Abstract
Neurogenesis in the developing central nervous system consists of the induction and proliferation of neural progenitor cells and their subsequent differentiation into mature neurons. External as well as internal cues orchestrate neurogenesis in a precise temporal and spatial way. In the last 20 years, the zebrafish has proven to be an excellent model organism to study neurogenesis in the embryo. Recently, this vertebrate has also become a model for the investigation of adult neurogenesis and neural regeneration. Here, we summarize the contributions of zebrafish in neural development and adult neurogenesis.
Background
Neurogenesis describes the process by which undifferentiated neural progenitor cells generate mature and functional neurons. The first steps in neurogenesis are the induction of neural progenitors and a phase of cell division that enlarges the pool of progenitors. This is followed by a sequence of specification to committed progenitors, and differentiation to post-mitotic neurons. Each of these steps is spatially and temporally orchestrated to generate the multiple neuronal and glial cell types that will eventually populate the mature central nervous system (CNS). The zebrafish as a vertebrate model organism has been used in numerous studies of the various aspects of neurogenesis. Similar to mice and fruit flies, zebrafish can be used for genetic analysis. In contrast with mammals, the development of fish larvae occurs externally, making the zebrafish CNS accessible for experimental manipulation. In addition, such optical methods as high-speed and high-resolution microscopy, as well as new manipulative tools in the emerging field of optogenetics can be applied to this model organism. Thus, a unique combination of genetics, embryology, and state-of-the art optical techniques makes the zebrafish a unique vertebrate model organism to study neurogenesis.
The majority of studies conducted so far focus on neurogenesis at embryonic stages. However, recent studies have shown that the mature zebrafish brain may also serve as a valuable model for the study of adult neurogenesis. Indeed, as early as the 1960s, first experiments suggested that new neurons are born in the hippocampus and the olfactory bulb of the adult mammalian brain by in-situ3H]-thymidine labeling of newly synthesized DNA [1,2]. It took, however, more than 20 years before neurogenesis in the adult mammalian brain became widely accepted [3-7]. In rodents and non-human primates, the formation of new neurons is particularly evident in two regions, the subventricular zone (SVZ) and the subgranular zone (SGZ) of the telencephalon. However, neurogenesis has equally been reported in several brain regions outside the SGZ and SVZ [8], such as the basal forebrain [9], striatum [10,11], amygdala [12], substantia nigra [13], subcortical white matter [14], and, more recently, the hypothalamus [15-17]. Also the human brain seems to form neurons in the adult state [18-22]. The evidence for neurogenesis of olfactory bulb neurons derived from the SGZ in the adult human brain is, however, still controversial [23,24].
In contrast with mammals, teleosts like the zebrafish exhibit a much greater proliferative potential [25-27]. Up to 16 different proliferating regions were detected in discrete areas of the brain of adult zebrafish, including the regions equivalent to the mammalian SVZ and SGZ [26-30]. Around 6000 cells are born within every 30 min period in the adult zebrafish brain, representing ≈ 0.06% of the estimated 107 cells of the zebrafish brain [31]. This widespread neurogenic activity has made the zebrafish an attractive model for the study of adult neurogenesis.
Here we summarize recent advances of neurogenesis in zebrafish both during development and in adulthood.
Early neurogenesis
Neural induction by extrinsic factors
The first step in the development of the vertebrate nervous system is the specification of the neuroectoderm. This process is called ‘neural induction’ and is initiated during early embryonic development. At the onset of gastrulation, the forming mesodermal layer involutes and comes into contact with the overlying ectoderm [32-34]. This presumptive mesodermal layer secretes important factors locally to induce or inhibit neural induction in the ectodermal layer.
Over the past years, important advances have been made showing that vertebrate neural induction relies on complex interactions between extrinsic signaling factors, such as members of the bone morphogenetic protein (BMP), wingless-integrated (Wnt) and fibroblast growth factor (Fgf) families [35,36], and the intrinsic transcription factor program, most importantly members of the SRY-box containing genes B1 (SoxB1) family [37-39]. According to the ‘default model’ described in studies in Xenopus, cells within the ectodermal layer have the tendency to differentiate into neural tissue [40]. Ventral secretion of BMPs, in particular BMP2, 4 and 7, blocks neural induction by inducing an epidermal fate [41,42]. BMP antagonists such as Noggin and Chordin are produced early in the dorsal pre-organizer region, which later forms the Spemann organizer, corresponding to the shield organizer in fish. These secreted proteins act permissively for the establishment of the neural fate in the dorsal ectoderm and allow the formation of the neural plate [32,33,42,43]. However, in rare cases low levels of BMP are required to maintain brain compartments such as protecting telencephalic fate in neural plate stage [44]. It turns out that inhibition of BMP signaling is required for acquisition of ‘neural plate’ fate but not sufficient [45]. Additional secreted molecules, such as Cerberus from the anterior endoderm [46] or Fgfs from the blastoderm margin [47-49] are needed to induce a complete neural fate. Indeed, Fgf signaling has been shown to induce a posterior neural ectodermal fate independent of BMP inhibition [50,51].
Intrinsic factors determine the neuronal fate
Recent data suggest that members of the SRY-box containing genes B1 (SoxB1) gene family are the transcriptional basis of this ‘default program’: The SoxB1 genes are important for specification of the embryonic ectoderm into Type A neuroectodermal lineage in vertebrates [38,39] (Figure 1).
Figure 1.
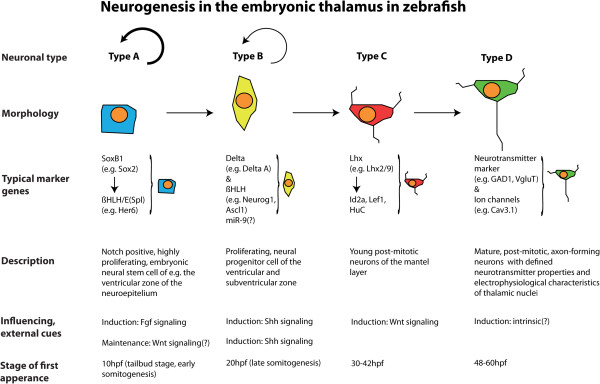
Neurogenetic cascade in the embryonic thalamus of zebrafish. Highly proliferating neural epithelial Type A cells are marked by Notch, SoxB1 and Her/Hes expression. After downregulation of SoxB1/Her/Hes, thalamic progenitors become proneural Type B cells. Type C cells are immature, post-mitotic neurons, which express transcription factors, such as lhx2b and lhx9. Mature, post-mitotic Type B neurons can be identified by, for example, neurotransmitter-specific factors.
