Abstract
Some Bantu languages spoken in southwestern Zambia and neighboring regions of Botswana, Namibia, and Angola are characterized by the presence of click consonants, whereas their closest linguistic relatives lack such clicks. As clicks are a typical feature not of the Bantu language family, but of Khoisan languages, it is highly probable that the Bantu languages in question borrowed the clicks from Khoisan languages. In this paper, we combine complete mitochondrial genome sequences from a representative sample of populations from the Western Province of Zambia speaking Bantu languages with and without clicks, with fine-scaled analyses of Y-chromosomal single nucleotide polymorphisms and short tandem repeats to investigate the prehistoric contact that led to this borrowing of click consonants. Our results reveal complex population-specific histories, with female-biased admixture from Khoisan-speaking groups associated with the incorporation of click sounds in one Bantu-speaking population, while concomitant levels of potential Khoisan admixture did not result in sound change in another. Furthermore, the lack of sequence sharing between the Bantu-speaking groups from southwestern Zambia investigated here and extant Khoisan populations provides an indication that there must have been genetic substructure in the Khoisan-speaking indigenous groups of southern Africa that did not survive until the present or has been substantially reduced.
Keywords: Zambia, Bantu, Khoisan, mtDNA, Y chromosome, clicks
Introduction
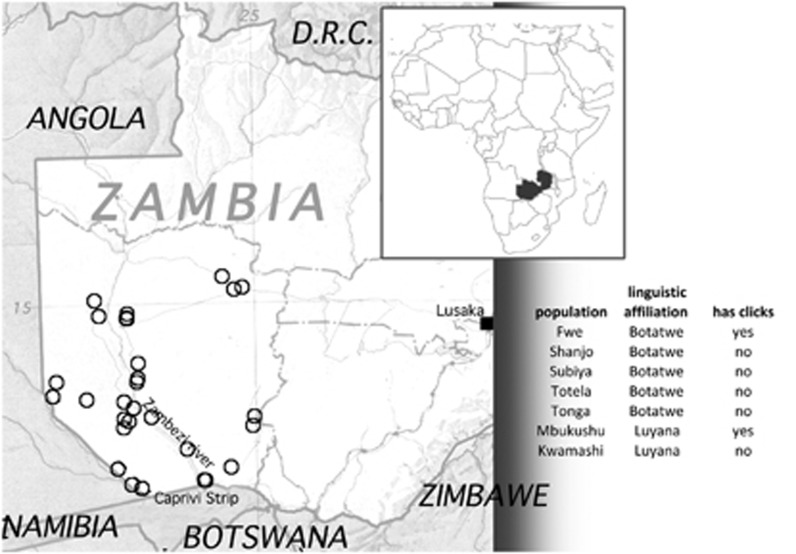
Although clicks are generally considered the hallmark of the so-called ‘Khoisan' languages, they have also been borrowed into some Bantu languages of southern Africa;1 best known among these are the South African Bantu languages Zulu and Xhosa. Less well known is the fact that some Bantu languages further north also have click consonants, though to a far lesser degree.2 These are spoken in a small contiguous area encompassing southeastern Angola, southwestern Zambia, northwestern Botswana, and northeastern Namibia (Figure 1), and belong to different subgroups of the Bantu family.3, 4 In the Botatwe subgroup, clicks are found only in Fwe, being absent from the closely related languages Shanjo, Totela, and Subiya and the more distantly related Tonga; in the Luyana subgroup, clicks are found in Mbukushu, but are absent from its close relative Kwamashi (cf. Figure 1).2
Figure 1.
Map showing the position of Zambia within the African continent and the location of the villages sampled. Thirty-one circles are plotted according to their registered latitude and longitude; populations sampled are listed with information on language affiliation and presence/absence of clicks.
From a genetic perspective, Khoisan-speaking populations are characterized by specific haplogroups both on the Y chromosome and the mtDNA, which are found in considerable frequencies only in these populations or in groups with a known history of contact with such populations.5, 6 Among Bantu-speaking populations of southern Africa, the amount of detectable intermarriage with Khoisan peoples varies between regions and populations and is not always correlated with the presence of click sounds in the languages they speak. For example, so-called ‘southeastern Bantu' populations from South Africa show ∼29% of Khoisan-specific mtDNA haplogroups L0d and L0k7 and ∼5% of Y-chromosomal haplogroup A-M51,8 while only some of their languages have clicks. Bantu-speaking groups from southern Angola also carry varying proportions of characteristic Khoisan haplogroups,9 with the pastoralist Herero-speaking Kuvale showing surprisingly high levels of intermarriage (∼22% of mtDNA haplogroup L0d and ∼12% of Y-chromosomal haplogroup B-M112), but none of them has clicks.
The presence of clicks in certain Bantu languages of southwestern Zambia, and their absence in close relatives, raises the question of the origin of these consonants. Apart from their independent innovation in the Bantu languages, which is highly unlikely, there are three probable pathways by which clicks might have entered the Southwest Bantu languages that have them: (1) through superficial ‘culture contact' in which Bantu speakers borrowed words containing clicks from Khoisan languages without further intimate contact; (2) through language shift, in which entire groups of Khoisan speakers, both men and women, gave up their original language in favor of a Bantu language, transferring some words and sounds to the new language in the process; or (3) through intermarriage between Bantu speakers and Khoisan speakers. If the sociocultural situation in prehistoric times was similar to that of the present,10 this intermarriage is likely to have been sex-biased, with Khoisan-speaking women marrying Bantu-speaking men, but not the opposite.
