|
| GG | GC | CC | ||||
| −108 Polymorphism | CC | 1.29 (0.87-1.42) | 0.42 (0.37-0.45) | 1.01 (0.89-1.11) | ||
| TC | 1.01 (0.83-1.09) | 0.98 (0.81-1.05) | 1.21 (0.92-1.32) | |||
| TT | 2.06 (0.98-2.24) | 1.06 (0.92-1.12) | 0.97 (0.89-1.07) | |||
Figures are odds (95% CI)
Discussion
Previous studies have shown a significant effect of the PON1 −108 and −909 promoter region polymorphisms of both PON1 activity towards phenylacetate and PON1 concentration [17, 18]. Using paraoxon as a substrate for PON1 we have been able to confirm a significant effect of the −108 polymorphism on PON1 activity in both controls and subjects with CHD, but we could only find an effect of the −909 polymorphism on PON1 activity in the group with CHD. The −108 polymorphism lies within a potential transcription factor Sp1 binding site and influences promoter activity resulting in different serum PON1 activities, the −108C polymorphism having greater promoter activity than the −108T polymorphism [22].
Neither the −108 nor the −909 promoter polymorphism was related to the presence of CHD. Results from other laboratories have reported a relationship between the presence of CHD and the PON1 −108 polymorphism [19, 20]. However, some contradictory results have been reported with elevated CHD risk being found in both −108C and −108T carriers in different populations [23, 24]. The reasons for these differences are unclear. However, the gene frequencies reported in this study are consistent with those previously published [17–24]. Differences in the results presented here and those of other researchers could be due to different methodologies employed between studies and the use of different substrates (paraoxon as opposed to phenyl-acetate). Differences in PON1 activity, concentration and genotype distribution have long been known to occur between different populations throughout the world [25]. We and others have shown that subtle but significant differences exist in all PON1 parameters between European populations [26]. Genetic epidemiological studies of the relationship between PON1 coding region polymorphisms and the presence of CHD have been inconsistent. However, a recent meta-analysis showed a small but significantly increased odds ratio for the association of the PON1-192R allele and CHD, a relationship which was confounded by reporting bias [27]. Therefore, differences between populations from differing geographical regions may be the major reason for any difference between this study and those reported earlier. It is interesting to note, in this context, that a recent study of the Han Chinese has indicated that the PON1-55L/M polymorphism does not exist in this population [28], although it is known to exist in other oriental populations [29]. It is also possible that the PON1 promoter polymorphisms only have a small effect on CHD development, which may be overwhelmed by competing influences such as dietary and environmental factors influencing CHD and PON1 activity more in some populations than in others. It is also worth noting that this (and the other studies) were cross sectional in design and as such have limitations such as the difficulty in proving causality, which may also have contributed to differences in results.
Given the putative role of PON1 in protecting against the development of atherosclerosis by hydrolysing lipid peroxides generated during the oxidation of LDL, it is surprising that such large differences in activity between the PON1 −108T/C alleles are not reflected in a genetic association with CHD. We were also unable to show any interactions between the promoter polymorphisms and plasma lipid/lipoprotein concentrations as has been recently suggested [30]. Correcting for any possible influence of the −909 polymorphism in the haplotype analysis failed to strengthen any association. This analysis did, however, show that in people with CHD, the –909 polymorphism negates the influence of the −108 polymorphism on PON1 activity (Figure 1), which may be the reason why there is no relationship between the PON1 promoter polymorphisms studied here and CHD. A similar analysis of the odds of having CHD by −108/55 and −108/192 haplotypes also showed no significant genetic association with CHD (result not shown). Such problems of inconsistency in results will only be clarified when large prospective studies of PON1 promoter polymorphisms and CHD are reported.
In conclusion, low PON1 activity (rather than promoter region polymorphisms) is associated with CHD presence. Developing strategies to raise PON1 activity in people at high risk of developing CHD may be beneficial in preventing/retarding the development of CHD.
Acknowledgments
Bharti Mackness and Wajdi Turkie collected and analysed the data and corrected the manuscript. Mike Mackness analysed data and wrote and corrected the manuscript.