The neural ectoderm is specified by members of the SoxB1 family [52-56]. So far, the following members of the SoxB1 have been characterized in zebrafish: sox1(a/b), 2, 3, and 19(a/b). All of these are induced during blastula stages and are redundantly required to specify neural ectodermal fate [57,58]. In fact, sox2 can be found in neural progenitors in the embryo as well as in neural stem cells in the adult zebrafish brain [29,30,58,59]. In combination with Pou2/Oct4, Sox2 activates repressors of neuronal differentiation, such as hesx1 and her3[58,60]. Hence, Sox2 is one of the most important factors required for the maintenance of neural progenitor properties and functions in the vertebrate lineage [61-63]. Prior to differentiation, SoxB1 members become downregulated, so that they are no longer expressed, neither in immature neurons nor in post-mitotic terminal neurons [39,64]. In addition, in the adult zebrafish brain, SoxB1 members label quiescent and proliferating glia as well as juvenile neurons.
However, one cannot entirely separate extrinsic cues from intrinsic factors. In zebrafish, the expression of the SoxB1 family member sox3 depends on early Fgf signaling from the blastoderm margin [50] and, in turn, regulates expression of early BMPs, such as BMP2 and 7 [58]. In summary, this suggests that in concert with the inhibition of BMP signaling and activation of Fgf signaling, SoxB1 members are important factors for maintaining the pool of neural stem cells in early gastrulation stages in the zebrafish embryo.
From neural plate to neural tube
Once specified, the neural ectoderm forms the neural plate, a pseudostratified epithelial structure in zebrafish. During early somitogenesis in fish, the neural plate converges and forms the neural keel, eventually fusing at the dorsal midline to form the neural rod. Starting from the anterior, cells in the emerging neural keel become polarized at the embryonic midline. A recent study suggests that midline polarization of structural proteins, such as Pard3 and Rab11a, occurs during cell interdigitation in the neural rod [65]. The subsequent apical localization of these proteins, including Zona Occludens protein ZO-1, aPKC, and the basolateral localization of Numb and Lgl2 and the subsequent mirror-symmetric cell divisions in the medial neural rod lead to the formation of its lumen, the neurocoel, which will give rise to the brain ventricles and is important for the expansion neural progenitor pool [66-68].
During the process of neurulation, the zebrafish differs from other vertebrates: instead of folding up the neural plate immediately into a tube with a lumen, it first forms a solid neural keel. However, the topological arrangement of cells in zebrafish during formation of the neural keel from the neural plate is similar to that of other vertebrates. In the so-called ‘secondary neurulation’, the fish neural rod inflates and forms a vertebrate typical tube. Thus, although there are differences, neurulation in fish and mammals leads to the formation of a highly similar structure in each case, the neural tube [69].
After lumen formation, we find an increasing number of asymmetrically dividing cells in the neural tube. The asymmetric inheritance of a subcellular membrane domain of the dividing progenitors is strongly correlated with the asymmetric fate of the daughter cells [70]. A more apically derived daughter cell becomes the neuron, whereas a more basal daughter replenishes the apical progenitor pool. It was suggested that Notch signaling is important for the fate decision: basal self-renewing daughter cells display high Notch activity and an apical differentiating daughter cell show a low Notch activity [71]. The directionality of Notch signaling involves Pard3-dependent asymmetric localization of Mindbomb to the apical daughter.
The dividing neuronal progenitors move from the apical side of the neural epithelium to the pial side in a process described as ‘interkinetic nuclear migration’ (reviewed in [72]). Cells divide at the apical side and differentiate in the basal zone. Experiments in the zebrafish retina have shown that if migration is speeded up and cells remain too long in the basal zone, progenitors exit the cell cycle prematurely. This leads to an increase of early-born projection neurons at the expense of late-born interneurons [73]. Thus, the establishment of apical-basal polarity of neuroepithelial cells is critical for CNS maturation.
Initiation of neurogenesis by bHLH factors, the Enhancer-of-split E(Spl) subfamily
The onset of neurogenesis in the zebrafish neural plate becomes apparent during late gastrulation by the expression of proneural genes [74]. The first proneural genes expressed encode transcription factors, such as the bHLH genes neurogenin1 (neurog1; [75,76]) and achaete-scute1 (asc1, Figure 1) [77]. Transcription of these genes is not ubiquitous but restricted to cell clusters that are Type B proneural clusters [75], from which the nuclei of the primary neuronal network will arise [78].
At this stage, one can still find large SoxB1 positive, non-neurogenic territories that separate the first proneural clusters (reviewed in [79]). These domains will be progressively recruited towards their neural fate during early neurogenesis. Members of the E(Spl) family, the Her/Hes genes, are transcriptional targets of SoxB1, that is, her3[58], and are expressed in various combinations in most domains of non-neurogenic territories, such as the other hairy genes her5, her6, her9, and her11[80,81]. Ectopic expression of these Her/Hes genes lead to a downregulation of neurog1 expression whereas loss-of-function experiments show an increase in neurog1 expression.
The function of hairy genes, for example, Her5, has been studied intensively during midbrain-hindbrain boundary (MHB) formation [79,82]. The progenitor pool at the MHB is located between the ventrocaudal cluster of the midbrain and presumptive neurons of anterior rhombomeres. At the onset of neurogenesis, cells of this progenitor pool give rise to differentiated cells within the MHB domain [80]. This progenitor pool is marked by her5 and her11 expression and combined blockage of these E(Spl) factors suggests that they are partially redundant in domains of co-expression [83-85]. Both factors block expression of several proneural genes such as neurog1, ascl1a, and cyclin-dependent kinase inhibitors, but their direct transcriptional targets remain unknown.
Taken together, SoxB1/Hairy-dependent, active inhibition of neurogenesis is the crucial mechanism expanding the pool of cells. Spatially controlled downregulation of Hairy gene activity defines progenitor pools within the zebrafish embryonic neural plate.