During the migration of Bantu speakers to southwestern Zambia, there would have been several opportunities for contact with local Khoisan speakers. The oldest Early Iron Age archeological sites in the Upper Zambezi valley, which are generally associated with the settlement of the first Bantu speech communities, date back ∼2200 years.11 These presumably Bantu-speaking communities reached areas already inhabited by hunter-gatherers who probably spoke languages related to modern-day Khoisan languages. As to the Western Bantu-Botatwe peoples (the Fwe, Shanjo, Totela, and Subiya), their ancestors were initially settled further to the east, in the Kafue plains, as indicated by linguistic and archeological data. From there, they spread to the southwest several hundred years ago, with a further migration south to the Zambezi river and beyond during the eighteenth and nineteenth centuries, to escape the pressures of the expanding Luyi/Lozi kingdom.12 Contact between the ancestors of the Fwe with groups speaking click languages could thus have taken place at different points in time: shortly after the arrival of Bantu speakers in southern Africa, after the split-off of the Western Botatwe languages from the ancestral nucleus in the east, or after the southward migration in the eighteenth/nineteenth century.
In this paper, we attempt to solve the puzzle of the origin of clicks in some of the languages of southwestern Zambia with the help of fine-scaled Y-chromosomal analyses and sequences of complete mtDNA genomes from Fwe- and Mbukushu-speakers as well as their closest linguistic relatives, the Shanjo, Totela, Subiya, and Tonga, and the Kwamashi, respectively (cf. Figure 1). We aim at investigating which of the three possible contact scenarios is the most likely, with the following hypotheses: culture contact is expected to take place in the absence of a significant influx of Khoisan lineages, language shift is expected to lead to an influx of both paternal and maternal lineages, while sex-biased intermarriage is expected to lead to an influx of mtDNA lineages, but not Y-chromosomal ones.
Materials and methods
Materials and DNA analysis
Saliva samples from various populations settled over the entire Western Province of Zambia were collected in August–September 2007.13 As reported in de Filippo et al,13 after DNA extraction the Y chromosomes were analyzed for 31 single nucleotide polymorphisms, plus 12 short tandem repeat (STR) loci by means of the Promega Y-Powerplex kit (http://www.promega.com). From the total West Zambian data set, only those 132 individuals whose father's father was affiliated with one of the seven populations included in this study were chosen: Fwe, Shanjo, Subiya, Totela, Tonga, Mbukushu, and Kwamashi (see Figure 1 for the approximate location of collection sites for these samples and Supplementary Table 1 for details). The subset of the data analyzed for this study is given in Supplementary Table 2.
mtDNA full genome sequences were generated for 169 individuals whose mother's mother was affiliated with one of the seven populations listed above. Genomic libraries were hybridized with the protocol described in Maricic et al,14 with in-solution capture on target mtDNA. Sequencing was performed on an Illumina GAIIx (Solexa) sequencer. Coverage ranged from an average minimum of 19 × to an average maximum of 438 × . The number of bases with missing data (gaps, sites with coverage <2 × or where the major base was not present at >70%) is <1%. The two poly-C regions (np 303–315, 16183–16194), which are prone to sequencing errors, were not considered in any of the analyses. All sequences were submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank) and given accession numbers JX303745 and JX303913.
Data analysis
Analysis of Molecular Variance (AMOVA) and standard diversity indices for the Y-chromosome haplogroups and Y-STR haplotypes, plus Φst and RST matrices of distances for the complete mtDNA sequences and the Y-STR haplotypes, respectively, were computed in Arlequin ver. 3.11.15 For the STR analyses in Arlequin and Network one Tonga sample was not considered because of non-integer numbers of repeats at two loci. Nucleotide diversity and variance for the mtDNA sequence data in single populations was calculated in R with the function ‘nuc.div' of the Pegas package.16 Y-chromosomal haplotype and mtDNA sequence sharing were estimated and plotted with in-house scripts for R. The patterns of mtDNA sequence variation and STR haplotype variation were further investigated with the help of Median Joining networks17 constructed with Network 4.11 (www.fluxus-engineering.com). For the STR networks, weights were assigned to each individual STR locus as inversely proportional to the variance observed in our data set.18
Multi-dimensional scaling analyses of matrices of genetic distances based on Y-chromosomal haplogroup frequencies and complete mtDNA sequences were plotted in Statistica ver. 10.19
Simulations were performed in R to assess the levels of migration rates compatible (at P=0.05) with the observed proportion of Khoisan-specific haplogroups in extant Bantu populations. Two possible Khoisan source populations were considered: the Ju as the prototypical ‘San' group (as an average of haplogroup frequencies of the !Kung and !Xun from Soodyall et al20 and the Tsumkwe San and Sekele/!Kung from Wood et al21), resulting in 90 and 75% Khoisan-specific mtDNA and Y-chromosome haplogroups, respectively; and the Khwe, with 60% and 16% Khoisan-specific mtDNA and Y-chromosome haplogroups, respectively (Soodyall et al 2008). Two of the Zambian populations were considered as recipient populations: the Fwe with 24 and 0%, and the Tonga with 0 and 3% Khoisan-specific maternal and paternal lineages, respectively. Contact was assumed to have taken place from 800 years ago (or 29 generations with a generation time of 28 years) until present, with constant effective population size of 10 000 for both source and recipient populations and a constant migration rate. The probability of seeing a proportion of Khoisan haplogroups within the 95% confidence intervals of the observed values (adjusted for the sample size of the recipient population) was calculated over 10 000 iterations and repeated for a range of migration rates, with the significant thresholds of migration taken from the final distribution of probabilities for each of the eight scenarios.