References
- 1.Primo-Parma SL, Sorenson RC, Teiber J, La Du BN. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics. 1996;33:498–509. doi: 10.1006/geno.1996.0225. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Sanabria F, Rull A, Beltran-Debon R, et al. Tissue distribution and expression of paraoxonases and chemokines in the mouse: the ubiquitous and joint localisation suggest a systemic and coordinated role. J Mol Histol. 2010;41:379–86. doi: 10.1007/s10735-010-9299-x. [DOI] [PubMed] [Google Scholar]
- 3.Mackness B, Durrington PN, Mackness MI. The paraoxonase gene family and coronary heart disease. Curr Opinion Lipidol. 2002;13:357–62. doi: 10.1097/00041433-200208000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Reddy ST, Devarajan A, Bourquard N, Shih D, Fogelman AM. Is it just paraoxonase 1 or are other members of the paraoxonase gene family implicated in atherosclerosis? Curr Opin Lipidol. 2008;19:405–8. doi: 10.1097/MOL.0b013e328304b64e. [DOI] [PubMed] [Google Scholar]
- 5.Rozenberg O, Shih DM, Aviram M. Paraoxonase 1 (PON1) attenuates macrophage oxidative status: studies in PON1 transfected cells and in PON1 transgenic mice. Atherosclerosis. 2005;181:9–18. doi: 10.1016/j.atherosclerosis.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 6.Mackness B, Quarck R, Verreth W, Mackness M, Holvoet P. Human paraoxonase-1 overexpression inhibits atherosclerosis in a mouse model of metabolic syndrome. Arterioscler Thromb Vasc Biol. 2006;26:1545–50. doi: 10.1161/01.ATV.0000222924.62641.aa. [DOI] [PubMed] [Google Scholar]
- 7.Guns PJ, Van Assche T, Verreth W, et al. Paraoxonase 1 gene transfer lowers vascular oxidative stress and improves vasomotor function in apolipoprotein E-deficient mice with pre-existing atherosclerosis. Br J Pharmacol. 2008;153:508–16. doi: 10.1038/sj.bjp.0707585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackness B, Durrington P, McElduff P, et al. Low paraoxonase activity predicts coronary events in the Caerphilly Prospective Study. Circulation. 2003;107:2775–9. doi: 10.1161/01.CIR.0000070954.00271.13. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharyya T, Nicholls SJ, Topol EJ, et al. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. 2008;299:1265–76. doi: 10.1001/jama.299.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Himbergen TM, van der Schouw YT, Voorbij HAM, et al. Paraoxonase (PON1) and the risk for coronary heart disease and myocardial infarction in a general population of Dutch women. Atherosclerosis. 2008;198:408–14. doi: 10.1016/j.atherosclerosis.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda Y, Inoue M, Suehiro T, Arii K, Kumon Y, Hashimoto K. Low human paraoxonase predicts cardiovascular events in Japanese patients with type 2 diabetes. Acta Diabetol. 2009;46:239–42. doi: 10.1007/s00592-008-0066-3. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda Y, Suehiro T, Itahara T, et al. Human serum paraoxonase concentration predicts cardiovascular mortality in hemodialysis patients. Clin Nephrol. 2007;67:358–65. doi: 10.5414/cnp67358. [DOI] [PubMed] [Google Scholar]
- 13.Troughton JA, Woodside JV, Yarnell JWG, et al. Paraoxonase activity and coronary heart disease in healthy middle-aged males: the PRIME study. Atherosclerosis. 2008;197:556–63. doi: 10.1016/j.atherosclerosis.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 14.Birjmohun RS, Vergeer M, Stroes ES, et al. Both paraoxonase-1 genotype and activity do not predict the risk of future coronary artery disease; the EPIC- Norfolk prospective population study. PLoS One. 2009;4:e6809. doi: 10.1371/journal.pone.0006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aviram M, Billecke S, Sorenson R, et al. Paraoxonase active site required for protection against LDL oxidation involves its free sulphydryl group and is different from that required for its arylesterase/paraoxonase activities: selective action of human paraoxonase alloenzymes Q and R. Arterioscler Thromb Vasc Biol. 1998;10:1617–24. doi: 10.1161/01.atv.18.10.1617. [DOI] [PubMed] [Google Scholar]
