Abstract
Background
The most important disease of dairy cattle is mastitis, caused by the infection of the mammary gland by various micro-organisms. Although the transcriptional response of bovine mammary gland cells to in vitro infection has been studied, the interplay and consequences of these responses in the in vivo environment of the mammary gland are less clear. Previously mammary gland quarters were considered to be unaffected by events occurring in neighbouring quarters. More recently infection of individual quarters with mastitis causing pathogens, especially Escherichia coli, has been shown to influence the physiology of neighbouring uninfected quarters. Therefore, the transcriptional responses of uninfected mammary gland quarters adjacent to quarters infected with two major mastitis causing pathogens, E. coli and Staphylococcus aureus, were compared.
Results
The bacteriologically sterile, within-animal control quarters exhibited a transcriptional response to the infection of neighbouring quarters. The greatest response was associated with E. coli infection, while a weaker, yet significant, response occurred during S. aureus infection. The transcriptional responses of these uninfected quarters included the enhanced expression of many genes previously associated with mammary gland infections. Comparison of the transcriptional response of uninfected quarters to S. aureus and E. coli infection identified 187 differentially expressed genes, which were particularly associated with cellular responses, e.g. response to stress. The most affected network identified by Ingenuity Pathway analysis has the immunosuppressor transforming growth factor beta 1 (TGFB1) at its hub and largely consists of genes more highly expressed in control quarters from S. aureus infected cows.
Conclusions
Uninfected mammary gland quarters reacted to the infection of neighbouring quarters and the responses were dependent on pathogen type. Therefore, bovine udder quarters exhibit interdependence and should not be considered as separate functional entities. This suggests that mastitis pathogens not only interact directly with host mammary cells, but also influence discrete sites some distance away, which will affect their response to the subsequent spread of the infection. Understanding the underlying mechanisms may provide further clues for ways to control mammary gland infections. These results also have implications for the design of experimental studies investigating immune regulatory mechanisms in the bovine mammary gland.
Keywords: Bovine, Mastitis, Escherichia coli, Staphylococcus aureus, Microarray
Background
Mastitis, the inflammation of the mammary gland, is the most important disease of dairy cattle, with respect to the frequency of occurrence, animal welfare and economic cost, which is estimated to approach $2 billion annually in the US alone (reviewed by [1]). Mastitis is most frequently caused by infection by a variety of bacteria, of which Staphylococcus aureus and Escherichia coli are amongst the most important gram-positive and gram-negative bacteria respectively (reviewed by [2]). There is considerable variation in the mastitic disease caused by these two pathogens. E. coli intramammary infections often result in acute mastitis with severe clinical manifestations, which usually resolves within a few days. In contrast, symptoms induced by S. aureus infection are usually less severe, even asymptomatic, and the infection can persist for long periods of time.
Current control strategies largely involve hygiene practices on the farm and the direct administration of antibiotics into udder quarters exhibiting clinical signs of infection. However, many infections are asymptomatic and there is a desire to minimize the use of antibiotics. Therefore, there is a need to develop new and improved control strategies. To achieve this goal our understanding of events that occur during infection must be improved. The use of global transcriptomic studies, either involving the use of microarrays or more recently Next Generation Sequencing, is ideally suited to investigate the complex host-pathogen interplay occurring during mastitis, by simultaneously quantifying the expression of thousands of genes. Several studies have used microarrays to investigate the bovine mammary gland response to infection with particular bacteria, including E. coli[3,4], Streptococcus uberis[5,6] and S. aureus[7]. A direct comparison of the induced transcriptional responses identified in these studies is difficult because of differences in the experimental design and the microarray platforms used. However, meta-analyses of such transcriptional studies have recently been reported [8,9]. The meta-analysis reported by Genini et al. [8] identified a common transcriptional response in the mammary glands of several species to different pathogens and identified several new pathways not previously identified in the individual studies. However, a considerable amount of information was excluded from this meta-analysis because of the limited overlap of probes included on the microarrays used in the original studies. Therefore, we designed an infection study to investigate and compare the early transcriptional response of the bovine mammary gland to challenge with S. aureus and E. coli using a single microarray platform.
The experimental model used in this study involved the sequential inoculation of udder quarters with bacteria over the course of the study period, leaving one quarter uninfected as a within animal control [10]. The use of within animal controls is a common and accepted practice because the udder quarters are generally considered to be separate, independent anatomical structures [3-7,11-16] and reduces the between animal variation seen in outbred species such as cattle, as well as addressing the 3Rs, Reduction, Replacement and Refinement, the principles behind the ethical framework for conducting animal experiments. However, analysis of the microarray experiment reported here revealed that E. coli infection of mammary gland quarters can have a profound effect on gene expression in other, uninfected quarters. This conclusion is supported by another transcriptional study comparing uninfected quarters from healthy and E. coli infected cattle [17]. Furthermore, analysis of the microarray data presented here suggests that a similar, but distinct, effect also occurs during S. aureus infection.
Results
Microarray analysis of the transcriptional response of the bovine mammary gland to E. coli and S. aureus infection
A strictly defined model for E. coli- and S. aureus-induced mastitis was developed involving the sequential infection of three udder quarters whilst the fourth quarter acted as a control [10]. RNA samples collected from each quarter of the twelve animals included in this study were interrogated using the ARK-Genomics B. taurus 20 K cDNA microarray. Initially, the intramammary transcriptional response to E. coli and S. aureus infection was investigated by comparing the expression of genes in infected quarters with those expressed in the uninfected, within animal control quarters. Only five clones exhibited differential expression during S. aureus infection (false discovery rate (FDR) ≤ 0.05, fold difference ≥ 2) (Table 1). The clone AJ813772, which is currently unannotated and only matches EST sequences and an intergenic region of bovine chromosome 6, was significantly down-regulated 2.6 fold at 6 hours post infection. Similarly, two of the four clones that exhibit differential expression at 12 hours post infection are currently unannotated. These clones, AJ819694 and BF775518, were up-regulated 2.2 and 3.7 fold respectively. The other clones match lipopolysaccharide binding protein (LBP) and superoxide dismutase 2 (SOD2), which were up-regulated 4.8 and 5.9 fold respectively. No clones were identified as being differentially expressed at 24 or 72 hours post infection with S. aureus.
Table 1.
Number of genes identified as being differentially expressed (FDR ≤ 0.05, fold change ≥ 2) during Escherichia coli and Staphylococcus aureus infection by comparing samples to respective within-animal control samples
| Hours post infection | E. coli | S. aureus |
|---|---|---|
| 6 |
0 |
1 |
| 12 |
6 |
4 |
| 24 |
1048 |
0 |
| 72 | - | 0 |
The E. coli infection induced clinical symptoms in cattle within 12 hours of infection, including; increased somatic cell count (SCC) in milk from infected quarters, decreased milk production, fever and decreased blood leukocyte counts [10]. It was, therefore, surprising that large numbers of differentially expressed genes were only detected in quarters infected for 24 hours (Table 1). No clones exhibited differential expression at 6 hours post infection, whilst only six were differentially expressed at 12 hours post infection (FDR ≤ 0.05, fold difference ≥ 2). The currently unannotated clone C0882029 was down-regulated 2.1 fold. The remaining five clones were up-regulated and represent; chemokine (C-C motif) ligand 2 (3.9 fold), R3H domain containing 1 (3.3 fold), BTG family, member 2 (3.0 fold), Tribbles homolog 1 (2.4 fold) and chromosome 17 open reading frame 42 (2.3 fold). In contrast, 1048 clones were differentially expressed at 24 hours post infection. The two most up-regulated clones represent S100A8 and S100A12, which exhibited 21.9 and 19.8 fold up-regulation respectively. The two most down-regulated clones represent lipin 1 and WD repeat domain, phosphoinositide interacting 1 (WIPI1), which exhibited 5.4 and 5.2 fold down-regulation respectively.
Preliminary comparison of control quarters
The low numbers of differentially expressed genes (DEG) detected in infected udder quarters, especially those infected with E. coli, were unexpected. Recently over 70 genes were found to be differentially expressed in the bovine mammary gland 12 hours after E. coli infection [3]. Furthermore, 183 genes were found to be differentially expressed 16 hours post S. aureus infection of the bovine mammary gland [7] using the smaller bovine innate immune microarray [18]. The discrepancy between these results may relate to the strain of bacterium used or the tissue collection procedure. Another potential explanation is that the low number of DEG may be due to the base-line provided by the control quarters. To investigate this possibility the transcriptomes of the E. coli control quarters (EC24T0) and control quarters from both S. aureus infection experiments (SAT0) were compared. Preliminary analysis using GeneSpring (Agilent) highlighted differences between the transcription profiles. Log intensity plots (sample/reference) of EC24T0 and SAT0 had different shapes (Figure 1). Whilst both had normal type distributions centred around one, EC24T0 had a broader distribution than SAT0. This suggests that the transcription profile of SAT0 is more similar to the reference sample, a mixed pool of all the RNA samples, than EC24T0.
Figure 1.
Differential expression in uninfected udder quarters from animals infected with Escherichia coli and Staphylococcus aureus. Preliminary analysis of the microarray data revealed different patterns of gene expression in control quarters from animals infected with Escherichia coli and Staphylococcus aureus. The log normalized intensity values (sample [Cy5]/reference [Cy3]) were plotted for the combined data from control quarters from animals infected with E. coli (EC24T0) and S. aureus (SAT0) using GeneSpring (Agilent). Both plots have a normal-type distribution around 1. EC24T0 have a broader peak of intensity values suggesting a greater range of expression compared to SAT0.
Genes exhibiting differential expression in control quarters from animals infected with E. coli and S. aureus
Due to the observed difference in the transcription profiles of RNA from the control quarters of E. coli and S. aureus infected animals, the transcriptomes of these samples were compared. Only the control quarters from the four animals infected with S. aureus for up to 24 hours (SA24T0) were included in this comparison. The analysis revealed that 255 clones exhibited significantly different expression levels in the control quarters (FDR ≤ 0.05, fold difference ≥ 2). The 255 clones, of which 212 are currently annotated, represent 187 genes and are listed in Additional file 1. Therefore, the uninfected quarters, which remained bacteriologically negative [10], exhibited significantly different transcriptional profiles, which were dependent on whether the animals had been infected in neighbouring quarters with E. coli or S. aureus. Ninety nine clones, representing 77 genes, were expressed at significantly higher levels in SA24T0 than in EC24T0. The clone AJ814393, which exhibited the highest expression level compared to EC24T0, a 4.6 fold difference, is currently unannotated, matching only EST sequences and an intergenic region of bovine chromosome 14. The remaining 156 clones, representing 110 genes, were expressed at significantly higher levels in EC24T0 than in SA24T0. The clone exhibiting the highest expression level compared to SA24T0, with a 6.6 fold difference, represents FK506 binding protein 5 (FKBP5).