Results
Y chromosome
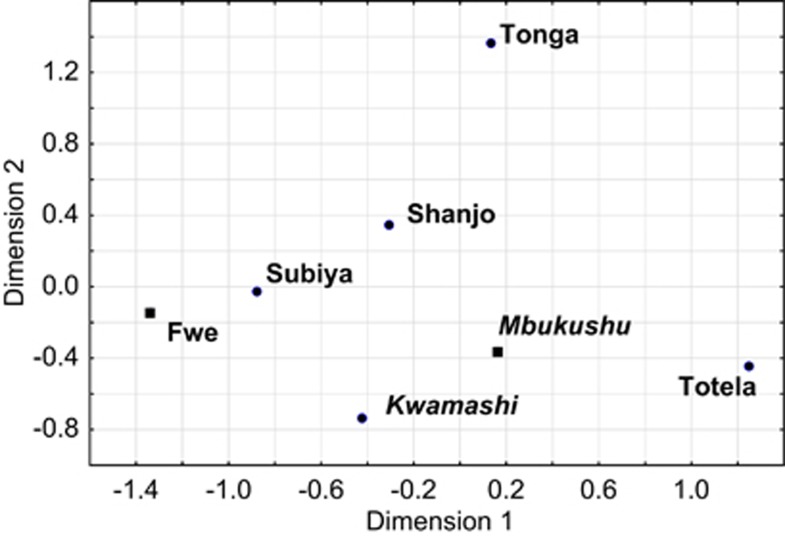
The seven populations included here show a fairly homogenous Y-chromosomal haplogroup composition that is very similar to surrounding groups from east Zambia, Angola, DRC, and Gabon13 (Table 1). Y-chromosomal haplogroups characteristic of Khoisan-speaking populations are found in only very low frequency and not at all in the Fwe and the Mbukushu, the two groups with clicks in their language: haplogroup A is entirely absent from the data set, and haplogroup B-M112 is found in only one individual each of the Subiya, Tonga, and Totela. The homogeneity of all the groups included in the analysis is apparent in the multi-dimensional scaling analysis (Figure 2), where no clear clusters emerge; only the Tonga, who are geographically the most distant population in the data set, are separated slightly from the other groups.
Table 1. Y-chromosomal diversity and haplogroup composition.
| STR Data | Haplogroup data | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Used for STR | n Htypes | Var | G Div | SD | HG Div | SD | B-M152 | B-M112 | E-M2 | E-U174 | E-U175 | E-M75 | R | |
| Fwe | 26 | 26 | 21 | 0.40 | 0.98 | 0.02 | 0.44 | 0.10 | 0.04 | 0.19 | 0.73 | 0.04 | |||
| Shanjo | 13 | 13 | 13 | 0.50 | 1.00 | 0.03 | 0.56 | 0.11 | 0.08 | 0.31 | 0.62 | ||||
| Subiya | 11 | 11 | 11 | 0.44 | 1.00 | 0.04 | 0.47 | 0.16 | 0.09 | 0.00 | 0.18 | 0.73 | |||
| Totela | 13 | 13 | 11 | 1.37 | 0.97 | 0.04 | 0.76 | 0.10 | 0.15 | 0.08 | 0.23 | 0.08 | 0.46 | ||
| Tonga | 32 | 31 | 28 | 0.94 | 0.99 | 0.01 | 0.65 | 0.06 | 0.03 | 0.03 | 0.06 | 0.38 | 0.47 | 0.03 | |
| Mbukushu | 11 | 11 | 9 | 0.59 | 0.96 | 0.05 | 0.69 | 0.12 | 0.09 | 0.18 | 0.55 | 0.18 | |||
| Kwamashi | 26 | 26 | 21 | 0.60 | 0.98 | 0.02 | 0.55 | 0.10 | 0.08 | 0.15 | 0.65 | 0.12 | |||
Abbreviations: G Div, gene diversity; Htypes, haplotypes; HG Div, haplogroup diversity; n, number of individuals; SD, standard deviation; Var, variance.
Figure 2.
Multi-dimensional scaling based on FST distances calculated from haplogroup frequencies. Stress value: 0.011. Bantu Luyana populations are indicated in italics, Bantu Botatwe in regular bold. Populations that speak languages with clicks are indicated with a square, the remaining populations with a dot.
This homogeneity is further confirmed by the Y-STR analyses, which demonstrate extensive haplotype sharing among the populations (Supplementary Figures 1 and 2), and by the non-significant pairwise RST values between the populations (Supplementary Table 3). Furthermore, an AMOVA analysis (Table 2a) shows that the seven populations cannot be differentiated at all on the basis of Y-chromosomal haplogroup frequencies: the variance among populations (1%) is not statistically significant. Although there is significant differentiation between the groups at the STR level, this can be shown to be due entirely to the distinctiveness of the Totela, as evidenced by the complete lack of differentiation between groups when this population is removed from the analysis (Table 2a). Grouping the populations by presence vs absence of clicks or by linguistic subgroup (Botatwe vs Luyana) does not lead to any significant proportion of the variation being apportioned to the between-group component (Table 2a).
