Abstract
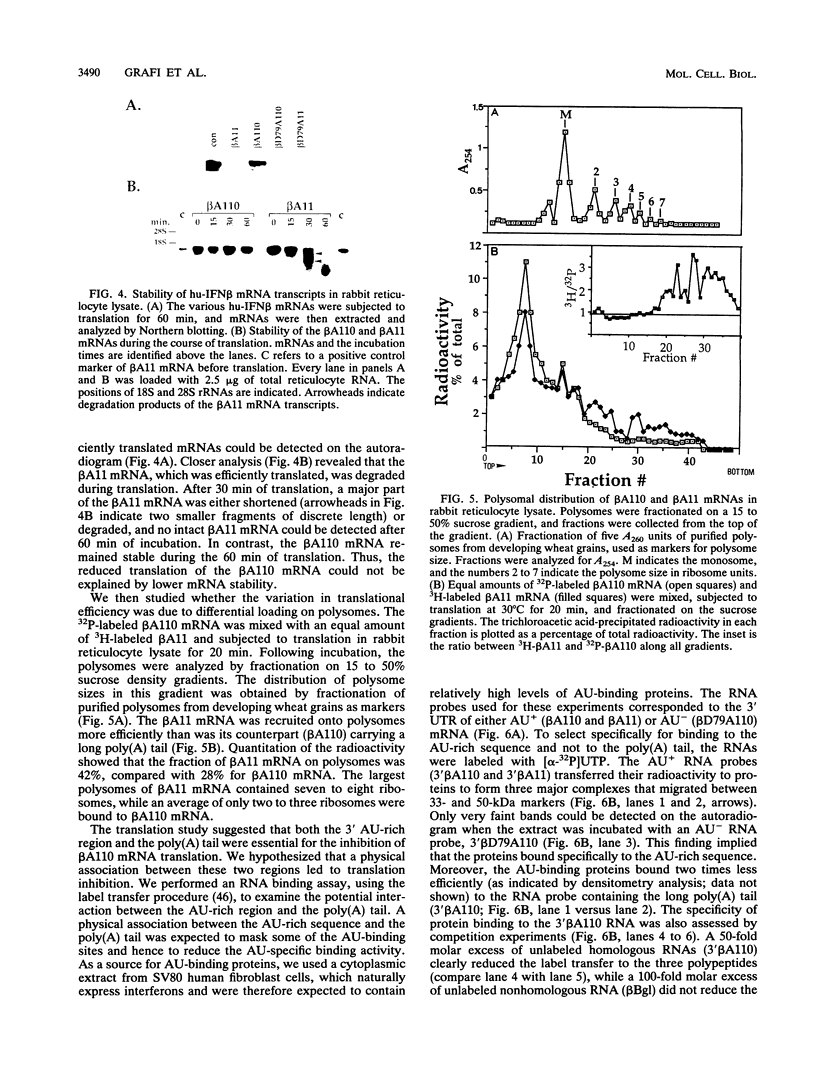
The 3' AU-rich region of human beta-1 interferon (hu-IFN beta) mRNA was found to act as a translational inhibitory element. The translational regulation of this 3' AU-rich sequence and the effect of its association with the poly(A) tail were studied in cell-free rabbit reticulocyte lysate. A poly(A)-rich hu-IFN beta mRNA (110 A residues) served as an inefficient template for protein synthesis. However, translational efficiency was considerably improved when the poly(A) tract was shortened (11 A residues) or when the 3' AU-rich sequence was deleted, indicating that interaction between these two regions was responsible for the reduced translation of the poly(A)-rich hu-IFN beta mRNA. Differences in translational efficiency of the various hu-IFN beta mRNAs correlated well with their polysomal distribution. The poly(A)-rich hu-IFN beta mRNA failed to form large polysomes, while its counterpart bearing a short poly(A) tail was recruited more efficiently into large polysomes. The AU-rich sequence-binding activity was reduced when the RNA probe contained both the 3' AU-rich sequence and long poly(A) tail, supporting a physical association between these two regions. Further evidence for this interaction was achieved by RNase H protection assay. We suggest that the 3' AU-rich sequence may regulate the translation of hu-IFN beta mRNA by interacting with the poly(A) tail.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bandyopadhyay R., Coutts M., Krowczynska A., Brawerman G. Nuclease activity associated with mammalian mRNA in its native state: possible basis for selectivity in mRNA decay. Mol Cell Biol. 1990 May;10(5):2060–2069. doi: 10.1128/mcb.10.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P., Ross J. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem Sci. 1989 Sep;14(9):373–377. doi: 10.1016/0968-0004(89)90011-x. [DOI] [PubMed] [Google Scholar]
- Blobel G. A protein of molecular weight 78,000 bound to the polyadenylate region of eukaryotic messenger RNAs. Proc Natl Acad Sci U S A. 1973 Mar;70(3):924–928. doi: 10.1073/pnas.70.3.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3' untranslated region of lymphokine mRNA. Mol Cell Biol. 1991 Jun;11(6):3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawerman G. mRNA decay: finding the right targets. Cell. 1989 Apr 7;57(1):9–10. doi: 10.1016/0092-8674(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991 May;11(5):2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doel M. T., Carey N. H. The translational capacity of deadenylated ovalbumin messenger RNA. Cell. 1976 May;8(1):51–58. doi: 10.1016/0092-8674(76)90184-7. [DOI] [PubMed] [Google Scholar]
- Farrell P. J., Balkow K., Hunt T., Jackson R. J., Trachsel H. Phosphorylation of initiation factor elF-2 and the control of reticulocyte protein synthesis. Cell. 1977 May;11(1):187–200. doi: 10.1016/0092-8674(77)90330-0. [DOI] [PubMed] [Google Scholar]
- Galili G., Kawata E. E., Cuellar R. E., Smith L. D., Larkins B. A. Synthetic oligonucleotide tails inhibit in vitro and in vivo translation of SP6 transcripts of maize zein cDNA clones. Nucleic Acids Res. 1986 Feb 11;14(3):1511–1524. doi: 10.1093/nar/14.3.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G., Kawata E. E., Smith L. D., Larkins B. A. Role of the 3'-poly(A) sequence in translational regulation of mRNAs in Xenopus laevis oocytes. J Biol Chem. 1988 Apr 25;263(12):5764–5770. [PubMed] [Google Scholar]
- Gillis P., Malter J. S. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine, and oncogene mRNAs. J Biol Chem. 1991 Feb 15;266(5):3172–3177. [PubMed] [Google Scholar]
- Graves R. A., Pandey N. B., Chodchoy N., Marzluff W. F. Translation is required for regulation of histone mRNA degradation. Cell. 1987 Feb 27;48(4):615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Grossi de Sa M. F., Standart N., Martins de Sa C., Akhayat O., Huesca M., Scherrer K. The poly(A)-binding protein facilitates in vitro translation of poly(A)-rich mRNA. Eur J Biochem. 1988 Oct 1;176(3):521–526. doi: 10.1111/j.1432-1033.1988.tb14309.x. [DOI] [PubMed] [Google Scholar]
- Hunter T., Hunt T., Jackson R. J., Robertson H. D. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J Biol Chem. 1975 Jan 25;250(2):409–417. [PubMed] [Google Scholar]