The DEG list was examined using the web-based Database of Annotation, Visualization and Integrated Discovery (DAVID) [19,20]. Human RefSeq accession numbers were used in the analysis due to the limitations in the current bovine annotation. Gene Ontology (GO) Biological Process classifications were assigned to 144 (77%) of the DEG. Gene enrichment analysis was carried out using the default human background and the probability of each functional category was calculated using the modified Fisher Extract test. The two GO terms exhibiting the greatest enrichment were; Cellular Compartment Organization and Biogenesis and Response to Stress (Table 2). Other enriched GO terms included Cell Death and Cell Activation (Table 2). These results indicate that a large cellular response to the infection of neighbouring quarters was occurring in EC24T0 and/or SA24T0.
Table 2.
Major gene ontology biological process classifications represented by genes differentially expressed between control quarters from cows infected with E. coli (EC24T0) and S. aureus (SA24T0)
| Clone accession No. | Gene symbol | Gene | Fold difference |
|---|---|---|---|
|
CELLULAR COMPONENT ORGANIZATION AND BIOGENESIS | |||
|
EC24T0 > SA24T0 |
|
|
|
|
AM017607 |
ARPC5L |
actin related protein 2/3 complex, subunit 5-like |
2.52 |
|
CN821828 |
AGT |
angiotensinogen |
3.73 |
|
AM034974 |
BZW2 |
basic leucine zipper and W2 domains 2 |
2.48 |
|
BG693016 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
2.67 |
|
BF076579 |
BAX |
BCL2-associated X protein |
2.02 |
|
AJ818096 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
2.19 |
|
CO888526 |
EMP1 |
epithelial membrane protein 1 |
2.30 |
|
CN824732 |
EIF1 |
eukaryotic translation initiation factor 1 |
2.08 |
|
CO894798 |
EIF1B |
eukaryotic translation initiation factor 1B |
2.03 |
|
CO888678 |
EIF3D |
eukaryotic translation initiation factor 3, subunit D |
2.03 |
|
AJ691100 |
EIF4A2 |
eukaryotic translation initiation factor 4A, isoform 2 |
2.52 |
|
CN823782 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
3.10 |
|
AJ818246 |
HSPD1 |
heat shock 60 kDa protein 1 |
2.34 |
|
AJ673268 |
HIST1H2AC |
histone cluster 1, H2ac |
3.30 |
|
CO884622 |
MID1IP1 |
MID1 interacting protein 1 |
2.18 |
|
AJ819252 |
MKI67IP |
MKI67 (FHA domain) interacting nucleolar phosphoprotein |
2.29 |
|
AJ673531 |
NDP |
Norrie disease |
3.44 |
|
AM037100 |
NUPR1 |
nuclear protein 1 |
2.27 |
|
CO888501 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
2.45 |
|
AM030492 |
PPP1R9B |
protein phosphatase 1, regulatory (inhibitor) subunit 9B |
2.63 |
|
CO890613 |
RAB43 |
RAB43, member RAS oncogene family |
2.29 |
|
AJ813402 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
2.40 |
|
AJ813146 |
SNRPG |
small nuclear ribonucleoprotein polypeptide G |
2.73 |
|
CO886482 |
SLC3A2 |
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
2.54 |
|
CO892251 |
SLC7A8 |
solute carrier family 7 (cationic amino acid transporter, y + system), member 8 |
4.39 |
|
CO890843 |
STOM |
stomatin |
3.65 |
|
CO880484 |
SNTA1 |
syntrophin, alpha 1 |
4.49 |
|
CO887666 |
UTP14A |
UTP14, U3 small nucleolar ribonucleoprotein, homolog A |
2.26 |
|
SA24T0 > EC24T0 |
|
|
|
|
CO889600 |
AMOT |
angiomotin |
2.12 |
|
CO889378 |
CNN1 |
calponin 1, basic, smooth muscle |
2.49 |
|
CN822971 |
CALR |
calreticulin |
2.36 |
|
BF604066 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
2.43 |
|
CO888048 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
3.23 |
|
CO202183 |
FEZ1 |
fasciculation and elongation protein zeta 1 |
3.13 |
|
CO873026 |
FLNA |
filamin A, alpha |
2.31 |
|
AJ692384 |
GLMN |
glomulin, FKBP associated protein |
2.01 |
|
CN824202 |
GSK3B |
glycogen synthase kinase 3 beta |
2.02 |
|
CO887046 |
LRRC4C |
leucine rich repeat containing 4C |
2.92 |
|
CO872154 |
LASP1 |
LIM and SH3 protein 1 |
2.09 |
|
AM025652 |
MRPL32 |
mitochondrial ribosomal protein L32 |
2.06 |
|
CO893984 |
MYO9B |
myosin IXB |
4.03 |
|
CO889939 |
MYH11 |
myosin, heavy chain 11, smooth muscle |
2.70 |
|
AM018723 |
PIGR |
polymeric immunoglobulin receptor |
2.03 |
|
AJ687977 |
SEC24D |
SEC24 related gene family, member D |
2.02 |
|
CO894656 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
3.32 |
|
CO876411 |
THY1 |
Thy-1 cell surface antigen |
2.10 |
| pTGFB1* |
TGFB1 |
transforming growth factor beta 1 |
2.32 |
|
AJ814326 |
B4GALT1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase polypeptide 1 |
2.45 |
|
AM033343 |
VIM |
vimentin |
2.36 |
|
CO880794 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
2.34 |
|
RESPONSE TO STRESS | |||
|
EC24T0 > SA24T0 |
|
|
|
|
AM006216 |
ATF4 |
activating transcription factor 4 |
2.07 |
|
CN821828 |
AGT |
angiotensinogen |
3.73 |
|
BF076579 |
BAX |
BCL2-associated X protein |
2.02 |
|
AJ818096 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
2.19 |
|
BF606419 |
CXCL2 |
chemokine (C-X-C motif) ligand 2 |
2.03 |
|
CO889145 |
CSDA |
cold shock domain protein A |
3.16 |
|
CN824732 |
EIF1 |
eukaryotic translation initiation factor 1 |
2.08 |
|
CO892245 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
2.91 |
|
AJ818246 |
HSPD1 |
heat shock 60 kDa protein 1 |
2.34 |
|
CN824191 |
HSP90AB1 |
heat shock protein 90 kDa alpha (cytosolic), class B member 1 |
2.25 |
|
AJ818172 |
HIF1A |
hypoxia-inducible factor 1, alpha subunit |
2.41 |
|
AM013900 |
LBP |
lipopolysaccharide binding protein |
2.79 |
|
CO888501 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
2.45 |
|
CN823413 |
SEPP1 |
selenoprotein P, plasma, 1 |
2.12 |
|
CO886480 |
STAT3 |
signal transducer and activator of transcription 3 |
2.25 |
|
AM025615 |
TRIB3 |
tribbles homolog 3 |
3.02 |
|
SA24T0 > EC24T0 |
|
|
|
|
CO888048 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
3.23 |
|
CO883340 |
CFD |
complement factor D |
2.31 |
|
CN824202 |
GSK3B |
glycogen synthase kinase 3 beta |
2.02 |
|
AJ820250 |
IDH1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
3.36 |
|
CO886661 |
RECQL5 |
RecQ protein-like 5 |
2.60 |
|
CO887968 |
TYMS |
thymidylate synthetase |
3.35 |
|
AJ695939 |
TFPI2 |
tissue factor pathway inhibitor 2 |
2.03 |
| pTGFB1* |
TGFB1 |
transforming growth factor beta 1 |
2.32 |
|
AJ814326 |
B4GALT1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase polypeptide 1 |
2.45 |
|
DEATH |
|
|
|
|
EC24T0 > SA24T0 |
|
|
|
|
CN821828 |
AGT |
angiotensinogen |
3.73 |
|
BG693016 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
2.67 |
|
BF076579 |
BAX |
BCL2-associated X protein |
2.02 |
|
AJ818096 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
2.19 |
|
CO888526 |
EMP1 |
epithelial membrane protein 1 |
2.30 |
|
CO892245 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
2.91 |
|
AJ818246 |
HSPD1 |
heat shock 60 kDa protein 1 |
2.34 |
|
AM037100 |
NUPR1 |
nuclear protein 1 |
2.27 |
|
CO883017 |
PHLPP |
PH domain and leucine rich repeat protein phosphatise |
2.26 |
|
CO885391 |
TAX1BP1 |
Tax1 (human T-cell leukemia virus type I) binding protein 1 |
3.05 |
|
AM025615 |
TRIB3 |
tribbles homolog 3 |
3.02 |
|
SA24T0 > EC24T0 |
|
|
|
|
CN822971 |
CALR |
calreticulin |
2.36 |
|
BF604066 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
2.43 |
| pCSF2* |
CSF2 |
colony stimulating factor 2 (granulocyte-macrophage) |
2.57 |
|
CN824202 |
GSK3B |
glycogen synthase kinase 3 beta |
2.02 |
| pTGFB1* |
TGFB1 |
transforming growth factor beta 1 |
2.32 |
|
AJ814326 |
B4GALT1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase polypeptide 1 |
2.45 |
|
CELL ACTIVATION | |||
|
EC24T0 > SA24T0 |
|
|
|
|
CN821828 |
AGT |
angiotensinogen |
3.73 |
|
BF076579 |
BAX |
BCL2-associated X protein |
2.02 |
|
AJ818096 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
2.19 |
|
SA24T0 > EC24T0 |
|
|
|
|
BF604066 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
2.43 |
| pCSF2* |
CSF2 |
colony stimulating factor 2 (granulocyte-macrophage) |
2.57 |
|
AJ692384 |
GLMN |
glomulin, FKBP associated protein |
2.01 |
|
CO876411 |
THY1 |
Thy-1 cell surface antigen |
2.10 |
| pTGFB1* | TGFB1 | transforming growth factor beta 1 | 2.32 |
A single accession number is listed for genes represented by multiple clones. Average fold differences were calculated when multiple clones represented a transcript. * pTGFB1 and pCSF2 are amplicons generated specifically for the Bov20K cDNA microarray.
Closer analysis of the gene list revealed that several genes previously associated with the mammary gland response to bacterial infection are differentially expressed between SA24T0 and EC24T0. These include signal transducer and activator of transcription 3 (STAT3), LBP, CXCL2, BCL2-associated X protein (BAX), B-cell translocation gene 1 (BTG1), metallothioneins and LCN2, which are all expressed at higher levels in EC24T0 samples. This suggests that the EC24T0 samples are responding to infection of neighbouring udder quarters and this accounts for the majority of the transcriptional differences observed between the control quarters.