Table 2. AMOVA analysis.
| (a) On Y-chromosome data | Percentage of variance | |||
|---|---|---|---|---|
| Criteria 1 group | Data set | Between populations | Within populations | |
| All seven populations | Y-haplogroup | 1.03 | 98.97 | |
| All seven populations | Y-STR | 6.65** | 93.35 | |
| Six populations (excluding Totela) | Y-STR | −1.47 | 101.47 | |
|
Criteria 2 groups |
Data set |
Between groups |
Within groups |
Within populations |
| Presence clicks vs absence clicks | Y-STR | −2.5 | 7.95** | 94.55** |
| Presence clicks vs absence clicks | Y-haplogroup | −0.75 | 1.4 | 99.35 |
| Botatwe vs Luyana | Y-STR | −2.42 | 7.91** | 99.59** |
| Botatwe vs Luyana | Y-haplogroup | 0.01 | 1.03 | 98.96 |
| (b) On mtDNA sequence data | ||||
|
Criteria 1 group |
|
Between populations |
Within populations |
|
| All seven populations | 1.91* | 98.09 | ||
|
Criteria 2 groups |
|
Between groups |
Within groups |
Within populations |
| Presence clicks vs absence clicks | 1.68 | 1.12 | 97.2* | |
| Botatwe vs Luyana | −0.22 | 2.01* | 98.21* | |
*Denotes values of significance <0.05. **Denotes values of significance <0.01.
Y-chromosomal haplogroup diversity (Table 1) is fairly low overall (0.44–0.76), and especially in the Fwe and Subiya, consistent with the restricted complement of haplogroups present. All populations have relatively high Y-STR gene diversity values; in contrast, the Fwe and the Subiya show reduced Y-STR variance, with the Fwe having the lowest value.
mtDNA
With regard to their mtDNA haplogroup composition, the groups included here are characterized by relatively high frequencies of haplogroups that are widespread in sub-Saharan Africa: L0a, L1b, L1c, L2a, and L3e (Table 3). L1c is typically associated with pygmy populations from Central Africa, but the sublineages to which the southwestern Zambian sequences belong (L1c2a and L1c2b) are characteristic of Bantu speakers rather than pygmies.5, 22 In contrast to the near absence of characteristic Khoisan Y-chromosomal lineages in the southwestern Bantu groups, mtDNA haplogroups L0d and L0k are found in several populations. The Fwe stand out with a very high frequency (24.3%) of these Khoisan haplogroups, in particular a high frequency of L0k (18.2%); in the linguistically closely related Shanjo L0d and L0k reach 16.7%. In the other populations, the Khoisan haplogroups are present in at most low frequency. Although the sequence diversity values are in general fairly high (Table 3), the Fwe stand out as having the lowest value (0.93±0.03) but relatively high mean pairwise differences and nucleotide diversity, demonstrating that whereas several sequences are shared between individuals, these are quite diverse.
Table 3. mtDNA diversity and haplogroup composition.
| Sequence data | Haplogroup data | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | n Htypes | MPD | SD | π | Var | HG Div | SD | L0a | L0d | L0k | L1b | L1c | L2 | L2a | L2b | L2c | L2d | L3b | L3d | L3e | L3f | L5a | |
| Fwe | 33 | 20 | 65.8 | 29.0 | 0.0039 | 0.0009 | 0.93 | 0.03 | 0.12 | 0.06 | 0.18 | 0.27 | 0.12 | 0.09 | 0.03 | 0.12 | |||||||
| Shanjo | 24 | 17 | 65.6 | 29.0 | 0.0039 | 0.0009 | 0.97 | 0.02 | 0.04 | 0.08 | 0.08 | 0.25 | 0.17 | 0.04 | 0.21 | 0.13 | |||||||
| Subiya | 17 | 15 | 62.7 | 29.0 | 0.0037 | 0.0009 | 0.99 | 0.03 | 0.06 | 0.06 | 0.12 | 0.29 | 0.24 | 0.24 | |||||||||
| Totela | 29 | 27 | 62.7 | 28.0 | 0.0037 | 0.0009 | 0.99 | 0.01 | 0.14 | 0.07 | 0.17 | 0.17 | 0.07 | 0.03 | 0.03 | 0.24 | 0.07 | ||||||
| Tonga | 22 | 22 | 66.5 | 30.0 | 0.0040 | 0.0010 | 1.00 | 0.01 | 0.14 | 0.09 | 0.23 | 0.27 | 0.09 | 0.14 | 0.05 | ||||||||
| Mbukushu | 12 | 11 | 64.3 | 28.0 | 0.0038 | 0.0010 | 0.98 | 0.04 | 0.08 | 0.08 | 0.25 | 0.33 | 0.17 | 0.08 | |||||||||
| Kwamashi | 32 | 26 | 64.4 | 29.0 | 0.0038 | 0.0009 | 0.98 | 0.02 | 0.16 | 0.03 | 0.09 | 0.16 | 0.03 | 0.06 | 0.06 | 0.19 | 0.19 | 0.03 | |||||
Abbreviations: Htypes, haplotypes; HG Div, haplogroup diversity; MPD, mean pairwise differences; n, number of individuals; π, nucleotide diversity; SD, standard deviation; Var, variance.