According to their dependency on active Notch signaling, hairy genes can be grouped into two families [86]. The expression of the Her genes mentioned above, her3, her5, her6, her9 and her11 are independent of Notch signaling [83,87]. This is in contrast with the important function of other hairy factors, such as her4. Within each cluster, cells expressing higher levels of proneural genes are selected as ‘neuroblasts’ for further commitment and differentiation, while concomitantly maintaining their neighbors as proliferating neural precursors available for a later round of neuroblast selection [88]. This process of ‘lateral inhibition’ relies on Notch-dependent Her4 signaling in precursors, and expression of the bHLH transcription factor Delta in the neighboring neuroblasts [89,90]. Her4 is required to maintain neural progenitor fate in the embryo as well as neural stem cell fate in later stages [91-93] The her4-positive proliferating neural precursors can be found within proneural clusters and fulfill an important function during the selection of neuronal versus glial fate by lateral inhibition [89].
Early anterior-posterior patterning of the neural plate
One of the most fascinating problems in developmental biology is how positional information is assigned along the anterior-posterior (AP) axis, especially in the neuroectoderm. Following neural induction, progenitor cells display an anterior neural plate fate and subsequently become transformed to adopt a more posterior character by signals originating from more caudal regions of the embryo [94,95]. Among the postulated posteriorizing signals in zebrafish are Wnt, Fgf and Nodal proteins, as well as retinoic acid [96,97]. Wnt proteins are likely to be the earliest patterning signals acting in the forming neural plate primordium. The zebrafish mutants headless (hdl) and masterblind (mbl) carry mutations in the genes encoding tcf3 and axin1, respectively, and exhibit severe anterior patterning defects [98-100]. In these embryos, the forebrain and the eye anlage are reduced or absent and the midbrain expands forward to the tip of the neural tube. Recent studies have shown that Wnt8, expressed in the non-axial mesendoderm, is important for acquisition of hindbrain fate [101]. Consistent with this, Wnt8 is required for induction of the hindbrain expression of gbx1 [102]. To establish a Wnt-gradient, a counteracting signal must be released from the most anterior neural structures. The anterior neural boundary (ANB) indeed acts as an organizer to pattern the anterior neural plate by the release of secreted frizzled-related protein (sFRP) Wnt antagonists, such as TLC [103,104]. Thus, graded Wnt activity acts to initiate AP patterning within the neural plate. Following this initial patterning step, further mechanisms maintain and refine regional identity in the neural tube, for example, the formation of local boundaries such as between the forebrain and midbrain [105]. These boundaries often co-localize, with local organizing centers within the neural tube mediating specific positional information along the AP axis from anterior to posterior [106,107].
One of these local signaling centers is the mid-diencephalic organizer (MDO), which is established at the intrathalamic boundary, the zona limitans intrathalamica [108]. The MDO is characterized by shh expression [109-112] and orchestrates the development of the thalamic complex in the diencephalon in zebrafish [113]. shh acts as the principal signal of this organizer and is required for the concentration-dependent induction of neurog1 and ascl1, leading to the establishment of the thalamic nuclei in vertebrates [81,114].
Within the neural tube, a further well-characterized organizing center is the MHB organizer at the isthmus rhombencephali (reviewed in [115,116]). The MHB is able to induce cellular fates in a nonautonomous manner by the secretion of the principal signal Fgf8 [117,118].
Initially, other signaling factors, such as the canonical Wnt proteins Wnt1, Wnt3, Wnt3a, and Wnt10b, are broadly expressed throughout the forebrain and midbrain, until their expressions become restricted to the MDO, the MHB, and the dorsal midline [119,120]. Wnts do not elicit inductive effects comparable to those of the principal signals Shh and Fgf8. However, blockage of Wnt signaling leads to a lack of local organizing centers [120,121]. Thus, Wnt signaling activity is a common theme upstream of many local brain organizers [113].
The thalamus in zebrafish – an example for controlled neurogenesis
Vertebrate neurogenesis within a proneural cluster is regulated with a high degree of temporal and spatial precision, with stereotypic patterns of neuronal differentiation and extensive neuronal migration [122,123]. Dynamic patterns of mitotically active neuronal precursors within the neuronal clusters have been described as ‘neurogenetic gradients’ [124]. In the thalamus, the progression of neuronal differentiation from posterior to anterior presents such a neurogenetic gradient [108]. Recently, we could show that this mechanism is used to guide neurogenesis in the developing caudal diencephalon, that is, the thalamus [81]. The thalamic primordium is initially a non-neurogenic territory. Shh from the MDO determines the neurogenic program of the thalamus by inducing proneural genes such as neurog1. However, neurogenesis does not start immediately after induction of Shh expression in the thalamic complex and is first suppressed [81]. E(Spl) factor-positive domains separate the proneural clusters. These domains exhibit delayed differentiation and become progressively neurogenic only in later developmental stages (reviewed in [79]). Neurogenesis in the thalamic complex is regulated by the E(Spl) factor Her6 [81]. The widespread expression of her6 in the anterior neural plate represses the Neurog1-mediated neurogenesis cell-autonomously. The subsequent posterior-to-anterior regression of her6 expression is accompanied by up-regulation of deltaA and neurog1 in the ventricular and subventricular zones of the neural tube in cells that formerly expressed her6. The mechanism of Her6 regression is not well understood; however, different explanations have recently been proposed. Firstly, Shh has been suggested to influence the stability of E(Spl) factors, such as Hes1, the murine homolog of Her6 [125], which could explain the maintenance of her6 expression in the vicinity of the MDO. Secondly, microRNAs have been reported to play an important role in the progression of progenitors to post-mitotic neurons [126]. Indeed, microRNA-9 defines an intermediate, ambivalent state by repression of both the progenitor marker Her6 as well as the post-mitotic marker Elavl3, formerly known as HuC. Thus, miR-9 facilitates the transition of progenitors toward cell-cycle exit at late stages of embryonic development [127], to enable the nervous system to generate an increased number of neurons at this stage [128]. Indeed, the dynamic of the Her6 regression from posterior to anterior is later reflected in the induction of neurog1 expression and the generation of Type C, post-mitotic thalamic neuronal markers such as id2a, lhx2b, lhx9 and HuC [129] (Figure 1). These transcription factors orchestrate the subsequent maturation to Type D neurons with defined neurotransmitter properties. Hence, a crucial mechanism that regulates neurogenetic gradients within the embryonic CNS is a process of spatially controlled de-repression of neurogenesis, mediated by the local inactivation of the Her/Hes proteins.