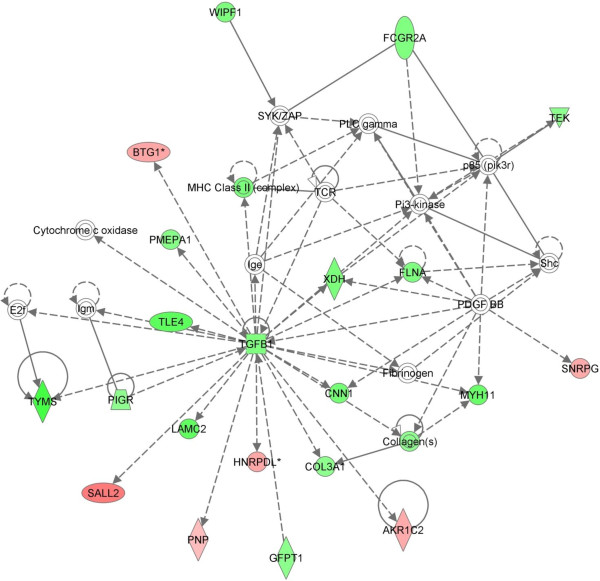
The DEG were further analyzed using the Ingenuity Pathway Analysis (IPA) software (V7.0, Ingenuity Systems) as described previously [21]. The most striking functional network found to be over-represented in the DEG list involved transforming growth factor beta 1 (TGFB1) and associated molecules (Figure 2). Of the 23 genes in this network that were in the DEG list, 17 were expressed at higher levels in SA24T0 samples than EC24T0 samples. It is unclear if this is due to up-regulation of these genes in SA24T0 or down-regulation in EC24T0, but the involvement of TGFB1 in immunosuppression suggests that regulation of these genes may have a profound effect on the ability of the mammary gland quarter to respond to infection.
Figure 2.
TGFB1 Nework. Ingenuity pathway analysis of the EC24T0 v SA24T0 DEG list identified an interaction network involving TGFB1 and associated molecules with a network score of 38. Green denotes molecules that were expressed at higher levels in SA24T0 samples and red denotes those more highly expressed in EC24T0 samples.
Validation of microarray results comparing EC24T0 and SA24T0
Ten genes that exhibited differential expression between EC24T0 and SA24T0 were chosen for validation studies by quantitative RT-PCR (RT-qPCR). Analysis of the microarray results suggested that five of these genes; BTG1, CXCL2, FKBP5, LBP and LCN2, were expressed at higher levels in EC24T0 samples. The RT-qPCR results for all five genes were statistically significant and agreed with the microarray results (Figure 3). The other five genes; ATP-binding cassette, sub-family G (WHITE), member 2 (ABCG2), CXCL12, isocitrate dehydrogenase 1 (NADP+), soluble (IDH1), TGFB1 and xanthine dehydrogenase (XDH), were chosen from those that exhibited greater expression in SA24T0 than EC24T0 samples. The RT-qPCR analysis revealed that all five are expressed at statistically significantly higher levels in SA24T0 than EC24T0 samples (Figure 3), including ABCG2 which was expressed on average 20.9 fold higher levels in SA24T0 than EC24T0 samples.
Figure 3.
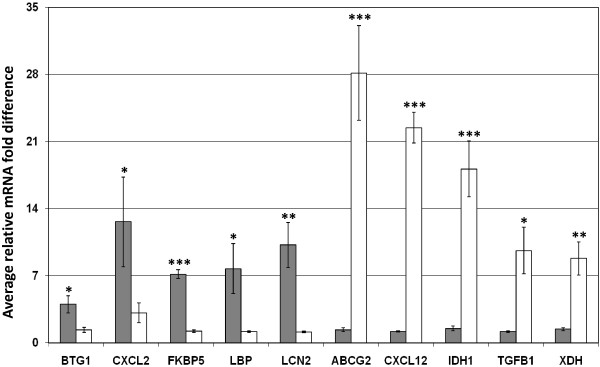
Validation of EC24T0 v SA24T0 differentially expressed genes. Summary of the quantitative RT-PCR (RT-qPCR) results for ten selected genes that were identified from the microarray data analysis as exhibiting differential expression between the control quarters from E. coli (EC24T0) and S. aureus (SA24T0) infected cattle. The results are represented as the average mRNA level detected in EC24T0 (grey bars) and SA24T0 (white bars) relative to the sample with the lowest expression. The bars denote standard error of the mean, *, ** and *** denote that the expression difference is statistically significant with P < 0.05, P < 0.005 and P < 0.0005 respectively.
Genes exhibiting differential expression in control quarters from animals infected with S. aureus for 24 and 72 hours
To confirm that the variation between udder quarters was due only to events occurring in EC24T0, the transcriptome of SA24T0 was compared with that of the control quarters from the 72 hours S. aureus infection (SA72T0). No clones exhibited differential expression using a FDR less than 0.05. However, using less stringent statistical analyses revealed 115 clones that exhibited differential expression (P value ≤ 0.01, fold difference ≥ 2), which are listed in Additional file 2. Eighty one of these clones are annotated, representing 76 genes. Twenty two annotated genes (24 clones) were expressed at significantly higher levels in SA72T0. The remaining 54 annotated genes (57 clones), over two thirds of the differentially expressed genes, were expressed at significantly higher levels in SA24T0. The clone exhibiting the highest expression level compared to SA72T0 represents the cell surface marker CD4, with 4.5 fold higher expression. This suggests that the cellular composition of the control udder quarters differed depending on the time of infection of neighbouring quarters. SOD2, which was identified in the earlier analysis as an up-regulated gene during S. aureus infection, was expressed 2.3 fold higher in SA24T0 than SA72T0. Similarly, other genes associated with the mammary gland response to infection are expressed at higher levels in SA24T0, e.g. S100A8, S100A12, CXCL10, interleukin (IL) 1B and lactotransferrin (LTF).
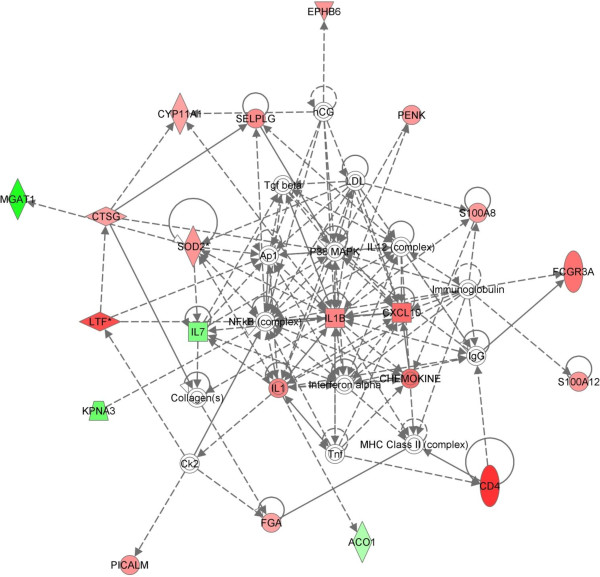
Gene enrichment analysis using DAVID revealed that several GO Biological Process terms are over-represented in the DEG list (Table 3). The GO term exhibiting the greatest enrichment was Response to Stress. Other over-represented GO terms include; Cell Proliferation, Cell Development, Immune Response and Defence Response. The majority of genes associated with these GO terms are expressed at higher levels in SA24T0 samples. Furthermore, subsequent IPA analysis revealed that the DEG list contained 21 genes that are involved in the functional network with the pro-inflammatory cytokine IL1 at its hub and 17 of these were expressed at higher levels in SA24T0 than SA72T0 samples (Figure 4). These results suggest that uninfected quarters respond to the infection of neighbouring quarters with S. aureus for at least 24 hours, but this response had waned by 72 hours post infection. Therefore, of the three sets of control samples generated in this study the transcriptome of SA72T0 samples most closely resembles that of an uninfected quarter. This explains the low number of DEG identified in the initial analysis of the transcriptional response of infected quarters to infection (Table 1). The EC24T0 and SA24T0 samples were used as the baseline to identify DEG, but the response of these quarters to the infection of neighbouring quarters was sufficient to mask the transcriptional response of the infected quarters.
Table 3.
Major gene ontology biological process classifications represented by genes differentially expressed between control quarters from cows infected with S. aureus for 24 hours (SA24T0) and 72 hours (SA72T0)
| Clone accession no. | Gene symbol | gene | Fold Difference |
|---|---|---|---|
|
RESPONSE TO STRESS | |||
|
SA24T0 > SA72T0 |
|
|
|
|
CO891789 |
ASF1A |
ASF1 anti-silencing function 1 homolog A |
2.53 |
| pCXCL10* |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
3.28 |
|
AJ693004 |
CYP11A1 |
cytochrome P450, family 11, subfamily A, polypeptide 1 |
2.03 |
|
AM018876 |
FGA |
fibrinogen alpha chain |
2.03 |
| pIL1B* |
IL1B |
interleukin 1, beta |
2.72 |
|
AM037229 |
PENK |
proenkephalin |
2.23 |
|
BM256666 |
S100A8 |
S100 calcium binding protein A8 (calgranulin A) |
2.19 |
|
BF655078 |
S100A12 |
S100 calcium binding protein A12 (calgranulin C) |
2.29 |
|
AM031467 |
SOD2 |
superoxide dismutase 2, mitochondrial |
2.30 |
|
CO875497 |
TP53BP1 |
tumor protein p53 binding protein 1 |
2.68 |
|
AM022673 |
XRCC2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
3.22 |
|
SA72T0 > SA24T0 |
|
|
|
|
CK847668 |
BRCA2 |
breast cancer 2, early onset |
3.47 |
|
CELL PROLIFERATION |
|
|
|
|
SA24T0 > SA72T0 |
|
|
|
|
AM009066 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
3.27 |
| pCXCL10* |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
3.28 |
|
CO874867 |
CSRP2 |
cysteine and glycine-rich protein 2 |
2.26 |
|
AM018876 |
FGA |
fibrinogen alpha chain |
2.03 |
| pIL1B* |
IL1B |
interleukin 1, beta |
2.72 |
|
AM024609 |
STIL |
SCL/TAL1 interrupting locus |
2.69 |
|
SA72T0 > SA24T0 |
|
|
|
|
CK847668 |
BRCA2 |
breast cancer 2, early onset |
3.47 |
|
BI680983 |
INHA |
inhibin, alpha |
2.63 |
|
AJ815547 |
IL7 |
interleukin 7 |
3.38 |
|
CELL DEVELOPMENT |
|
|
|
|
SA24T0 > SA72T0 |
|
|
|
|
AM009066 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
3.27 |
|
CO874867 |
CSRP2 |
cysteine and glycine-rich protein 2 |
2.26 |
|
BM256725 |
GZMH |
granzyme H |
2.35 |
| pIL1B* |
IL1B |
interleukin 1, beta |
2.72 |
|
CO887046 |
LRRC4C |
leucine rich repeat containing 4C |
2.46 |
|
AM031467 |
SOD2 |
superoxide dismutase 2, mitochondrial |
2.30 |
|
CO892784 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
2.84 |
|
SA72T0 > SA24T0 |
|
|
|
|
AJ817391 |
AFF4 |
AF4/FMR2 family, member 4 |
3.20 |
|
CK847668 |
BRCA2 |
breast cancer 2, early onset |
3.47 |
|
BI680983 |
INHA |
inhibin, alpha |
2.63 |
|
AJ815547 |
IL7 |
interleukin 7 |
3.38 |
|
IMMUNE RESPONSE |
|
|
|
|
SA24T0 > SA72T0 |
|
|
|
|
AM011262 |
CTSG |
cathepsin G |
2.09 |
|
BF706666 |
CD4 |
CD4 molecule |
4.50 |
| pCXCL10* |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
3.28 |
| pFCGR3A* |
FCGR3A |
Fc fragment of IgG, low affinity IIIa, receptor (CD16) |
3.09 |
|
AM023116 |
GBP4 |
guanylate binding protein 4 |
2.16 |
| pIL1B* |
IL1B |
interleukin 1, beta |
2.72 |
|
AM011258 |
LTF |
lactotransferrin |
3.48 |
|
SA72T0 > SA24T0 |
|
|
|
|
AJ815547 |
IL7 |
interleukin 7 |
3.38 |
|
DEFENSE RESPONSE |
|
|
|
|
SA24T0 > SA72T0 |
|
|
|
| pCXCL10* |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
3.28 |
| pIL1B* |
IL1B |
interleukin 1, beta |
2.72 |
|
AM011258 |
LTF |
lactotransferrin |
3.48 |
|
AM037229 |
PENK |
proenkephalin |
2.23 |
|
BM256666 |
S100A8 |
S100 calcium binding protein A8 (calgranulin A) |
2.19 |
| BF655078 | S100A12 | S100 calcium binding protein A12 (calgranulin C) | 2.29 |
A single accession number is listed for genes represented by multiple clones. Average fold differences were calculated for superoxide dismutase 2, mitochondrial and lactotransferrin, which are represented by multiple clones. * pCXCL10, pFCGR3A and pIL1B are amplicons generated specifically for the Bov20K cDNA microarray.