- Jackson R. J., Standart N. Do the poly(A) tail and 3' untranslated region control mRNA translation? Cell. 1990 Jul 13;62(1):15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Kruys V. I., Wathelet M. G., Huez G. A. Identification of a translation inhibitory element (TIE) in the 3' untranslated region of the human interferon-beta mRNA. Gene. 1988 Dec 10;72(1-2):191–200. doi: 10.1016/0378-1119(88)90144-8. [DOI] [PubMed] [Google Scholar]
- Kruys V., Marinx O., Shaw G., Deschamps J., Huez G. Translational blockade imposed by cytokine-derived UA-rich sequences. Science. 1989 Aug 25;245(4920):852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- Kruys V., Wathelet M., Poupart P., Contreras R., Fiers W., Content J., Huez G. The 3' untranslated region of the human interferon-beta mRNA has an inhibitory effect on translation. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6030–6034. doi: 10.1073/pnas.84.17.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laird-Offringa I. A., de Wit C. L., Elfferich P., van der Eb A. J. Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Mol Cell Biol. 1990 Dec;10(12):6132–6140. doi: 10.1128/mcb.10.12.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malter J. S. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989 Nov 3;246(4930):664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- McGrew L. L., Richter J. D. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 1990 Nov;9(11):3743–3751. doi: 10.1002/j.1460-2075.1990.tb07587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe D., Jacobson A. Tales of poly(A): a review. Gene. 1990 Jul 16;91(2):151–158. doi: 10.1016/0378-1119(90)90082-3. [DOI] [PubMed] [Google Scholar]
- Munroe D., Jacobson A. mRNA poly(A) tail, a 3' enhancer of translational initiation. Mol Cell Biol. 1990 Jul;10(7):3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., Taniguchi T. Structure of a chromosomal gene for human interferon beta. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5305–5309. doi: 10.1073/pnas.78.9.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris J., Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early Xenopus development. Dev Biol. 1990 Jul;140(1):221–224. doi: 10.1016/0012-1606(90)90070-y. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Pestka S., Langer J. A., Zoon K. C., Samuel C. E. Interferons and their actions. Annu Rev Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- Raj N. B., Pitha P. M. Analysis of interferon mRNA in human fibroblast cells induced to produce interferon. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7426–7430. doi: 10.1073/pnas.78.12.7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Deardorff J. A. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992 Sep 18;70(6):961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- Sachs A. The role of poly(A) in the translation and stability of mRNA. Curr Opin Cell Biol. 1990 Dec;2(6):1092–1098. doi: 10.1016/0955-0674(90)90161-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenborn E. T., Mierendorf R. C., Jr A novel transcription property of SP6 and T7 RNA polymerases: dependence on template structure. Nucleic Acids Res. 1985 Sep 11;13(17):6223–6236. doi: 10.1093/nar/13.17.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Todaro G. J., Green H., Swift M. R. Susceptibility of human diploid fibroblast strains to transformation by SV40 virus. Science. 1966 Sep 9;153(3741):1252–1254. doi: 10.1126/science.153.3741.1252. [DOI] [PubMed] [Google Scholar]
- Vakalopoulou E., Schaack J., Shenk T. A 32-kilodalton protein binds to AU-rich domains in the 3' untranslated regions of rapidly degraded mRNAs. Mol Cell Biol. 1991 Jun;11(6):3355–3364. doi: 10.1128/mcb.11.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken P., Raué H. A. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3'-terminal part of the coding region. Mol Cell Biol. 1992 Jul;12(7):2986–2996. doi: 10.1128/mcb.12.7.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore L. A., Maniatis T. Postinduction turnoff of beta-interferon gene expression. Mol Cell Biol. 1990 Apr;10(4):1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T., Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988 Nov 24;336(6197):396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- Wilusz J., Shenk T. A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell. 1988 Jan 29;52(2):221–228. doi: 10.1016/0092-8674(88)90510-7. [DOI] [PubMed] [Google Scholar]
- Wisdom R., Lee W. Translation of c-myc mRNA is required for its post-transcriptional regulation during myogenesis. J Biol Chem. 1990 Nov 5;265(31):19015–19021. [PubMed] [Google Scholar]