Adult neurogenesis
Neurogenesis in the adult zebrafish telencephalon
The mammalian telencephalon develops by evagination of the neural tube. In contrast, the telencephalon of teleosts forms by outward folding (eversion) so that the proliferative periventricular zones are not only present internally but also cover the outer surface of the telencephalon [130-133] (see Figure 2). Proliferative zones form discrete areas that differ in the architecture of the stem cell niche [134]. In mammals, the ventricular surface is covered by an epithelium, the so-called ependyma or ependymal layer [135-138]. In zebrafish, the ependymal layer is restricted to the roof of the telencephalic ventricle and to dorsolateral regions of the proliferative zones (D and Dm; see Figure 2) [134]. The other proliferative zones are devoid of an ependymal layer. In this respect, the cellular environment of the zebrafish stem cell niche resembles more closely the fragmented ependymal lining of the stem cell niches of reptiles and birds (reviewed in [139]). Proliferative areas also differ with respect to the presence of ciliated cell types. The D, Vd, Dm, and Dl areas (Figure 2) contain multiciliated cells whereas the stem cells at the ventricular cavity in the Vv are devoid of cilia [134]. Furthermore, proliferation is modulated along the rostrocaudal axis of the telencephalon: While cell proliferation can be detected in the entire periventricular zone at rostral levels, proliferation is confined to distinct subdomains in more caudal areas of the telencephalon [134].
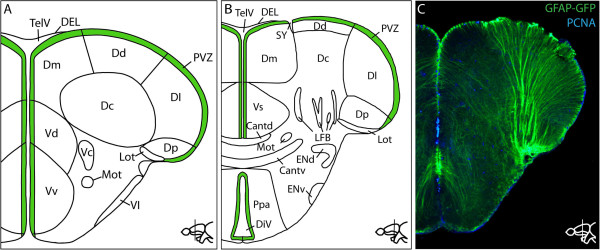
Figure 2.
Anatomy of the telencephalon of the adult zebrafish. Schematic of transverse sections through the anterior (A) and posterior (B) zebrafish telencephalon indicating the different anatomical subdomains. Proliferative periventricular zones are indicated in green. Schematic was redrawn from [191]. C: Radial glial cells have long processes that reach all the way to the pial surface. Immunostaining against GFP in the Tg(GFAPGFP) line marks the radial glial cells in the telencephalon of an adult zebrafish. The section was co-stained with the proliferation marker PCNA (proliferating cell nuclear antigen) to distinguish between PCNA-negative Type I and PCNA-positive Type II cells (described in [30]). Cantd: commissural anterior, pars dorsalis; Cantv: commissural anterior, pars ventralis; Dc: central zone of the dorsal telencephalic area (D); Dd: dorsal zone of D; DEL: dorsal ependymal lining; DiV: diencephalic ventricle; Dl: lateral zone of D; Dm: medial zone of D; Dp: posterior zone of D; Lot: lateral olfactory tract; ENd: entopeduncular nucleus, dorsal part; ENv: entopeduncular nucleus, ventral part; LFB: lateral forebrain bundle; Mot: medial olfactory tract; Ppa: anterior part of parvocellular preoptic nucleus; PVZ: periventricular zone; SY: sulcus ypsiloniformis; TelV: telencephalic ventricle; Vc: ventral nucleus of the ventral telencephalic area (V); Vd: dorsal nucleus of V; Vl: lateral nucleus of V; Vp: postcommissural nucleus of V; Vs: supracommisural nucleus of V; Vv: ventral nucleus of V.
Proliferative zones differ also in their rate of formation of new cells [26]. The most proliferative region is found in the medial subpallium (Vv) [26,30,140]. This region, in contrast to the other proliferative zones, consists almost exclusively of bromodeoxyuridine-positive (BrdU), S100 calcium-binding protein β-negative (S100β), non-ciliated cells, with neurons forming the deeper layer of the niche. In contrast with the other stem cell niches, no quiescent cells are present in this region [30,134]. Furthermore, only cells in the very dorsal part of Vv express S100β. The region is homologous to the SVZ of mammals and generates mostly neurons that migrate via the rostral migratory stream into the olfactory bulb [26,27,141-143]. In contrast with mammals, however, this region also generates neurons that settle in the adjacent parenchyma [26,141]. Lower proliferative rates were scored in other ventricular regions with somewhat elevated levels in the posterior zone of the dorsal telencephalic area (Dp), equivalent to the SGZ of the mammalian hippocampus [25,27]. Some proliferating cells were also detected throughout the parenchyma [26,144]. Marker analysis showed that these parenchymal cells are dividing oligodendrocyte precursor cells (OPCs) that both self-propagate and give rise to mature oligodendrocytes [144].
Cellular key players in adult zebrafish neurogenesis are radial glia cells. These cells, whose cell bodies are located close to the ventricle, have long processes that reach all the way to the pial surface of the telencephalon, where they terminate with their endfeet on the walls of blood vessels or at the pial surface itself [59,134].
Radial glial cells in the adult zebrafish telencephalon proliferate and, in addition to self-renewal, are able to generate new neurons [145]. Radial glial cells have also been observed in the developing mammalian telencephalon, where they act as embryonic neural stem cells. Mammalian radial glia disappear shortly after birth, giving rise to ependymal cells and astrocytes, some of which retain stem cell potential in the SVZ and SGZ [146-148]. Thus, the adult zebrafish appears to have retained embryonic features. Indeed, the adult zebrafish radial glial cells express similar genes to the embryonic counterparts, such as the glial acidic fibrillar protein (GFAP), S100β and the brain lipid-binding protein (BLBP) [26,59,149,150]. With the expression of proliferation markers, such as PCNA, or by detecting incorporation of the thymidine analog BrdU, one can distinguish two classes of radial glia cells: those which are in a quiescent state (Type I cells, BrdU−, PCNA−, GFAP+, S100β+, BLBP+) and slowly cycling cells (Type II cells, BrdU+, PCNA+, GFAP+, S100β+, BLBP+). Furthermore, Her/Hes genes, such as her3, her4, her5, her6, her8a, and her15, generally characterize quiescent rather than actively proliferating radial glial cells [93,140]. The majority of radial glial cells are in a resting state. Type II cells give rise to neuroblasts (Type III cells) that continue to proliferate, turn on neural marker genes, such as PSA-NCAM and the proneural gene ascl1, and eventually enter the rostral migratory stream or leave the periventricular zone to move deeper into the parenchyma. Proliferating Type II stem cells can divide symmetrically to self-renew or asymmetrically to generate Type III cells [145]. The relative proportion of proliferating and resting stem cells can be shifted by interfering with Notch signaling: Activation of the Notch pathway drives stem cells into quiescence, suggesting that cross talk between cells controls the maintenance of the quiescent stem cell population [151] (Figure 3). Hence, maintenance of neural stem cells in the adult appears to employ a similar mechanism as that seen in the embryo, where Notch-mediated lateral inhibition maintains a pool of embryonic neural stem cells for later production of neurons, such as the secondary motor neurons [152]. In the periventricular domain (Vv in Figure 2), but not in dorsal proliferative domains, Fgf signals are regulators of cell proliferation [140]. This highlights the differences of the stem cell niches in the ventral and dorsal telencephalon and points to Fgf signals as potential causes of the higher proliferation rate in the ventral proliferative zone.