Figure 4.
IL1 Network. Ingenuity pathway analysis of the SA24T0 v SA72T0 DEG list identified an interaction network involving the pro-inflammatory cytokine IL1 with a network score of 42. Red denotes molecules that were expressed at higher levels in SA24T0 samples and green denotes those more highly expressed in SA72T0 samples.
Validation of microarray results comparing SA24T0 and SA72T0
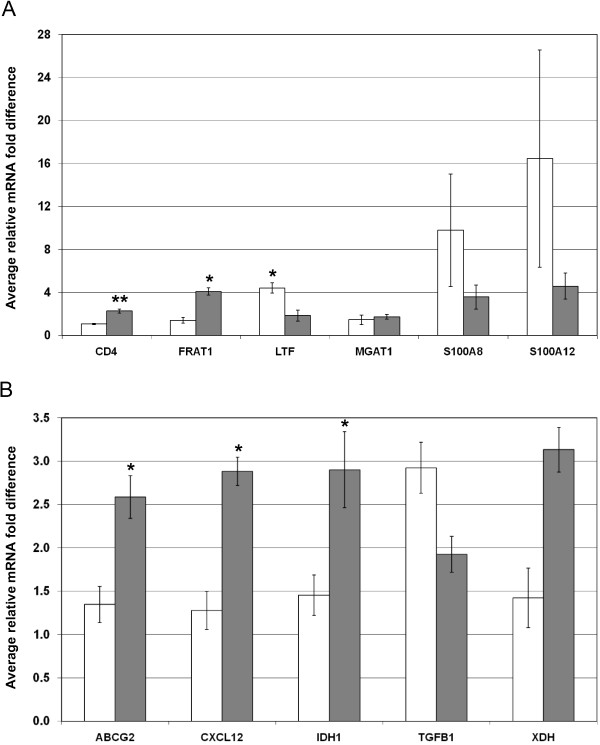
The validity of the DEG identified from the comparison of SA24T0 and SA72T0 using less stringent statistical analysis was investigated by RT-qPCR analysis of the expression levels of six genes in the RNA samples. Analysis of the microarray results suggested that CD4 was the most highly expressed gene in SA24T0 compared to SA72T0. However, the RT-qPCR revealed the opposite pattern of expression, with 2.1 fold higher levels in SA72T0 samples (P < 0.005) (Figure 5A). Furthermore, mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (MGAT1), which according to the microarray analysis was the most highly expressed gene in SA72T0 compared to SA24T0, exhibited no differential expression by RT-qPCR analysis (Figure 5A). However, frequently rearranged in advanced T-cell lymphomas (FRAT1) and LTF exhibited the same pattern of expression by RT-qPCR and microarray analyses. FRAT1 was expressed at on average 2.9 fold higher levels in SA72T0 samples (P < 0.05), whilst LTF was expressed at on average 2.4 fold higher levels in SA24T0 samples (P < 0.05) (Figure 5A). Furthermore, although S100A8 and S100A12 did not exhibit statistically significant differential expression by RT-qPCR analysis, the results followed the same trend as observed with the microarray results, with 2.7 fold and 3.6 fold higher expression in SA24T0 samples respectively (Figure 5A). The disparity between the microarray and RT-qPCR analyses may relate to the use of different RNA samples from the same udder quarters suggesting that there are localized differences in the response to infection of neighbouring quarters. Alternatively, the disparity may be due to the small number of out-bred animals used in this study.
Figure 5.
Validation of SA24T0 v SA72T0 differentially expressed genes. Summary of the quantitative RT-PCR (RT-qPCR) results for selected genes between the control quarters of animals infected with S. aureus for 24 hours (SA24T0) or 72 hours (SA72T0). The results are represented as the average mRNA level detected in SA24T0 (white bars) and SA72T0 (grey bars) relative to the sample with the lowest expression. The bars denote standard error of the mean, * and ** denote that the expression difference is statistically significant with P < 0.05 and P < 0.005 respectively. A. RT-qPCR results for six selected genes that were identified from the microarray data analysis as exhibiting differential expression between SA24T0 and SA72T0. B. RT-qPCR results comparing the differential expression of ABCG2, CXCL12, IDH1, TGFB1 and XDH between SA24T0 and SA72T0 samples.
In addition, the gene expression of the ten genes used to validate the microarray results for the comparison of EC24T0 and SA24T0 were also measured in the SA72T0 samples. No significant difference in expression was observed in the five genes more highly expressed in EC24T0 samples than SA24T0 samples (data not shown). However, four of the five genes more highly expressed in SA24T0 than EC24T0 samples were found to be more highly expressed in SA72T0 than SA24T0 samples and three showed statistical significance (Figure 5B). None of these four genes were identified from the microarray analysis comparison of SA24T0 and SA72T0. Levels of ABCG2, CXCL12 and IDH1 were all statistically significantly higher in SA72T0 than SA24T0 samples, with on average 1.9, 2.3 and 2.0 fold greater expression. Although not statistically significant XDH was on average 2.2 fold higher in SA72T0 compared to SA24T0 samples (P = 0.053). In contrast, TGFB1 levels were higher in SA24T0 samples compared to SA72T0 samples, albeit not to a statistically significant level. Therefore, although not in complete agreement with the microarray data, the RT-qPCR results confirm that the transcriptional profiles of uninfected mammary gland quarters differ during the course of S. aureus infection of neighbouring quarters.
Discussion
Studies involving farm animals require the use of a large number of animals to obtain statistically meaningful data because these out-bred populations exhibit considerable genetic variation. Therefore, it is appealing to use intra-animal controls where possible, since this reduces genetic variation and fewer biological replicates are required. The quarters of the bovine mammary gland have been considered as separate physiological entities and therefore the use of one or more quarters as controls is a generally accepted experimental design [3-7,11-16]. However, the results reported here demonstrate that the mammary gland quarters are interdependent during infection, with a profound change in the transcriptome of one quarter when neighbouring quarters are infected with S. aureus and especially E. coli. The control quarters remained bacteriologically negative throughout the study period [10] and therefore the observed differences are not due to contamination. The transcriptional response of the uninfected quarters was sufficiently high that it resulted in the masking of the response in infected quarters, when they were used as the base-line for DEG analysis (Table 1). In contrast, previous studies investigating the bovine mammary gland response to infection which have used within-animal controls have identified large numbers of differentially expressed genes [4-7]. However, in the earlier studies the animals were only challenged once, whilst in this study the animals were inoculated three times over 24 hours, which may have resulted in a greater signal triggering the response in the uninfected quarters.
Although a direct comparison between uninfected quarters from healthy and uninfected animals was not made in this study, the effect of infection in neighbouring quarters can be inferred from the differences observed between the various control samples. Recently, a microarray experiment comparing the transcriptome of uninfected quarters from healthy and E. coli infected animals was reported [17]. A total of 476 DEG were identified, confirming that E. coli infection alters the transcriptional response of neighbouring mammary gland quarters. Over twenty eight percent (53) of the DEG identified in our comparison of E. coli and S. aureus control quarters were also found in this study, which utilized the bovine Affymetrix platform, including ABCG2 and FKBP5. The direction of the transcriptional change observed in these 53 genes was the same in both studies, i.e. those up-regulated during E. coli infection of neighbouring quarters [17] were found at higher levels in EC24T0 samples than SA24T0 samples, suggesting that the differential expression of at least these 53 genes is associated with E. coli infection and not S. aureus infection. The broader intensity plots of the EC24T0 samples illustrated that there was a greater range of transcriptional change in these samples compared to the reference than observed in the SAT0 samples. This may suggest that the vast majority of the differential expression is due to the E. coli infection of neighbouring quarters. Furthermore, the transcriptome of the uninfected quarters was indistinguishable from that of quarters infected with E. coli for six hours.
However, the design of the experiment reported here does not allow us to conclusively say that the transcriptional differences are all due to changes in expression in the E. coli control quarters. Indeed, the comparison of control quarters from animals infected with S. aureus for 24 and 72 hours has revealed that S. aureus also induces transcriptional changes in neighbouring quarters 24 hours post infection, which may account for some of the differences observed between SA24T0 and EC24T0. Furthermore, XDH, which is involved in the generation of the bactericidal agent peroxynitrate and is believed to play an important role in the mammary gland immune response (reviewed by [22]), is expressed at significantly higher levels in SA24T0 than EC24T0 and therefore may be induced during S. aureus infection of neighbouring quarters. Moreover, additional evidence that S. aureus infection induces a response in neighbouring quarters has recently been reported. The expression of the acute phase proteins serum amyloid A3 (SAA3) and haptoglobin (HP) as well as the β-defensin DEFB5 was elevated in uninfected control quarters from animals infected with a low virulence strain of S. aureus compared to tissue from healthy animals [23].