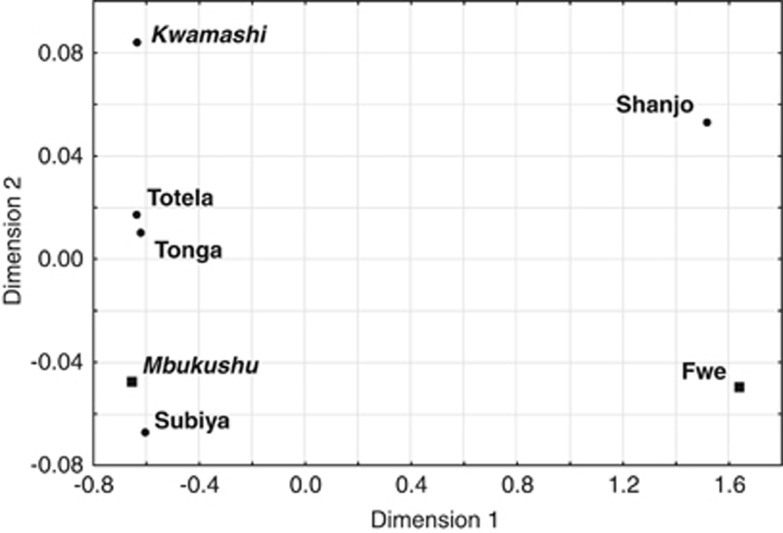
An AMOVA performed on the mtDNA sequence data (Table 2b) shows only a low, though statistically significant, differentiation. This is probably due to the distinctiveness of the Fwe, who are significantly different from all populations except for the Shanjo and Mbukushu (though not after Bonferroni correction), while none of the other populations differ significantly from each other, as demonstrated by pairwise ΦST values (Supplementary Table 4). Grouping the populations by linguistic subgroup or according to the presence vs absence of clicks again has no significant effect on the apportionment of variation (Table 2b). The distinctiveness of the Fwe and the Shanjo, the two groups with the highest amount of Khoisan lineages, becomes apparent in the multi-dimensional scaling plot based on ΦST distances (Figure 3), where the first dimension separates these two populations from all the others.
Figure 3.
Multi-dimensional scaling based on ΦST distances between mtDNA sequences. Stress value: 0.007. Bantu Luyana populations are indicated in italics, Bantu Botatwe in regular bold. Populations that speak languages with clicks are indicated with a square, the remaining populations with a dot.
Haplotype sharing patterns among the populations show an overall fairly high level of sharing (Supplementary Figure 3), and a more fine-scaled analysis of the shared sequences in a network (Supplementary Figure 4) highlights some interesting points. The Fwe and the Shanjo, who are both separated from the other populations in the multi-dimensional scaling plot and who are united in their high frequencies of Khoisan lineages, share only two haplotypes, both on the background of the Khoisan-specific haplogroups: one belonging to L0d and the other belonging to L0k, with another Shanjo L0k sequence only one mutational step away from a Fwe sequence. The results of a resampling test computed in R, where we drew two subsets of 20 and 24 individuals, respectively, from the total number of non-Khoisan sequences in the data set demonstrate that this complete lack of sharing of sequences belonging to non-Khoisan haplogroups is significant (P=0.04, calculated over 10 000 repeats).
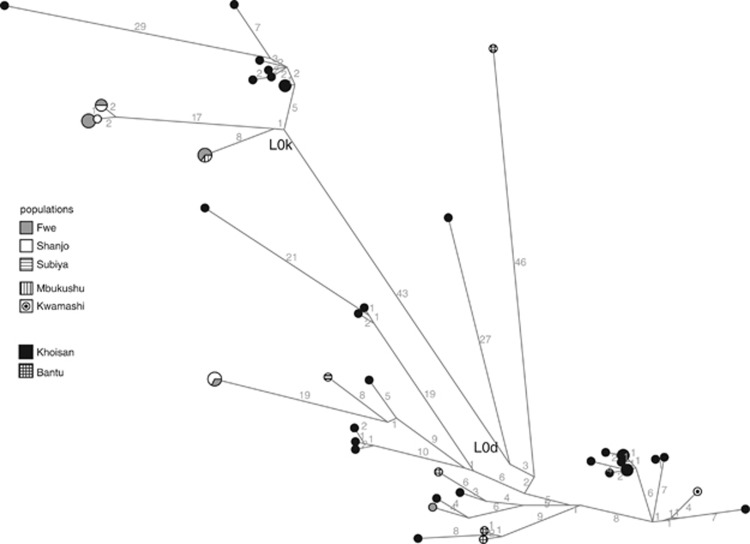
A comparison of the southwestern Zambian individuals belonging to haplogroups L0d and L0k with published sequences from southern Africa belonging to these haplogroups5, 23, 24, 25 shows a surprising lack of sequence sharing between the Zambians and others, be they Khoisan- or Bantu-speaking (Figure 4). Even though this is certainly due at least in part to a lack of available comparative data, it is still noticeable that the Zambian sequences are not even located close to any published Khoisan sequences, but at the end of very long branches. While any two non-Zambian sequences belonging to haplogroup L0d or L0k are on average separated by 10 mutations, the Zambian L0k sequences are separated from the closest non-Zambian sequences by 27, 26, and 15 mutations; the distance between the Zambian L0d sequences and the closest non-Zambian L0d sequences is 25, 14, 12, and 8 mutations.