Figure 3.
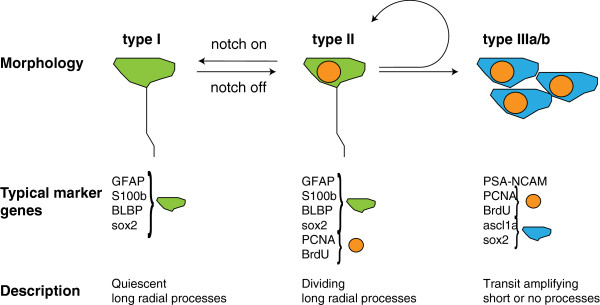
The major proliferative cell types in the telencephalon stem cell niche. The markers expressed by the three major cell types are indicated [30]. Notch signaling transforms proliferating Type II radial glial cells into quiescent Type I radial glial cells [151]. Note that there exists heterogeneity in the cellular composition and marker expression of stem cell niches in the telencephalon [30,140].
The long processes of radial glial cells have been proposed to serve as a scaffold for newborn neurons to migrate out of the ventricular zone [59,149,150]. However, most newborn neurons appear to settle in subperiventricular areas [92], suggesting that the majority of newborn neurons will not migrate to the pial surface as many types of newborn neurons do in the embryonic brains of fish [129] or mouse [153]. The cellular architectures of the stem cell niches are more complex than summarized in the schematic of Figure 3. For example, in addition to S100β/glutamine synthetase (GS)/GFAP/aromatase B-positive radial glial cells also S100β/GS/aromatase B-positive but GFAP negative cells were found at a smaller proportion in the dorsal and dorsolateral regions [30,140]. Moreover, the electron microscopic study of [134] detected additional cell types that differ in cellular morphology and marker expression. The Type IIa cells reported in [134] have shorter processes than the radial glia cells (the Type I and II cells described by [30]) and contact blood vessels in the vicinity of the stem cell niche [134]. This suggests the existence of additional cell types that may be involved in neurogenesis, either directly by being specialized progenitors or indirectly by contributing to the architecture of the niche.
Other neurogenic sites in the adult zebrafish brain
While the stem cell niches of the adult telencephalon are the most intensively investigated, several studies also reported on other proliferative regions in the brain. The cerebellum shows highly abundant neurogenesis [27]. The dividing cerebellar neural progenitors express the progenitor markers nestin, Sox2, Meis homeobox 2 (Meis) and Musashi homolog 1 (Drosophila) (Msi1). They also exhibit neuroepithelial properties such as expression of ZO-1, β-catenin, γ-tubulin, and protein kinase C (aPKC) [29]. These dividing cells do not express radial glial markers. The cerebellar niche, however, also contains radial glia-like cells expressing GFAP, vimentin, and BLBP. These cells appear to act as scaffolds for migrating progenitor cells rather than as stem cells per se. As found in the periventricular Vv, Fgf signaling is an important regulator of stem cell activity in the cerebellum. Blocking Fgf signaling with a heat-shock inducible dominant-negative Fgf receptor leads to a significant reduction of proliferating cells [29].
Another neurogenic site is located at the boundary between the midbrain and the hindbrain [86]. This region expresses the her5:gfp transgene in cells that form a restricted cluster at the ventricle of the MHB. These her5:gfp positive cells fulfill several general hallmarks of neural stem cells. A subpopulation of the her5:gfp cells cycle slowly, express the stem cell markers GFAP, BLBP, Sox2 and Musashi, and can differentiate into neurons and glia [86]. her5:gfp-expressing cells resemble the Type II cells of the telencephalic niche. However, evidence for the presence of an intermediate fast proliferating cell type such as the transit amplifying cells of the mammalian stem cell niches or the Type III cells of the telencephalic niches of the zebrafish brain is lacking.
In the optic tectum of adult zebrafish, proliferating cells exist in the medial, lateral, and caudal margins of the PGZ [25,27,154-156]. Proliferating cells in these regions express the neural progenitor markers PCNA, Sox2, Msi1 [157-159]. These cells do not express glial markers. However, they show neuroepithelial characteristics, such as the expression of ZO-1, γ-tubulin, and aPKC [73,160]. BrdU lineage tracing showed that these cells can differentiate into glutamatergic or GABAergic neurons, oligodendrocytes and radial glia, and hence represent a multipotent progenitor pool [156,161-163]. Similar to the architecture of the cerebellar stem cell niche, non-dividing radial glia-like cells expressing glial markers (GFAP, BLBP, S100β) reside adjacent to the proliferating neuroepithelial-like progenitors. In addition, proliferating radial glia-like cells can be detected in the deeper layer of the PGZ [156].
Although the analysis of these stem cell niches in comparison to the telencephalic niches is more limited, results suggest that the architecture of the different stem cell niches in the zebrafish brain differ significantly, which may reflect differences in the neurogenic potential of the sites.
Regeneration of the adult brain
Regeneration in the mammalian brain is very limited. It seems that the local environment in most parts of the mammalian brain is not suited for the long-term survival of newly formed neurons [164,165]. In contrast, zebrafish have a remarkable ability to regenerate injured organs, including the CNS [166-170]. Zebrafish regenerate axons [171] and whole neuronal assemblies with full restoration of function after injury [28,172-180].