The initial statistical analysis of the microarray data comparing the S. aureus control quarters from the different time courses failed to detect any significantly differentially expressed genes. Although this could be a genuine result, S. aureus infection generally induces a milder immune response than E. coli, modulating gene expression to a lesser extent at the site of infection, which would impact on the signals being detected by the neighbouring quarters. This variation, compounded with the well-known fact that individual cattle differ greatly in their response to udder infections (reviewed by [24]), would result in considerable variability in the response of neighbouring quarters to S. aureus infection and could account for the lack of statistical significance, which would be further exacerbated by the low number of biological replicates included in this study. Indeed, lowering the stringency of the microarray analysis allowed the identification of over 70 genes, including several genes known to be associated with the mammary gland response to infection, e.g. SOD2, IL1B, LTF, S100A8 and S100A12. Nearly one sixth of the differentially expressed genes are associated with the response of cells to stress. Furthermore, IPA revealed that a network of IL1 and associated molecules were expressed at higher levels in SA24T0 samples than in the SA72T0 samples, which are a well-known trigger for inflammation. Paradoxically, the cattle infected with S. aureus did not show signs of inflammation, even in the infused udder quarters, 24 hours post infection [10], when the elevated levels of IL1 and related molecules were detected in this study. Conversely, at 72 hours, when the IL1 response was lower in uninfected quarters, the infected quarters showed up-regulation of inflammation-associated genes [10]. Therefore, S. aureus infection may initially elicit a slight systemic inflammatory response that induces IL1 expression in uninfected quarters, which disappears during sustained infection. It is unclear if S. aureus actively contributes to the down-regulation of the systemic response, which may contribute to the typically sub-clinical disease pattern observed in S. aureus mastitis.
Some of the transcriptional changes may relate to changes in the cellular composition of the mammary gland. Bacterial infections induce the influx of immune cells, predominantly neutrophils, into the infected mammary gland quarter, which leads to increasing SCC in milk. The SCC measured in the control quarters from animals included in this study increased during the time course of both E. coli and S. aureus infection [10]. This is in agreement with previous studies which found significantly higher SCC in milk from uninfected quarters of naturally infected animals compared to those from healthy animals [25-27]. Moreover, the percentage of neutrophils was significantly higher, whilst the percentage of macrophages was significantly lower, in milk from uninfected quarters from animals with mastitis compared to those from healthy animals [27]. Differential expression of cell surface markers, e.g. CD74, Fc receptors and CD4, are indicative of differences in the cellular composition of the control quarters from E. coli and S. aureus infected animals. Levels of CD4+ T lymphocytes increase in milk during S. aureus infection [26,28] and the results reported here suggest that some cells may originate from the uninfected quarters.
The influx of immune cells may not account for all the transcriptional changes of uninfected quarters. The response of the first quarter to infection with E. coli was significantly different from that of the second infected quarter after the same period of infection. The profound increase in SCC observed in milk from the first quarter 12 hours post infection was not observed in milk samples from the second quarter infected 12 hours later [10]. The earlier infection appears to have primed the neighbouring mammary gland quarters to respond differently to infection. Similar results have been reported recently in an experimental infection study where one animal showed signs of infection in one udder quarter before the experiment started [7]. This animal was still included in the study, using the other, uninfected udder quarters. The proinflammatory cytokine response, i.e. up-regulation of IL1B, IL6, IL8 and tumour necrosis factor (TNF), of this animal was more pronounced than that observed in other animals included in the study [7]. The priming of the mammary gland may persist after infection. Experimental E. coli infection of individual quarters fourteen days after infection of neighbouring quarters resulted in significantly less bacteria being isolated from the milk than observed with the initial infection [29]. Furthermore, it was recently reported that priming udders with a low dose of E. coli lipopolysaccharide (LPS) protects against experimental infection with E. coli pathogens [30]. Moreover, LPS priming induces sustained expression of bactericidal factors (e.g. β-defensins) by bovine mammary epithelial cells (MEC), whilst reducing the over expression of pro-inflammatory cytokines which may be harmful to the animal [31]. However, there is little overlap in DEG from primed MEC and those observed in the uninfected quarters included in this study, suggesting that different mechanisms may be involved in priming neighbouring quarters.
TGFB1 levels were observed to be much lower in the control quarters from E. coli infected animals than those from animals infected with S. aureus. Therefore, it is interesting to hypothesize that the priming of the udder quarters involves the suppression of levels of TGFB1 and associated molecules which was revealed by IPA. Moreover, the same pattern of TGFB3 expression has previously been reported using the same experimental samples [32]. The TGFBs are pleiotropic cytokines which play an important role in multiple biological processes including regulating the adaptive and innate immune responses. TGFB1 inhibits inflammation by, amongst other mechanisms, suppressing the activation of several immune cells, e.g. NK cells and macrophages, and although chemotactic, can inhibit neutrophil transmigration (reviewed by [33]). Further analysis has shown that significantly lower levels of TGFB1 mRNA is present in the uninfected quarters of E. coli infected animals included in this study than in the quarters of healthy animals involved in another study [17] (Additional file 3). It appears that animals down regulate the expression of TGFB1 in the udder when a neighbouring quarter has been challenged with E. coli pathogens. This reduction in the level of the immunosuppressor TGFB1 would lower the threshold for triggering the induction of a variety of immunologically relevant genes, resulting in a faster and more robust response to infection. In contrast, the weaker stimulus elicited by invading S. aureus pathogens maintains, or possibly enhances (Additional file 3), the relatively high levels of TGFB1 present in uninfected quarters, thereby buffering immune stimuli in the healthy, uninfected udder quarter.
Response to Stress was one of the major GO Biological Processes categories identified in both the comparisons of EC24T0 and SA24T0 and that of S. aureus control quarters. Many of the genes within this category have previously been identified as being involved in the immune response to mastitis, including LBP, STAT3, S100A8 and SOD2. However, many of the most differentially regulated genes, e.g. ABCG2 and FKBP5, have not, to our knowledge, previously been associated with mastitis. Both ABCG2 and FKBP5 were also identified as DEG in the recent transcriptional comparison of uninfected quarters from healthy and mastitic animals [17], suggesting that their differential expression was due to the infection of neighbouring quarters with E. coli. Furthermore, the expression of both genes was modulated in E. coli infected mammary gland quarters [17].
ABCG2, also known as breast cancer resistance gene (BCRP), is a xenobiotic transporter that is expressed at high levels in lactating mammary glands of cows, humans and mice [34,35]. ABCG2 plays an important role in the transport of nutrients into milk, especially riboflavin (vitamin B2) [36] and has been identified as the candidate gene underlying a quantitative trait locus on bovine chromosome 6 that affects milk yield and composition [37,38]. Therefore, the down-regulation of ABCG2 in quarters neighbouring E. coli infected quarters may be associated with the decrease in milk yield and casein synthesis, which was observed in these animals [10,39] and may imply a nutrient deficiency in milk from these animals.
The immunophilin FKBP5 has several functions, including mediating the action of the immunosuppressive drug FK506. The FKBP5-FK506 complex binds protein phosphatase 3, catalytic subunit, alpha isozyme (PPP3CA), also known as calcineurin, thus preventing it from phosphorylating and activating NFAT transcription factors, which in turn prevents T cell activation and proliferation [40]. FKBP5 is highly expressed in murine T cells [40] and therefore the up-regulation observed during E. coli infection may be due to lymphocyte migration into mammary gland tissue. In addition, FKBP5 is induced by glucocorticoids and is believed to be involved in the negative feedback loop for glucocorticoid receptor signalling [41]. FKBP5, along with heat shock protein 90 (HSP90), is a component of the glucocortcoid receptor complex. The HSP90 family member HSP90AB1 exhibits a similar expression profile as FKBP5 in both this study and Mitterheumer et al. [17]. Glucocorticoids are able to modulate NF-κB activity by a FKBP5-dependent pathway [42]. FKBP5 interacts with conserved helix-loop-helix ubiquitous kinase (IKKα), which phosphorylates nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (IκBα), the inhibitor of NF-κB, resulting in IκBα proteosome-mediated degradation and the nuclear translocation and activation of NF-κB [43].
The mechanism by which the infection of neighbouring quarters affects the transcriptome of uninfected quarters is unknown. Mastitis induces a systemic effect, inducing transcriptional changes in the liver [44], including the production of acute phase proteins which are released into the blood that circulates around all the udder quarters. This has been postulated to explain the transcriptional profile in healthy quarters of animals infected with E. coli[17]. Furthermore, it has become clear that endogenous damage or danger signals [45] also lead to inflammation [46] and evidence now supports their role in the response to pathogens [47]. Therefore, it is possible that the uninfected udder quarters are responding to such danger or damage signals released into the extracellular environment following infection in the neighbouring quarters. However, analysis of TLR and β-defensin expression in the local and peripheral lymph nodes of animals included in this study revealed that a response was only observed in the lymph nodes draining the quarters infected with E. coli for 12 and 24 hours [10], suggesting that the systemic effect is quite limited. Local cross-talk between the udder quarters must, therefore, be responsible for priming the neighbouring udder quarters. Indeed, lymphocytes have been shown to be capable of migrating between mammary gland quarters [48] and therefore the presence of a more localized interaction between quarters is probable and requires further investigation.
Conclusions
It has previously been shown that during E. coli infection the immune status of neighbouring, sterile quarters is modified [17]. The results presented here provide further evidence of this and also illustrate that a similar phenomenon occurs during S. aureus infection. However, the details of the transcriptional response induced by these pathogens appear to be quite distinct, but are difficult to separate out fully in this study due to the experimental design used. However, the data suggest that the induced transcriptional changes prime the uninfected quarters to prevent or limit the spread of infection. The outcome of mastitis infections is influenced by multiple factors, including the virulence of the pathogen and host genetics. It is clear that the previous exposure of animals to pathogens is also important. Considering the frequency of mastitic infections on dairy farms the pathogen primed state may be the predominant status of uninfected quarters and therefore worthy of further investigation. Furthermore, the results described here illustrate that the four quarters of the bovine mammary gland cannot be considered as separate experimental units during infection studies. This has important ramifications for the design of experimental challenge studies, as the use of inappropriate controls can result in the masking of transcriptional changes during infection. Therefore caution should be taken when using within-animal controls.