Figure 4.
Median joining network of mtDNA sequences belonging to haplogroup L0k and L0d, including individuals from Zambia and sequences retrieved from the literature; the latter are here lumped as ‘Khoisan' and ‘Bantu', respectively. Numbers on the branches indicate the number of mutations having taken place along that branch.
Simulations
As can be seen from Table 4, the observed proportions of Khoisan-specific haplogroup frequencies in the Fwe are compatible with female-biased gene flow from a Ju-like source population, or unbiased female and male gene flow from a Khwe-like source population. The observed frequencies in the Tonga (and other populations similar to them) are compatible with at most low levels of female gene flow, but potentially high amounts of male gene flow from a Khwe-like population.
Table 4. Migration rates suggested to explain the proportion of ‘Khoisan' haplogroups, with significance >0.05.
| %HG source population | %HG receiver population | n receiver | CI for receiver population | Minimum migration rate | Migration rate associated with maximum probability | Maximum migration rate | |
|---|---|---|---|---|---|---|---|
| Ju into Fwe, mtDNA | 90 | 24 | 33 | 0.1–0.39 | 0.002 | 0.01 | 0.03 |
| Ju into Fwe, Y chromosome | 75 | 0 | 26 | 0–0.11 | 0 | 0.012 | |
| Khwe into Fwe, mtDNA | 60 | 24 | 33 | 0.1–0.39 | 0.0025 | 0.017 | 0.07 |
| Khwe into Fwe, Y chromosome | 16 | 0 | 26 | 0–0.11 | 0 | >0.5 (always P>0.05) | |
| Ju into Tonga, mtDNA | 90 | 0 | 22 | 0–0.13 | 0 | 0.012 | |
| Ju into Tonga, Y chromosome | 75 | 3 | 33 | 0–0.09 | 0 | 0.009 | |
| Khwe into Tonga, mtDNA | 60 | 0 | 22 | 0–0.13 | 0 | 0.02 | |
| Khwe into Tonga, Y chromosome | 16 | 3 | 33 | 0–0.09 | 0 | >0.5 (always P>0.05) |
Discussion
Clicks in the Bantu languages Fwe and Mbukushu, spoken in southwest Zambia and adjacent areas, may have arisen in three different ways: through mere culture contact without intensive physical interaction, hypothesized to correlate with the absence of large amounts of Khoisan genetic admixture; through language shift of entire groups of Khoisan speakers to a Bantu language, hypothesized to lead to an influx of both paternal and maternal Khoisan lineages to the Bantu gene pool; or through (presumably sex-biased) intermarriage hypothesized to lead to admixture only in the maternal line. At first glance, the results of the Y chromosome and mtDNA analyses appear to indicate sex-biased interactions between the Bantu-speaking populations and Khoisan groups, with a noticeable influx of mtDNA haplogroups L0d and L0k without corresponding levels of introgression of characteristic Y-chromosomal haplogroups. Especially for the Fwe, who speak a Bantu language with clicks, the results appear to indicate that borrowing of click consonants was associated with the incorporation of Khoisan women, as has also been argued for the southeast Bantu Xhosa and Zulu:26 nearly one quarter (24%) of the Fwe mitochondrial gene pool is of Khoisan origins, whereas no characteristic Khoisan Y-chromosome haplogroups were found in this population (Table 1 and Table 3). However, the simulations show that the observed haplogroup frequencies are compatible with two different scenarios, depending on whether the source population had a haplogroup composition more similar to Ju or to Khwe. Thus, should the Fwe have interacted with a Khwe-like source population, even large amounts of gene flow in the paternal line could have gone undetected in our approach, making it impossible to exclude the hypothesis of a language shift from Khwe-speakers to Fwe. On the other hand, should the source population have had a haplogroup composition similar to the Ju, our results are more compatible with sex-biased gene flow in the maternal line, with at most low levels of paternal gene flow. Two factors are in favor of the latter scenario of female-biased gene flow: first of all, the Fwe have a Y-STR variance of only 0.4 in combination with a haplotype diversity of 0.98 (±0.02; Table 1). This clearly shows that no very divergent Y-chromosomal haplotypes, such as one would expect to be present in a hunter-gatherer population long separated from the Bantu-speaking immigrants, have entered the Fwe gene pool, and argues against a large proportion of undetected male gene flow. Furthermore, the linguistic data show stronger affinities with a Ju language rather than with Khwe,2 lending greater weight to our estimates of plausible migration rates based on a Ju source population.
Yet a further hypothesis is that the Fwe and Shanjo shared a common ancestor with Khoisan groups before shifting to their current Bantu language in a process of intermarriage with Bantu, thereby incorporating Bantu genetic lineages and, in the case of the Fwe, carrying over some of the Khoisan click consonants. However, as this scenario would involve the replacement with Bantu lineages of up to 100% of the original Khoisan Y chromosomes and up to 90% of the original Khoisan mtDNA lineages (cf. Table 4), it appears less plausible than the scenario proposed above, namely intermarriage of a Bantu-speaking group with a Khoisan-speaking group restricted to the maternal line.