After wounding the telencephalon by stabbing with a needle, neurogenesis is stimulated along the entire ventricular surface (Figure 4), with many more Type II stem cells expressing the proliferation marker PCNA within 3 days after wounding [179]. This proliferation response is restricted to the injured hemisphere and persists at least up to 14 days [92,179]. In contrast with the predominant settlement of newborn neurons at the subventricular zone during constitutive neurogenesis, newborn neurons in the injured brain migrate over longer distances into the area of damaged tissue and differentiate into mature neurons [92,180]. Precisely which long-range signals mediate the increase of proliferation in the periventricular stem cell zones even at sites distal to the lesion remains unknown as yet. Chemokine signaling via Cxcr5 may play a role in this process [181]. It is also unclear why the activation remains restricted to the lesioned hemisphere [179], even though the proliferative zones in the medial telencephalon are separated only by a narrow ventricular gap.
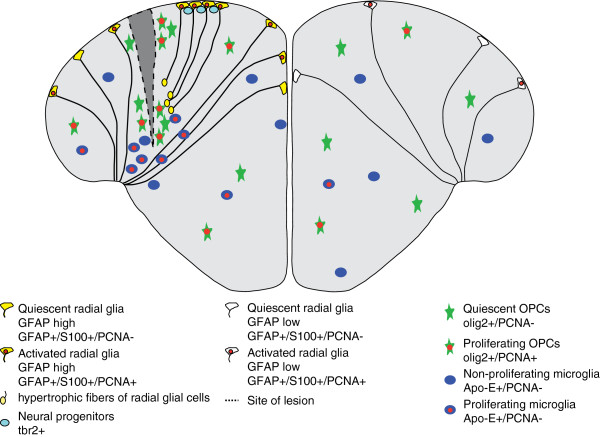
Figure 4.
Regenerative responses to stab wound injury. Lesions introduced by a syringe needle induce a proliferative response in the periventricular region of the stabbed hemisphere. Oligodendrocytes and microglial cells accumulate at the site of lesion. The glial marker GFAP and the proliferation marker PCNA are upregulated in the lesioned hemisphere. The number of T-box brain gene 2-positive (Tbr2+) cells is increased upon injury [179]. The injury is totally healed after 30 days without traces of a glial scar. Schematic shows transverse section through the medial telencephalon.
Some proliferation is also detectable at the site of lesion, which has rapidly filled with blood cells after the injury. However, these proliferating cells are mostly microglia involved in removal of the cellular debris [92,179,180]. In addition, OPCs accumulate at the site of lesion. These OPCs proliferate only to a limited extend and, hence, most of them appear to have migrated in from adjacent regions [179]. The accumulation of microglia and OPCs at the site of lesion is transient [179]. There is no scar formation in the zebrafish brain, as is seen in injured mammalian neural tissue [92,179,180]. In mammals, glial scars are thought to act as mechanical and biochemical barriers that prevent ingrowth of new axons or migration of cells into the lesion site [182]. By three weeks after injury, the proliferation rate at the ventricular zone has returned to the base line seen in uninjured brains, and the damaged tissue has recovered totally, without any signs of the traumatic impact remaining [179,180]. Interestingly, the molecular mechanism underlying constitutive and reactive neurogenesis in response to injury seems to differ: reactive neurogenesis depends on the zinc-finger transcription factor gata3, which is not expressed in the uninjured brain [183,184].
A fundamental question is why fish have this extraordinary ability to regenerate adult neural tissues. Zebrafish also grow in adulthood, with generation of new neurons that need to be incorporated into existing neuronal circuits, suggesting that this generates a favorable physiological situation for the repair of injured neural tissue. In lesions of the adult fish cerebellum, damaged cells seem to be removed by apoptosis [185]. In contrast, necrotic cell death is predominant in injured mammalian neural tissue [186,187]. Necrosis triggers inflammatory responses much more strongly than apoptosis, gradually leading to loss of neural cells and scar formation [188]. Thus, the difference in the mode of cell death may be one of the factors that contribute to the extraordinary recovery of neural tissue in fish after injury. The oligodendrocyte marker Olig2, which suppresses neurogenesis in mammals [189], is massively upregulated in injured mammalian neural tissues. By contrast, in the zebrafish telencephalon, olig2:EGFP-expressing cells accumulate transiently at the site of lesion [179]. It was demonstrated recently that inflammatory signals trigger reactive neurogenesis in the zebrafish telencephalon, pointing at yet another fundamental difference in the regenerative responses in teleost and mammal brains [190]. The central questions are whether we can learn from zebrafish how, for example, scar formation is suppressed and what the causes of the remarkable plasticity of neural tissue are in the zebrafish. Most of all, a burning challenge will be to use this knowledge from the fish brain to modulate the behavior of stem cells, neurons, glia, and immune cells in the injured human brain.
Conclusion
The regulatory cascades in the embryo and adult have similarities in the sequence of events underlying formation of functional neurons. However, the tissue settings are fundamentally different. In the embryo, neurons are derived from the neuroectodermal epithelium, which becomes structured in a hierarchy of increasing complexity. In contrast, adult neurons originate mainly from glia cells and need to be incorporated into a preexisting, fully functional tissue. Moreover, in case of repair, immune responses, cell death in addition to the complexity of a mature tissue are factors that contribute to the successful restoration of structure and function. This may explain the usage of different genetic regulators in the cascades. Elucidating these specific differences will provide additional insights into how neurogenesis is mediated in the embryo and adult. Detailed knowledge will enable us to develop tools that might be used to increase the neurogenic potential of neural stem cells in terms of their ability to undergo neuronal determination and differentiation. In the future, these insights may form the basis for the design of therapeutic strategies that will allow eventually the activation of quiescent progenitors and their successful incorporation in functional neuronal circuits in the mammalian brain.
Abbreviations
ANB: Anterior neural boundary; AP: anterior-posterior; BLBP: Brain lipid-binding protein; BMP: Bone morphogenetic protein; BrdU: Bromodeoxyuridine; CNS: Central nervous system; Fgf: Fibroblast growth factor; GFAP: Glial acidic fibrillar protein; KIT: Karlsruhe Institute of Technology; MDO: Mid-diencephalic organizer; MHB: Midbrain-hindbrain boundary; OPC: Oligodendrocyte precursor cells; PCNA: Proliferating cell nuclear antigen; sFRP: Secreted frizzled-related protein; SGZ: Subgranular zone; SVZ: Subventricular zone; Wnt: Wingless-integrated.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SS summarized neurogenesis in the embryo, whereas RS and US focused on neurogenesis in the adult. All authors read and approved the final manuscript.
Contributor Information
Rebecca Schmidt, Email: rebecca.schmidt@kit.edu.
Uwe Strähle, Email: uwe.straehle@kit.edu.