Methods
Experimental design of the infection study
The design of the infection study has been described in detail previously in accordance with ARRIVE guidelines [10]. This work was carried out under the approval of the ethics committee of the regional government in Hannover, Germany (No. 509.6-42502-03/678). Briefly, twelve Holstein cows in the middle of their first lactation were challenged over 24 or 72 hours with 500 cfu E. coli strain 1303 or 10,000 cfu S. aureus strain 1027. The animals were randomly divided into three groups. Four animals were inoculated intramammary with E. coli in the front right, hind right and hind left udder quarters at 0 (T24), 12 (T12) and 18 (T6) hours after the trial started respectively. The front left quarter received 2 ml sterile saline at the start of the experiment. The animals were euthanized after 24 hours, with a penetrating captive bolt gun followed by exsanguination, and tissue samples were collected asceptically from each udder quarter. The second group of four animals were inoculated with S. aureus over the same 24 hour period. The third group of four animals were inoculated with S. aureus in both right quarters (T72) and inoculated with 2 ml sterile saline in the front left quarter at 0 hours. The hind left quarter was inoculated at 60 hours (T12) after the start of the experiment. The cows were euthanized after 72 hours as described above and tissue samples were collected from each udder quarter. The control quarters from animals in these three groups are referred to as EC24T0, SA24T0 and SA72T0 respectively in this paper. All animals infected with E. coli developed acute mastitis within 12 hours, although the major clinical symptoms, e.g. udder swelling and elevated SCC, where only observed in the inoculated mammary gland quarters [10]. In contrast, clinical signs of mastitis were only observed in animals infected with S. aureus for 72 hours [10].
RNA samples
A 5 cm3 tissue sample were removed asceptically from the inner area of each udder quarter, 10 cm dorsal of the milk cistern, within 5–10 minutes of culling. A small section of the centre of this sample (1.0 x 0.5 x 0.5 cm) was stored in RNAlater (Ambion) for RNA isolation. Total RNA was extracted as described previously [49] by caesium chloride gradient followed by phenol/chloroform extraction and sodium acetate precipitation. One hundred micrograms of each RNA sample was purified further using RNeasy mini columns (Qiagen) using the manufacturer’s protocols. The quality and quantity of the resulting RNA was determined by gel electrophoresis and Agilent 2100 Bioanalyzer.
Microarray experiment
Experimental design
The RNA samples from each mammary gland quarter were analyzed separately and hybridized in competition with a pooled reference sample, made up of all the RNA samples. A common reference design was used to allow the multifactorial analysis of the complete experiment. The reference sample was labelled with cyanine (Cy) 3 and the treatment sample with Cy5 on each microarray slide. The microarray experiment therefore consisted of a total of 48 microarray slides. The microarray data has been submitted to ArrayExpress and assigned the accession number E-TABM-704.
RNA labelling & hybridizations
Fluorescently labelled cDNA was generated from 20 μg total RNA using the FairPlay II microarray labelling kit (Stratagene), following the manufacturer’s protocol. The labelled cDNA was purified using a DyeEx spin column (Qiagen) and the labelling efficiency was determined by running 0.5 μl of each sample on a 1% agarose gel. Labelled cDNA was hybridized onto the ARK-Genomics Bos taurus 20 K v1.0 microarray (ArrayExpress accession no. A-MEXP-1402). Hybridizations were carried out in a GeneTac automated hybridization station (Genomic Solutions) to control for reactions conditions and washing of the arrays. The Cy3 and Cy5 labelled cDNA were mixed and added to 125 μl hybridization solution (ARK-Genomics protocols: http://www.ark-genomics.org) and hybridized onto the microarray for 12 hours. The microarray slides were then washed in wash buffers of increasing stringency (Genomic Solutions). After removal from the hybridization station the microarray slides were sequentially washed in post-wash buffer and isopropanol for one minute before being dried by centrifugation at 220 g for six minutes. The dried slides were scanned in a Scanarray 5000 XL scanner (GSI Lumonics) at constant laser power of 75% and 73% for Cy3 and Cy5 respectively.
Data analysis
Microarray spot intensity and quality data were extracted from the scanned images using the BlueFuse software version II (BlueGnome). Preliminary analysis was carried out using the GeneSpring software package (Agilent). Further analysis was carried out using modifications of the Limma package of Bioconductor [50], with additional plotting from the Marray package. The microarray data exhibited low signal to noise ratios, which resulted in excessive variability at low intensity values. To reduce this each channel was augmented by 256. The normalization and analysis steps were weighted using the BlueFuse quality measure, confidence values between 0 and 1 [51], and manually excluded spots were given the minimum confidence value. Each slide was normalized separately, using the log2-ratios of treatment to reference intensities for all the non-control spots. The data was normalized using a 2-step process of spatial followed by intensity dependent bias correction for each print-tip, as described previously [52]. The spatial bias was corrected by subtracting corresponding row and column means from each spot [53]. The intensity dependent bias was removed by block-lowess [54].
Genes that were differentially expressed between the infected and control samples were identified by analyzing the means of replicate spots. The Limma eBayes correction [55] was used to shrink the residual variances of each gene towards their median value. Comparisons between the infected and control samples were assessed by 1-way ANOVA followed by t-tests. FDR values for each comparison were calculated using the method of Benjamini and Hochberg [56]. Genes with FDR values less than or equal to 0.05 were considered as significantly different. To refine the list of differentially expressed genes further to those of probable biological relevance, only those genes exhibiting a 2 fold or greater average difference were considered.
Validation of microarray experiments
RT-qPCR analysis was used to validate the microarray results for sixteen differentially expressed genes. Oligonucleotides were designed for each gene using Primer3 [57] and Netprimer (Biosoft International) software (Table 4). Total RNA was isolated from a different sample of mammary gland tissue than that used for the microarray experiment. The tissue was powdered under liquid nitrogen and mixed with the chaotropic RLT lysis buffer (Qiagen). RNA was extracted with Phenol (Ambion) and purified using RNeasy spin columns (Qiagen) including a DNase digestion step (Qiagen) according to the manufacturer’s instructions. The RNA quality was assessed by agarose gel electrophoresis and NanoDrop ND-1000 spectrophotometer (Thermo Scientific).
Table 4.
Details of the oligonucleotides used for the quantitative RT-PCR analysis
| Gene | Gene Symbol | Direction | Sequence (5′-3′) | Product size |
|---|---|---|---|---|
| ATP-binding cassette, sub-family G |
ABCG2 |
F |
CCTTCGGCTTCCAACAACT |
129 |
| (WHITE), member 2 |
|
R |
CCAGACACACCACGGATAAA |
|
| B-cell translocation gene 1, anti- |
BTG1 |
F |
TGAAAGTAGCAAGTGACCAGAA |
192 |
| proliferative |
|
R |
CAAGGAGAGTTACAAACCAGACC |
|
| CD4 molecule |
CD4 |
F |
GGCAGAACCACAGACTCACA |
133 |
| |
|
R |
GACAAACAAGCCCAAAGGAA |
|
| Chemokine (C-X-C motif) ligand 2 |
CXCL2 |
F |
GCCAAACCGAAGTCATAGCC |
213 |
| |
|
R |
TGGAAACCAGCCATTCTCTTC |
|
| Chemokine (C-X-C motif) ligand 12 |
CXCL12 |
F |
TTGAAAGCCTGACCCATAAA |
146 |
| |
|
R |
GACAGTGGCAGCAGAGAAG |
|
| FK506 binding protein 5 |
FKBP5 |
F |
CCACAGCAGCATCACACAC |
189 |
| |
|
R |
GGGAAGGCTAATCCAGAACC |
|
| Frequently rearranged in advanced T- |
FRAT1 |
F |
GCCCAAAGGACAAGGATG |
186 |
| cell lymphomas |
|
R |
CCAAGAACAAGCACCTCAAA |
|
| Isocitrate dehydrogenase 1 (NADP+), |
IDH1 |
F |
CTCTCAAGGGTAAAGGCAAA |
113 |
| soluble |
|
R |
TTCACAAAGGTGGCATAACTG |
|
| Lactotransferrin |
LTF |
F |
CTGTGGCTAAATTCTTCTCTGC |
187 |
| |
|
R |
TTAACAAAAGCCACGTCTCCAG |
|
| Lipocalin 2 |
LCN2 |
F |
CCAACTACGAGCTGAAGGAAGAC |
103 |
| |
|
R |
TGGGAGCTTGGGACAAAAGT |
|
| Lipopolysaccharide binding protein |
LBP |
F |
AGGGCAAGGTGAAAGACAGG |
178 |
| |
|
R |
TGGAGTCAGAGAGGGTGTGG |
|
| Mannosyl (alpha-1,3-)-glycoprotein beta- |
MGAT1 |
F |
TCTCCATCCAGTCCTTTCCA |
191 |
| 1,2-N-acetylglucosaminyltransferase |
|
R |
ACATTGCTCTCCAACCCATC |
|
| S100 calcium binding protein A8 |
S100A8 |
F |
TCTATTTTGGGGAGACCTGGTG |
203 |
| |
|
R |
CCAAGTGTCCGCATCCTTTT |
|
| S100 calcium binding protein A12 |
S100A12 |
F |
AGGGAATCATCAACATCTTCCAC |
172 |
| |
|
R |
TCTTTATCGGCATCCAGGTCTT |
|
| Transforming growth factor, beta 1 |
TGFB1 |
F |
AATGGTGGAATACGGCAACA |
121 |
| |
|
R |
CCGAGAGAGCAACACAGGTTC |
|
| Xanthine dehydrogenase |
XDH |
F |
TCAGGATGATGGTTGGAAGA |
193 |
| |
|
R |
GGGAGTTAGGACATAGCACGA |
|
| Chromosome alignment maintaining |
CHAMP1 |
F |
AGCAGTGACCAAGAGCAGGT |
205 |
| phosphoprotein 1 | R | TCATAGCACGACAGCAACAA |
F and R denote forward and reverse primers respectively.
The RT-qPCRs for twelve genes were carried out as described previously [58]. The relative quantities of mRNA were calculated using the method described by Pfaffl [59]. The RT-qPCR results for chromosome alignment maintaining phosphoprotein 1 (CHAMP1) were used to calculate differences in the template RNA levels and thereby standardize the results for the genes of interest. The RT-qPCR for the remaining four genes; chemokine (C-X-C motif) ligand (CXCL) 2, lipocalin 2 (LCN2), S100 calcium bind proteins (S100) A8 and S100A12, were carried out as described previously [60] and relative copy numbers were calculated. These results were converted to fold differences to standardize the results for all sixteen genes. All samples were measured twice from two independent cDNA preparations. The quality and authenticity of the resulting RT-qPCR products were assessed by agarose gel electrophoresis and sequencing. The relative quantity values were transformed on the log2 scale before statistical analyses to stabilize the variance and make them comparable to the log (intensity ratios) from the microarray analysis. The effect of infection in neighbouring quarters was examined by t-test analysis (Genstat 8.1, Lawes Agricultural Trust, Rothamsted).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KJ designed the microarray experiment, analyzed the differentially expressed genes and drafted the manuscript. RT constructed the microarray and oversaw the microarray experiments. KJ and JG carried out the RT-qPCR analysis. JG analyzed the gene lists using Ingenuity. HMS, WP, HZ and HJS designed and carried out the infection study and collected the samples. HMS conceived the study and coordinated the use of the infection samples in the microarray experiment. EJG was involved in the microarray design and coordination of the study. All authors read and approved the final manuscript.