In agreement with the estimates of migration rates (Table 4), the contact between the Khoisan and the ancestors of the Fwe appears to have been intense, as at least four of the five L0d and L0k mtDNA haplotypes found in the Fwe are so divergent that it is unlikely that they could have evolved from only a couple of ancestral sequences of Khoisan origins. Thus, two of the L0k sequences are separated by 27 mutations; the two L0d sequences are separated by 56 mutations. To accumulate this amount of divergence from a single shared ancestor per haplogroup would take more than a thousand generations,27 whereas Bantu speakers arrived in Zambia only around 40 generations ago.
These data are therefore compatible with a scenario of intense contact with relatively high levels of intermarriage in the maternal line leading to the borrowing of click phonemes into these languages. However, this apparently straightforward conclusion is complicated by the puzzling lack of haplotype sharing between the Fwe and Khoisan populations, and the long branches, which lead to the Zambian Bantu L0k and L0d haplotypes (Figure 4). This is clearly due in part to the lack of comparative data, as the few complete mtDNA genomes available from Khoisan populations were sampled in a non-random fashion and stem from a highly restricted number of populations. Indeed, a comparison with preliminary data from a more representative range of Khoisan populations shows fewer numbers of mutations separating the Zambian L0d sequences and those from Khoisan populations (Barbieri et al, unpublished data); nevertheless, there is still no direct haplotype sharing between extant Khoisan populations and the southwest Zambian groups. Furthermore, the Zambian L0k sequences remain completely distinct, even when more data are included in the analysis (data not shown).
One possible explanation for the lack of sequence sharing between the Bantu and extant Khoisan populations might be that subsequent drift has erased lineages in the Khoisan groups that were retained in the Bantu populations through admixture. An alternative explanation might be that the ancestral hunter-gatherer groups living in the area at the time of the Bantu immigration have since been replaced by the immigrants. A third possibility would be that there was genetic structure among the ancient Khoisan-speaking hunter-gatherer groups, and that the Fwe intermarried with a Khoisan group whose genetic composition differed from that of the populations included in molecular anthropological investigations to date. This assumption is supported to a certain degree by the presence of higher frequencies of L0k than L0d in the Fwe. This differs from what is found in Khoisan-speaking populations7, 28, 29 and in populations that have experienced admixture with Khoisan groups,7, 9, 26, 30 where the proportion of L0d far outweighs that of L0k. More data on both Khoisan and Bantu-speaking groups of southwestern Africa are needed to shed light on this puzzle. Of course, these different explanations are not mutually exclusive, and it is plausible that the Fwe ancestors interacted with a Khoisan community that differed genetically from those still settled in southern Africa today, which was ultimately replaced by the newcomers.
Although the large proportion of Khoisan maternal lineages in the Fwe is in good accordance with the click consonants they have incorporated into their language, the high frequency of haplogroups L0d and L0k in the Shanjo is unexpected from a linguistic perspective, as their language did not incorporate clicks. It is of course quite possible that the Shanjo intermarried to the same extent with Khoisan-speaking women as the Fwe, but for sociocultural reasons did not borrow clicks. If the Khoisan mtDNA lineages in the Fwe and the Shanjo should indeed be the result of independent admixture events, the admixture would arguably have taken place with the same Khoisan-speaking population, as the Khoisan lineages found in the Fwe and the Shanjo are shared. Another possibility, however, is that the Khoisan mtDNA haplogroups found in the Shanjo did not originate directly from admixture with Khoisan communities, but through intermarriage with Fwe. This appears all the more plausible as two of the Shanjo L0d/L0k haplotypes are shared with the Fwe, with the third one being only one mutational step distant from a Fwe sequence type. What is extremely puzzling, however, is the significant lack of sharing of non-Khoisan lineages between the Fwe and Shanjo. This would appear to indicate that the possible intermarriage was biased specifically toward Fwe women with Khoisan maternal ancestry – a bias that is very hard to explain, though some form of social or physical preference may have been at play.
Similar to the Shanjo, it is possible that the Mbukushu did not interact directly with Khoisan communities, as their single L0k haplotype is shared with the Fwe. Unfortunately, however, due to the small sample size available for the Mbukushu, it is not possible to come to any definitive conclusions concerning their prehistory. Nevertheless, the linguistic data, too, is compatible with a possible influx of the click words in this language not through direct interaction with Khoisan speakers, but through borrowing from a Bantu language with clicks belonging to a different subgroup.2
In summary, although we cannot exclude substantial amounts of paternal gene flow from a Khwe-like source population and thus language shift, the genetic and linguistic data are in favor of admixture in the maternal line between some of the Bantu groups from Zambia and Khoisan-speaking populations. The amount of this intermarriage does not correlate with the presence of clicks in the languages of the groups concerned, as the Shanjo show a high frequency of Khoisan mtDNA haplotypes in the absence of clicks. The precise modality of the contact between the ancestors of the Fwe and Khoisan-speaking populations is hard to elucidate, but ultimate replacement of the Khoisan group by the Bantu-speaking community coupled with some female-biased admixture is the most plausible scenario. Furthermore, our results show that the mtDNA composition of the ancestral Khoisan population is most likely to have been distinct from that found in Khoisan groups investigated to date, pointing to the existence of deep genetic structure in the ancestral Khoisan groups of southern Africa. This demonstrates that it will be possible to gain insights into the genetic structure of pre-Bantu Khoisan groups that may no longer exist by looking for their genetic traces in Bantu groups that they admixed with. However, our conclusions are hampered by a lack of comparative data, and in order to obtain further insights into the history of interactions between the immigrating Bantu speech communities and the autochthonous Khoisan groups, more data from populations speaking Khoisan languages as well as Bantu-speaking groups of southern and central Africa are needed.