Steffen Scholpp, Email: steffen.scholpp@kit.edu.
Acknowledgement
We would like to thank Corinne Houart (King’s College London), Thomas Dickmeis and Sepand Rastegar (both KIT), Martin März (Max-Planck-Institut für molekulare Biomedizin, Münster), and the Scholpp group for valuable comments on the manuscript. RS and US are supported by the EU IP ZF Health and the Helmholtz Association and SS is supported by a DFG Emmy Noether fellowship 847/2.
References
- Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319–335. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- Altman J. Autoradiographic and histological studies of postnatal neurogenesis. IV. Cell proliferation and migration in the anterior forebrain, with special reference to persisting neurogenesis in the olfactory bulb. J Comp Neurol. 1969;137:433–457. doi: 10.1002/cne.901370404. [DOI] [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc Natl Acad Sci USA. 1993;90:2074–2077. doi: 10.1073/pnas.90.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH, Coates PW, Palmer TD, Kuhn HG, Fisher LJ, Suhonen JO, Peterson DA, Suhr ST, Ray J. Survival and differentiation of adult neuronal progenitor cells transplanted to the adult brain. Proc Natl Acad Sci USA. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson PS, Perfilieva E, Björk-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- Kukekov VG, Laywell ED, Suslov O, Davies K, Scheffler B, Thomas LB, O’Brien TF, Kusakabe M, Steindler DA. Multipotent stem/progenitor cells with similar properties arise from two neurogenic regions of adult human brain. Exp Neurol. 1999;156:333–344. doi: 10.1006/exnr.1999.7028. [DOI] [PubMed] [Google Scholar]
- Gould E. How widespread is adult neurogenesis in mammals? Nat Rev Neurosci. 2007;8:481–488. doi: 10.1038/nrn2147. [DOI] [PubMed] [Google Scholar]
- Palmer TD, Ray J, Gage FH. FGF-2-responsive neuronal progenitors reside in proliferative and quiescent regions of the adult rodent brain. Mol Cell Neurosci. 1995;6:474–486. doi: 10.1006/mcne.1995.1035. [DOI] [PubMed] [Google Scholar]
- Pencea V, Bingaman KD, Wiegand SJ, Luskin MB. Infusion of brain-derived neurotrophic factor into the lateral ventricle of the adult rat leads to new neurons in the parenchyma of the striatum, septum, thalamus, and hypothalamus. J Neurosci. 2001;21:6706–6717. doi: 10.1523/JNEUROSCI.21-17-06706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie DC, Dziewczapolski G, Willhoite AR, Kaspar BK, Shults CW, Gage FH. The adult substantia nigra contains progenitor cells with neurogenic potential. J Neurosci. 2002;22:6639–6649. doi: 10.1523/JNEUROSCI.22-15-06639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MC, Roy NS, Keyoung HM, Goodman RR, McKhann G, Jiang L, Kang J, Nedergaard M, Goldman SA. Identification and isolation of multipotential neural progenitor cells from the subcortical white matter of the adult human brain. Nat Med. 2003;9:439–447. doi: 10.1038/nm837. [DOI] [PubMed] [Google Scholar]
- Kokoeva MV, Yin H, Flier JS. Neurogenesis in the hypothalamus of adult mice: potential role in energy balance. Science. 2005;310:679–683. doi: 10.1126/science.1115360. [DOI] [PubMed] [Google Scholar]
- Migaud M, Batailler M, Segura S, Duittoz A, Franceschini I, Pillon D. Emerging new sites for adult neurogenesis in the mammalian brain: a comparative study between the hypothalamus and the classical neurogenic zones. Eur J Neurosci. 2010;32:2042–2052. doi: 10.1111/j.1460-9568.2010.07521.x. [DOI] [PubMed] [Google Scholar]
- Lee DA, Bedont JL, Pak T, Wang H, Song J, Miranda-Angulo A, Takiar V, Charubhumi V, Balordi F, Takebayashi H, Aja S, Ford E, Fishell G, Blackshaw S. Tanycytes of the hypothalamic median eminence form a diet-responsive neurogenic niche. Nat Neurosci. 2012;15:700–702. doi: 10.1038/nn.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones-Hinojosa A, Sanai N, Soriano-Navarro M, Gonzalez-Perez O, Mirzadeh Z, Gil-Perotin S, Romero-Rodriguez R, Berger MS, Garcia-Verdugo JM, Alvarez-Buylla A. Cellular composition and cytoarchitecture of the adult human subventricular zone: a niche of neural stem cells. J Comp Neurol. 2006;494:415–434. doi: 10.1002/cne.20798. [DOI] [PubMed] [Google Scholar]
- Boldrini M, Underwood MD, Hen R, Rosoklija GB, Dwork AJ, John Mann J, Arango V. Antidepressants increase neural progenitor cells in the human hippocampus. Neuropsychopharmacology. 2009;34:2376–2389. doi: 10.1038/npp.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaysse L, Labie C, Canolle B, Jozan S, Béduer A, Arnauduc F, Vieu C, Sol JC, Loubinoux I. Adult human progenitor cells from the temporal lobe: another source of neuronal cells. Brain Inj. 2012;26:1636–1645. doi: 10.3109/02699052.2012.700084. [DOI] [PubMed] [Google Scholar]
- Sanai N, Tramontin AD, Quiñones-Hinojosa A, Barbaro NM, Gupta N, Kunwar S, Lawton MT, McDermott MW, Parsa AT, Manuel-García Verdugo J, Berger MS, Alvarez-Buylla A. Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature. 2004;427:740–744. doi: 10.1038/nature02301. [DOI] [PubMed] [Google Scholar]
- Curtis MA, Kam M, Nannmark U, Anderson MF, Axell MZ, Wikkelso C, Holtås S, Van Roon-Mom WMC, Björk-Eriksson T, Nordborg C, Frisén J, Dragunow M, Faull RLM, Eriksson PS. Human neuroblasts migrate to the olfactory bulb via a lateral ventricular extension. Science. 2007;315:1243–1249. doi: 10.1126/science.1136281. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Liebl J, Bernard S, Alkass K, Yeung MSY, Steier P, Kutschera W, Johnson L, Landén M, Druid H, Spalding KL, Frisén J. The age of olfactory bulb neurons in humans. Neuron. 2012;74:634–639. doi: 10.1016/j.neuron.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Macklis JD. Human adult olfactory bulb neurogenesis? Novelty is the best policy. Neuron. 2012;74:595–596. doi: 10.1016/j.neuron.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Zupanc GKH, Hinsch K, Gage FH. Proliferation, migration, neuronal differentiation, and long-term survival of new cells in the adult zebrafish brain. J Comp Neurol. 2005;488:290–319. doi: 10.1002/cne.20571. [DOI] [PubMed] [Google Scholar]