Supplementary Material
List of clones exhibiting statistically significant differential expression between control quarters from E. coli and S. aureus infected cows. An excel file listing the 255 clones, representing 187 known genes, exhibiting differential expression between control quarters from E. coli and S. aureus infected cows (fold difference ≥ 2, FDR < 0.05). The fold differences between the control quarters from E. coli (EC24T0) and S. aureus (SA24T0) infected cattle are displayed as a positive number when EC24T0>SA24T0 (highlighted in orange) and a negative number when SA24T0>EC24T0 (highlighted in green). Genes shown in bold are represented by more than one clone. The calculated FDR values are included.
List of clones exhibiting statistically significant differential expression between control quarters from cows infected with S. aureus for 24 and 72 hours. An excel file listing the 115 clones, representing 76 known genes, exhibiting differential expression between control quarters from cows infected with S. aureus for 24 (SA24T0) and 72 hours (SA72T0) (fold difference ≥ 2, P < 0.01). The fold differences between the control quarters from animals infected with S. aureus for 24 hours (SA24T0) or 72 hours (SA72T0) are displayed as a positive number when SA24T0>SA72T0 (highlighted in orange) and a negative number when SA72T0>SA24T0 (highlighted in green). Genes shown in bold are represented by more than one clone. The calculated P values are included.
Infection of neighbouring quarters affects the levels of TGFB1 mRNA in uninfected quarters. Comparison of TGFB1 mRNA levels in uninfected quarters of cattle infected for 24 hours with S. aureus (SA24T0), E. coli (EC24T0) or from healthy, uninfected (HEALTHY) cattle. TGFB1 levels are expressed as relative copy numbers and the bars denote standard error of the mean. The statistical significance is indicated as P values and n.s. denotes not significant. The HEALTHY samples were included in a previously published study [17]. Animals in both studies were selected using the same criteria and hormonally synchronized using the same procedure.
Contributor Information
Kirsty Jensen, Email: kirsty.jensen@roslin.ed.ac.uk.
Juliane Günther, Email: j.guenther@fbn-dummerstorf.de.
Richard Talbot, Email: richard.talbot@roslin.ed.ac.uk.
Wolfram Petzl, Email: wpetzl@lmu.de.
Holm Zerbe, Email: h.zerbe@lmu.de.
Hans-Joachim Schuberth, Email: hans-joachim.schuberth@tiho-hannover.de.
Hans-Martin Seyfert, Email: seyfert@fbn-dummerstorf.de.
Elizabeth J Glass, Email: liz.glass@roslin.ed.ac.uk.
Acknowledgements
This work was supported by the European Animal Disease Genomics Network of Excellence for Animal Health and Food Safety (EADGENE) (EU Contract No. FOOD-CT-2004-506416). EJG was also supported by the Biotechnology and Biological Sciences Research Council (Institute Strategic Programme Grant). HMS was also supported by the Deutsche Forschungsgemeinschaft, through the Researcher Group FOR585 (grant Se 326/14-3). We would like to thank Dave Waddington from The Roslin Institute & Royal (Dick) School of Veterinary Studies, University of Edinburgh for the statistical analysis of the microarray data. We would also like to thank Mark Fell from the ARK-Genomics facility for helping with the submission of the experimental data to ArrayExpress.
References
- Viguier C, Arora S, Gilmartin N, Welbeck K, O’Kennedy R. Mastitis detection: current trends and future perspectives. Trends Biotech. 2009;27:486–493. doi: 10.1016/j.tibtech.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Schukken YH, Günther J, Fitzpatrick J, Fontaine MC, Goetze L, Holst O, Leigh J, Petzl W, Schuberth HJ, Sipka A, Smith DGE, Quesnell R, Watts J, Yancey R, Zerbe H, Gurjar A, Zadoks RN, Seyfert HM. Host-response patterns of intramammary infections in dairy cows. Vet Immunol Immunopath. 2011;144:270–289. doi: 10.1016/j.vetimm.2011.08.022. [DOI] [PubMed] [Google Scholar]
- Rinaldi M, Li RW, Bannerman DD, Daniels KM, Evock-Clover C, Silva MVB, Paape MJ, Van Ryssen B, Burvenich C, Capuco AV. A sentinel function for teat tissues in dairy cows: dominant innate immune response elements define early response to E. coli mastitis. Funct Integr Genomics. 2010;10:21–38. doi: 10.1007/s10142-009-0133-z. [DOI] [PubMed] [Google Scholar]
- Buitenhuis B, Røntved CM, Edwards SM, Ingvartsen KL, Sørensen P. In depth analysis of genes and pathways of the mammary gland involved in the pathogenesis of bovine Escherichia coli-mastitis. BMC Genomics. 2011;12:130. doi: 10.1186/1471-2164-12-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes KM, Drackley JK, Morin DE, Bionaz M, Rodriguez-Zas SL, Everts RE, Lewin HA, Loor JJ. Gene network and pathway analysis of bovine mammary tissue challenged with Streptococcus uberis reveals induction of cell proliferation and inhibition of PPARγ signalling as potential mechanism for the negative relationships between immune response and lipid metabolism. BMC Genomics. 2009;10:542. doi: 10.1186/1471-2164-10-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KM, Stelwagen K, Dobson J, Henderson HV, Davis SR, Farr VC, Singh K. Transcriptome profiling of Streptococcus uberis-induced mastitis reveals fundamental differences between immune gene expression in the mammary gland and in a primary cell culture model. J Dairy Sci. 2009;92:117–129. doi: 10.3168/jds.2008-1382. [DOI] [PubMed] [Google Scholar]
- Lutzow YCS, Donaldson L, Gray CP, Vuocolo T, Pearson RD, Reverter A, Byrne KA, Sheehy PA, Windon R, Tellam RL. Identification of immune genes and proteins involved in the response of bovine mammary tissue to Staphylococcus aureus infection. BMC Vet Res. 2008;4:18. doi: 10.1186/1746-6148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genini S, Badaoui B, Sclep G, Bishop SC, Waddington D, van der Pinard LMH, Klopp C, Cabau C, Seyfert HM, Petzl W, Jensen K, Glass EJ, de Greeff A, Smith HE, Smits MA, Olsaker I, Boman GM, Pisoni G, Moroni P, Castiglioni B, Cremonesi P, Del Corvo M, Foulon E, Foucras G, Rupp R, Giuffra E. Strengthening insights into host reponses to mastitis infection in ruminants by combining heterogeneous microarray data sources. BMC Genomics. 2011;12:225. doi: 10.1186/1471-2164-12-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loor JJ, Moyes KM, Bionaz M. Functional adaptations of the transcriptome to mastitis-causing pathogens: the mammary gland and beyond. J Mammary Gland Biol Neoplasia. 2011;16:305–322. doi: 10.1007/s10911-011-9232-2. [DOI] [PubMed] [Google Scholar]
- Petzl W, Zerbe H, Günther J, Yang W, Seyfert H-M, Nürnberg G, Schuberth HJ. Escherichia coli, but not Staphylococcus aureus, triggers an early increased expression of factors contributing to the innate immune defense in the udder of the cow. Vet Res. 2008;39:18. doi: 10.1051/vetres:2007057. [DOI] [PubMed] [Google Scholar]
- Chang CC, Winter AJ, Norcross NL. Immune response in the bovine mammary gland after intestinal, local and systemic immunization. Infect Immun. 1981;31:650–659. doi: 10.1128/iai.31.2.650-659.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrzad J, Dosogne H, Meyer E, Burvenich C. Local and systemic effects of endotoxin mastitis on the chemiluminescence of milk and blood neutrophils in dairy cows. Vet Res. 2001;32:131–144. doi: 10.1051/vetres:2001100. [DOI] [PubMed] [Google Scholar]
- Bannerman DD, Paape MJ, Lee JW, Zhao X, Hope JC, Rainard P. Escherichia coli and Staphylococcus aureus elicit differential innate immune responses following intramammary infection. Clin Diagn Lab Immunol. 2004;11:463–472. doi: 10.1128/CDLI.11.3.463-472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz S, Pfaffl MW, Meyer HHD, Bruckmaier RM. Short-term changes of mRNA expression of various inflammatory factors and milk proteins in mammary tissue during LPS-induced mastitis. Domest Anim Endocrin. 2004;26:111–126. doi: 10.1016/j.domaniend.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Silva LFP, Liesman JS, Etchbarne BE, Nielsen MSW, VandeHaar MJ. Intramammary infusion of IGF-1 increases bromodeoxyuridine labelling in mammary epithelial cells of prepubertal heifers. J Dairy Sci. 2005;88:2771–2773. doi: 10.3168/jds.s0022-0302(05)72956-8. [DOI] [PubMed] [Google Scholar]
- Grönlund U, Johannisson A, Waller KP. Changes in blood and milk lymphocyte sub-populations during acute and chronic phases of Staphylococcus aureus induced bovine mastitis. Res Vet Sci. 2006;80:147–154. doi: 10.1016/j.rvsc.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Mitterhuemer S, Petzl W, Krebs S, Mehne D, Klanner A, Wolf E, Zerbe H, Blum H. Escherichia coli infection induces distinct local and systemic transcriptome responses in the mammary gland. BMC Genomics. 2010;11:138. doi: 10.1186/1471-2164-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson L, Vuocolo T, Gray C, Strandberg Y, Reverter A, McWilliam S, Wang Y, Byrne K, Tellam R. Construction and validation of a bovine innate immune microarray. BMC Genomics. 2005;6:135. doi: 10.1186/1471-2164-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Günther J, Esch K, Poschadel N, Petzl W, Zerbe H, Mitterhuemer S, Blum H, Seyfert HM. Comparative kinetics of Escherichia coli- and Staphylococcus aureus-specific activation of key immune pathways in mammary epithelial cells demonstrates that S. aureus elicits a delayed response dominated by interleukin-6 (IL-6) but not IL-1A or tumor necrosis factor alpha. Infect Immun. 2011;79:695–707. doi: 10.1128/IAI.01071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainard P, Riollet C. Innate immunity of the bovine mammary gland. Vet Res. 2006;37:369–400. doi: 10.1051/vetres:2006007. [DOI] [PubMed] [Google Scholar]
- Whelehan CJ, Meade KG, Eckersall PD, Young FJ, O’Farrelly CO. Experimental Staphylococcus aureus infection of the mammary gland induces region-specific changes in innate immune gene expression. Vet Immunol Immunopathol. 2011;140:181–189. doi: 10.1016/j.vetimm.2010.11.013. [DOI] [PubMed] [Google Scholar]
- Burvenich C, Van Merris V, Mehrzad J, Diez-Fraile A, Duchateau L. Severity of E. coli mastitis is mainly determined by cow factors. Vet Res. 2003;34:521–564. doi: 10.1051/vetres:2003023. [DOI] [PubMed] [Google Scholar]