- 16.Mackness B, Mackness MI, Arrol S, Turkie W, Durrington PN. Effect of the human serum paraoxonase 55 and 192 genetic polymorphisms on the protection by high density lipoprotein against low density lipoprotein oxidative modification. FEBS Letts. 1998;423:57–60. doi: 10.1016/s0014-5793(98)00064-7. [DOI] [PubMed] [Google Scholar]
- 17.Brophy VH, Jampsa RL, Clendenning JB, McKinstry LA, Jarvik GP, Furlong CE. Effects of 5’ regulatory region polymorphisms on paraoxonase-gene (PON1) expression. Am J Hum Genet. 2001;68:1428–36. doi: 10.1086/320600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leviev I, James RW. Promoter polymorphisms of human paraoxonase PON1 gene and serum paraoxonase activities and concentrations. Arterioscler Thromb Vasc Biol. 2001;20:516–21. doi: 10.1161/01.atv.20.2.516. [DOI] [PubMed] [Google Scholar]
- 19.James RW, Leviev I, Ruiz J, Passa P, Froguel P, Blatter Garin MC. Promoter polymorphism T(-107)C of the paraoxonase PON1 gene is a risk factor for coronary heart disease in type 2 diabetic patients. Diabetes. 2000;49:1390–3. doi: 10.2337/diabetes.49.8.1390. [DOI] [PubMed] [Google Scholar]
- 20.Leviev I, Righetti A, James AW. Paraoxonase promoter polymorphism (T-107)C and relative paraoxonase deficiency as determinants of risk of coronary artery disease. J Mol Med. 2001;79:457–63. doi: 10.1007/s001090100240. [DOI] [PubMed] [Google Scholar]
- 21.Mackness B, Davies GK, Turkie W, et al. Paraoxonase status in coronary heart disease. Are activity and concentration more important than genotype? Arterioscler Thromb Vasc Biol. 2001;21:1451–7. doi: 10.1161/hq0901.094247. [DOI] [PubMed] [Google Scholar]
- 22.Deakin S, Leviev I, Brulhart-Meynet MC, James RW. Paraoxo nase-1 promoter haplotype sans serum paraoxonase: a predominant role for polymorphic position -107, implicating Sp1 transcription factor. Biochem J. 2003;372:643–9. doi: 10.1042/BJ20021670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campo S, Sardo MA, Trimarchi M, et al. The paraoxonase promoter polymorphism (-107)T > C is not associated with carotid intimal media thickness in Sicilian hypercholesterolemic patients. Clin Biochem. 2004;37:388–94. doi: 10.1016/j.clinbiochem.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Najafi M, Gohari LH, Firoozrai M. Paraoxonase 1 gene promoter polymorphisms are associated with the extent of stenosis in coronary arteries. Thromb Res. 2009;123:503–10. doi: 10.1016/j.thromres.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Furlong CF. Paraoxonase: an historical perspective. In: Mackness B, et al., editors. The paraoxonases: their role in disease development and xenobiotics metabolism. Dordrecht: Springer; 2008. pp. 3–31. [Google Scholar]
- 26.Mackness B, Mackness MI, Durrington PN, et al. Paraoxonase activity in two healthy populations with differing rates of coronary heart disease. Eur J Clin Invest. 2002;30:4–10. doi: 10.1046/j.1365-2362.2000.00580.x. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler JG, Keavney BD, Watkins H, Collins R, Danesh J. Four paraoxonase gene polymorphisms in 11212 cases of coronary heart disease and 12786 controls: meta-analysis of 43 studies. Lancet. 2004;363:689–95. doi: 10.1016/S0140-6736(04)15642-0. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Fan Z, Huang J, et al. Extensive association analysis between polymorphisms of PON gene cluster with coronary heart disease in Chinese Han population. Arterioscler Thromb Vasc Biol. 2003;23:328–34. doi: 10.1161/01.atv.0000051702.38086.c1. [DOI] [PubMed] [Google Scholar]
- 29.Sanghera DK, Saha N, Kamboh MI. The codon 55 polymorphism of the paraoxonase 1 gene is not associated with risk of coronary heart disease in Asian Indians and Chinese. Atherosclerosis. 1998;136:217–23. doi: 10.1016/s0021-9150(97)00206-2. [DOI] [PubMed] [Google Scholar]
- 30.James RW, Kalix B, Biobetto S, Brulhart-Maynet MC. Paraoxo nase-1 promoter polymorphisms C-107T and serum apolipoprotein A1 interact to modulate serum paraoxonase-1 status. Pharmacogenet Genomics. 2005;15:441–6. doi: 10.1097/01213011-200506000-00011. [DOI] [PubMed] [Google Scholar]