Acknowledgments
We are grateful to all sample donors for their participation in this study. We thank Mark Stoneking for helpful comments, the Livingstone Museum, Cesare de Filippo, Terry Nyambe, Ellen Gunnarsdóttir, and Mark Stoneking for help with sample collection, Antje Müller for help in the lab, Serena Tucci for help with sequence alignment, and Cesare de Filippo for organizing sample information, extracting DNA, writing R scripts and commenting on the analysis. This research was supported by the Max Planck Society and the Belgian Science Policy; CB was supported by a grant from the Deutsche Forschungsgemeinschaft (to BP).
Author contributions
KB and BP conceived the study, AB prepared the sequencing libraries, and CB performed phylogenetic analyses. The paper was written by BP and CB with detailed input from KB.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Güldemann T, Stoneking M. A historical appraisal of clicks: a linguistic and genetic population perspective. Annu Rev Anthropol. 2008;37:93–109. [Google Scholar]
- Bostoen K, Sands B.Clicks in south-western Bantu languages: contact-induced vs. language-internal lexical changein Brenzinger M (ed).Proceedings of the 6th World Congress of African Linguistics Cologne: Köln: Rüdiger Köppe Verlag; 2012(in press), pp129–140. [Google Scholar]
- Fortune G. The languages of the Western Province of Zambia. J Lang Assoc East Africa. 1970;1:31–38. [Google Scholar]
- Bostoen K.Shanjo and Fwe as part of Bantu Botatwe: a diachronic phonological approachin Ojo A, Moshi L (eds).Selected proceedings of the 39th Annual Conference on African Linguistics: Linguistic Research and Languages in Africa Sommerville, MA, USA: Cascadilla Press; 2009110–130. [Google Scholar]
- Behar DM, Villems R, Soodyall H, et al. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batini C, Ferri G, Destro-Bisol G, et al. Signatures of the preagricultural peopling processes in Sub-Saharan Africa as revealed by the phylogeography of early Y chromosome Lineages. Mol Biol Evol. 2011;28:2603–2613. doi: 10.1093/molbev/msr089. [DOI] [PubMed] [Google Scholar]
- Schlebusch CM, Naidoo T, Soodyall H. SNaPshot minisequencing to resolve mitochondrial macro-haplogroups found in Africa. Electrophoresis. 2009;30:3657–3664. doi: 10.1002/elps.200900197. [DOI] [PubMed] [Google Scholar]
- Naidoo T, Schlebusch CM, Makkan H, et al. Development of a single base extension method to resolve Y chromosome haplogroups in sub-Saharan African populations. Investig Genet. 2010;1:6. doi: 10.1186/2041-2223-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M, Sequeira F, Luiselli D, Beleza S, Rocha J. On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola. BMC Evol Biol. 2009;9:80. doi: 10.1186/1471-2148-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretorius JL. The Fwe of the East Caprivi. Unpublished MA thesis University of Stellenbosch. 1975.
- Phillipson DW. African archaeology. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- de Luna K. Classifying Botatwe: M60 languages and the settlement chronology of south central Africa. Afr Linguist. 2010;16:65–96. [Google Scholar]
- de Filippo C, Barbieri C, Whitten M, et al. Y-chromosomal variation in sub-Saharan Africa: insights into the history of Niger-Congo groups. Mol Biol Evol. 2011;28:1255–1269. doi: 10.1093/molbev/msq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic T, Whitten M, Pääbo S. Multiplexed DNA Sequence Capture of Mitochondrial Genomes Using PCR Products. PLoS ONE. 2010;5:e14004–e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;1:47–47. [PMC free article] [PubMed] [Google Scholar]
- Paradis E. pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics. 2010;26:419–419. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Gonzalez-Neira A, et al. Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genet. 2006;70:459–487. doi: 10.1111/j.1469-1809.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- StatSoft, Inc STATISTICA (data analyisis software system), version 10,2011
- Soodyall H, Heeran M, Philip H, Thijessen N. The genetic prehistory of the Khoe and San. S Afr Humanit. 2008;20:37–48. [Google Scholar]
- Wood ET, Stover DA, Ehret C, et al. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur J Hum Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- Batini C, Lopes J, Behar DM, et al. Insights into the Demographic History of African Pygmies from Complete Mitochondrial Genomes. Mol Biol Evol. 2011;28:1099–1110. doi: 10.1093/molbev/msq294. [DOI] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Arnason U, Gullberg A, Janke A, Kullberg M. Mitogenomic analyses of caniform relationships. Mol Phylogenet Evol. 2007;45:863–874. doi: 10.1016/j.ympev.2007.06.019. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Thieme M, Nanduri LK, et al. Validation of microarray-based resequencing of 93 worldwide mitochondrial genomes. Hum Mutat. 2009;30:115–122. doi: 10.1002/humu.20816. [DOI] [PubMed] [Google Scholar]
- Salas A, Richards M, De la Fe T, et al. The making of the African mtDNA landscape. Am J Hum Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- Henn BM, Gignoux CR, Jobin M, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci USA. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Harmant C, Quach H, et al. Strong Maternal Khoisan Contribution to the South African Coloured Population: A Case of Gender-Biased Admixture. Am J Hum Genet. 2010;86:611–620. doi: 10.1016/j.ajhg.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.