- Bansal BK, Hamann J, Grabowski NT, Singh KB. Variation in the composition of selected milk fraction samples from healthy and mastitic quarters and its significance for mastitis diagnosis. J Dairy Res. 2005;72:144–152. doi: 10.1017/s0022029905000798. [DOI] [PubMed] [Google Scholar]
- Hamann J, Schröder A, Merle R. In: Mastitis in dairy production: current knowledge and future solutions. Hogeveen H, editor. The Netherlands: Wageningen Academic Publishers; 2005. Differential cell count and interdependence of udder quarters; pp. 190–195. [Google Scholar]
- Merle R, Schröder A, Hamann J. Cell function in the bovine mammary gland: a preliminary study on interdependence of healthy and infected udder quarters. J Dairy Res. 2007;74:174–179. doi: 10.1017/S002202990600238X. [DOI] [PubMed] [Google Scholar]
- Riollet C, Rainard P, Poutrel B. Cell subpopulations and cytokine expression in cow milk in response to chronic Staphylococcus aureus infection. J Dairy Sci. 2001;84:1077–1084. doi: 10.3168/jds.S0022-0302(01)74568-7. [DOI] [PubMed] [Google Scholar]
- Suojala L, Orro T, Järvinen H, Saatsi J, Pyörälä S. Acute phase response in two consecutive experimentally induced E. coli intramammary infections in dairy cows. Acta Vet Scand. 2008;50:18. doi: 10.1186/1751-0147-50-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzl W, Günther J, Pfister T, Sauter-Louis C, Goetze L, von Aulock S, Hafner-Marx A, Schuberth H-J, Seyfert H-M, Zerbe H. Lipopolysaccharide pretreatment of the udder protects against experimental Escherichia coli mastitis. Innate Immun. 2012;18:467–477. doi: 10.1177/1753425911422407. [DOI] [PubMed] [Google Scholar]
- Günther J, Petzl W, Zerbe H, Schuberth H-J, Koczan D, Goetze L, Seyfert H-M. Lipopolysaccharide priming enhances expression of effectors of immune defence while decreasing expression of pro-inflammatory cytokines in mammary epithelia cells from cows. BMC Genomics. 2012;13:17. doi: 10.1186/1471-2164-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zerbe H, Petzl W, Brunner RM, Gunther J, Draing C, von Aulock S, Schuberth HJ, Seyfert HM. Bovine TLR2 and TLR4 properly transduce signals from Staphylococcus aureus and E. coli, but S. aureus fails to both activate NF-κB in mammary epithelial cells and to quickly induce TNFα and interleukin-8 (CXCL8) expression in the udder. Mol Immunol. 2008;45:1385–1397. doi: 10.1016/j.molimm.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Li MO, Wan YY, Sanjabi S, Robertson AKL, Flavell RA. Transforming growth factor-β regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- Jonker JW, Merino G, Musters S, van Herwaarden AE, Bolscher E, Wagenaar E, Mesman E, Dale TC, Schinkel AH. The breast cancer resistance protein BCRP (ABCG2) concentrates drugs and carcinogenic xenotoxins into milk. Nat Med. 2005;11:127–129. doi: 10.1038/nm1186. [DOI] [PubMed] [Google Scholar]
- Mani O, Sorensen MT, Sejrsen K, Bruckmaier RM, Albrecht C. Differential expression and localization of lipid transporters in the bovine mammary gland during the pregnancy-lactation cycle. J Dairy Sci. 2009;92:3744–3756. doi: 10.3168/jds.2009-2063. [DOI] [PubMed] [Google Scholar]
- van Herwaarden AE, Wagenaar E, Merino G, Jonker JW, Rosing H, Beijnen JH, Schinkel AH. Multidrug transporter ABCG2/breast cancer resistance protein secretes riboflavin (vitamin B2) into milk. Mol Cell Biol. 2007;27:1247–1253. doi: 10.1128/MCB.01621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Zinder M, Seroussi E, Larkin DM, Loor JJ, Loor JJ, van der Everts WA, Lee J-H, Drackley JK, Band MR, Hernandez AG, Shani M, Lewin HA, Weller JI, Ron M. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 2005;15:936–944. doi: 10.1101/gr.3806705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikard R, Widmann P, Buitkamp J, Emmerling R, Kuehn C. Revisiting the quantitative trait loci for milk production traits on BTA6. Anim Genet. 2011;43:318–323. doi: 10.1111/j.1365-2052.2011.02258.x. [DOI] [PubMed] [Google Scholar]
- Vanselow J, Yang W, Herrmann J, Zerbe H, Schuberth H-J, Petzl W, Tomek W, Seyfert H-M. DNA-remethylation around a STAT5-binding enhancer in the αS1-casein promoter is associated with abrupt shutdown of αS1-casein synthesis during acute mastitis. J Mol Endocrinol. 2006;37:463–477. doi: 10.1677/jme.1.02131. [DOI] [PubMed] [Google Scholar]
- Baughman G, Wiederrecht GJ, Campbell NF, Martin MM, Bourgeois S. FKBP51, a novel T-cell-specific immunophilin capable of calcineurin inhibition. Mol Cell Biol. 1995;15:4395–4402. doi: 10.1128/mcb.15.8.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG. Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology. 2000;141:4107–4113. doi: 10.1210/endo.141.11.7785. [DOI] [PubMed] [Google Scholar]
- Park J, Kim M, Na G, Jeon I, Kwon Y, Kim J, Youn H, Koo Y. Glucocorticoids modulate NF-κB-dependent gene expression by up-regulating FKBP51 expression in Newcastle disease virus-infected chickens. Mol Cell Endo. 2007;278:7–17. doi: 10.1016/j.mce.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand P-O, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schleg J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Naubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- Jiang L, Sørensen P, Røntved C, Vels L, Ingvartsen KL. Gene expression profiling of liver from dairy cows treated intra-mammary with lipopolysaccharide. BMC Genomics. 2008;9:443. doi: 10.1186/1471-2164-9-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- Manson J, Thiemermann C, Brohi K. Trauma alarmins as activators of damage-induced inflammation. Brit J Surg. 2012;99(Suppl 1):12–20. doi: 10.1002/bjs.7717. [DOI] [PubMed] [Google Scholar]
- Fontana MF, Vance RE. Two signal models in innate immunity. Immunol Rev. 2011;243:26–39. doi: 10.1111/j.1600-065X.2011.01037.x. [DOI] [PubMed] [Google Scholar]
- Kimura K, Harp JA, Goff JP, Olsen SC. Lymphocytes from one side of the bovine mammary gland migrate to the contra lateral gland and lymph node tissue. Vet Immunol Immunopathol. 2005;108:409–415. doi: 10.1016/j.vetimm.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Smyth GK. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editor. New York: Springer; 2005. Bioinformatics and Computational Biology Solutions using R and Bioconductor; pp. 397–420. [Google Scholar]
- Jaffrézic F, de Koning DJ, Boettcher PJ, Bonnet A, Buitenhuis B, Closset R, Déjean A, Delmas C, Detilleux JC, Dovč P, Duval M, Foulley J-L, Hedegaard J, Hornshøj H, Hulsegge I, Janss L, Jensen K, Jiang L, Lavrič M, Lê Cao K-A, Lund MS, Malinverni R, Marot G, Nie H, Petzl W, Pool MH, Robert-Granié C, San Cristobal M, van Schothorst EM, Schuberth H-J. et al. Analysis of the real EADGENE dataset: comparison of methods and guidelines for data normalisation and selection of differentially expressed genes. Genet Sel Evol. 2007;39:633–650. doi: 10.1186/1297-9686-39-6-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K, Talbot R, Paxton E, Waddington D, Glass EJ. Development and validation of a bovine macrophage specific cDNA microarray. BMC Genomics. 2006;7:224. doi: 10.1186/1471-2164-7-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D, Johnstone P, Wilson T. Normalization of microarray data using a spatial mixed model analysis which includes splines. Bioinformatics. 2004;20:3196–3205. doi: 10.1093/bioinformatics/bth384. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed TP. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editor. Totowa: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Jensen K, Paxton E, Waddington D, Talbot R, Darghouth M, Glass EJ. Differences in the transcriptional responses induced by Theileria annulata infection in bovine monocytes derived from resistant and susceptible cattle breeds. Int J Parasitol. 2008;38:313–325. doi: 10.1016/j.ijpara.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldammer T, Zerbe H, Molenaar A, Schuberth HJ, Brunner RM, Kata SR, Seyfert HM. Mastitis increases mammary mRNA abundance of beta-defensin 5, toll-like-receptor 2 (TLR2), and TLR4 but not TLR9 in cattle. Clin Diagn Lab Immunol. 2004;11:174–185. doi: 10.1128/CDLI.11.1.174-185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of clones exhibiting statistically significant differential expression between control quarters from E. coli and S. aureus infected cows. An excel file listing the 255 clones, representing 187 known genes, exhibiting differential expression between control quarters from E. coli and S. aureus infected cows (fold difference ≥ 2, FDR < 0.05). The fold differences between the control quarters from E. coli (EC24T0) and S. aureus (SA24T0) infected cattle are displayed as a positive number when EC24T0>SA24T0 (highlighted in orange) and a negative number when SA24T0>EC24T0 (highlighted in green). Genes shown in bold are represented by more than one clone. The calculated FDR values are included.
List of clones exhibiting statistically significant differential expression between control quarters from cows infected with S. aureus for 24 and 72 hours. An excel file listing the 115 clones, representing 76 known genes, exhibiting differential expression between control quarters from cows infected with S. aureus for 24 (SA24T0) and 72 hours (SA72T0) (fold difference ≥ 2, P < 0.01). The fold differences between the control quarters from animals infected with S. aureus for 24 hours (SA24T0) or 72 hours (SA72T0) are displayed as a positive number when SA24T0>SA72T0 (highlighted in orange) and a negative number when SA72T0>SA24T0 (highlighted in green). Genes shown in bold are represented by more than one clone. The calculated P values are included.
Infection of neighbouring quarters affects the levels of TGFB1 mRNA in uninfected quarters. Comparison of TGFB1 mRNA levels in uninfected quarters of cattle infected for 24 hours with S. aureus (SA24T0), E. coli (EC24T0) or from healthy, uninfected (HEALTHY) cattle. TGFB1 levels are expressed as relative copy numbers and the bars denote standard error of the mean. The statistical significance is indicated as P values and n.s. denotes not significant. The HEALTHY samples were included in a previously published study [17]. Animals in both studies were selected using the same criteria and hormonally synchronized using the same procedure.