- Adolf B, Chapouton P, Lam CS, Topp S, Tannhäuser B, Strähle U, Götz M, Bally-Cuif L. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev Biol. 2006;295:278–93. doi: 10.1016/j.ydbio.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Grandel H, Kaslin J, Ganz J, Wenzel I, Brand M. Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate. Dev Biol. 2006;295:263–277. doi: 10.1016/j.ydbio.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Zupanc GKH, Zupanc MM. New neurons for the injured brain: mechanisms of neuronal regeneration in adult teleost fish. Regen Med. 2006;1:207–216. doi: 10.2217/17460751.1.2.207. [DOI] [PubMed] [Google Scholar]
- Kaslin J, Ganz J, Geffarth M, Grandel H, Hans S, Brand M. Stem cells in the adult zebrafish cerebellum: initiation and maintenance of a novel stem cell niche. J Neurosci. 2009;29:6142–6153. doi: 10.1523/JNEUROSCI.0072-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- März M, Chapouton P, Diotel N, Vaillant C, Hesl B, Takamiya M, Lam CS, Kah O, Bally-Cuif L, Strähle U. Heterogeneity in progenitor cell subtypes in the ventricular zone of the zebrafish adult telencephalon. Glia. 2010;58:870–888. doi: 10.1002/glia.20971. [DOI] [PubMed] [Google Scholar]
- Hinsch K, Zupanc GKH. Generation and long-term persistence of new neurons in the adult zebrafish brain: a quantitative analysis. Neuroscience. 2007;146:679–696. doi: 10.1016/j.neuroscience.2007.01.071. [DOI] [PubMed] [Google Scholar]
- Doniach T, Musci TJ. Induction of anteroposterior neural pattern in Xenopus: evidence for a quantitative mechanism. Mech Dev. 1995;53:403–413. doi: 10.1016/0925-4773(95)00457-2. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Spemann H, Mangold H. Induction of embryonic primordia by implantation of organizers from a different species. 1923. Int J Dev Biol. 2001;45:13–38. [PubMed] [Google Scholar]
- Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406:74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Rydström A, Trimborn T, Willert K, Nusse R, Jessell TM, Edlund T. The status of Wnt signalling regulates neural and epidermal fates in the chick embryo. Nature. 2001;411:325–330. doi: 10.1038/35077115. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A, Sockanathan S, Pérez L, Rex M, Scotting PJ, Sharpe PT, Lovell-Badge R, Stern CD. Preventing the loss of competence for neural induction: HGF/SF, L5 and Sox-2. Development. 1997;124:1191–1202. doi: 10.1242/dev.124.6.1191. [DOI] [PubMed] [Google Scholar]
- Sasai Y. Identifying the missing links: genes that connect neural induction and primary neurogenesis in vertebrate embryos. Neuron. 1998;21:455–458. doi: 10.1016/S0896-6273(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Hemmati-Brivanlou A. Neural induction. Annu Rev Cell Dev Biol. 1999;15:411–433. doi: 10.1146/annurev.cellbio.15.1.411. [DOI] [PubMed] [Google Scholar]
- Sasai Y, De Robertis EM. Ectodermal patterning in vertebrate embryos. Dev Biol. 1997;182:5–20. doi: 10.1006/dbio.1996.8445. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Lagna G, Suzuki A, Hemmati-Brivanlou A. Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development. 1997;124:3177–3184. doi: 10.1242/dev.124.16.3177. [DOI] [PubMed] [Google Scholar]
- Wessely O, Agius E, Oelgeschläger M, Pera EM, De Robertis EM. Neural induction in the absence of mesoderm: β-catenin-dependent expression of secreted BMP antagonists at the blastula stage in Xenopus. Dev Biol. 2001;234:161–173. doi: 10.1006/dbio.2001.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielen H, Houart C. BMP signaling protects telencephalic fate by repressing eye identity and its Cxcr4-dependent morphogenesis. Dev Cell. 2012;23:812–22. doi: 10.1016/j.devcel.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern CD. Neural induction: 10 years on since the “default model”. Curr Opin Cell Biol. 2006;18:692–697. doi: 10.1016/j.ceb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann’s organizer. Nature. 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Harland RM. Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development. 1995;121:3627–3636. doi: 10.1242/dev.121.11.3627. [DOI] [PubMed] [Google Scholar]
- Alvarez IS, Araujo M, Nieto MA. Neural induction in whole chick embryo cultures by FGF. Dev Biol. 1998;199:42–54. doi: 10.1006/dbio.1998.8903. [DOI] [PubMed] [Google Scholar]
- Storey KG, Goriely A, Sargent CM, Brown JM, Burns HD, Abud HM, Heath JK. Early posterior neural tissue is induced by FGF in the chick embryo. Development. 1998;125:473–484. doi: 10.1242/dev.125.3.473. [DOI] [PubMed] [Google Scholar]
- Rentzsch F, Bakkers J, Kramer C, Hammerschmidt M. Fgf signaling induces posterior neuroectoderm independently of Bmp signaling inhibition. Dev Dyn. 2004;231:750–757. doi: 10.1002/dvdy.20244. [DOI] [PubMed] [Google Scholar]
- Londin ER, Niemiec J, Sirotkin HI. Chordin, FGF signaling, and mesodermal factors cooperate in zebrafish neural induction. Dev Biol. 2005;279:1–19. doi: 10.1016/j.ydbio.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Penzel R, Oschwald R, Chen Y, Tacke L, Grunz H. Characterization and early embryonic expression of a neural specific transcription factor xSOX3 in Xenopus laevis. Int J Dev Biol. 1997;41:667–677. [PubMed] [Google Scholar]
- Rex M, Uwanogho DA, Orme A, Scotting PJ, Sharpe PT. cSox21 exhibits a complex and dynamic pattern of transcription during embryonic development of the chick central nervous system. Mech Dev. 1997;66:39–53. doi: 10.1016/S0925-4773(97)00086-5. [DOI] [PubMed] [Google Scholar]
- Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197–201. doi: 10.1016/S0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Jensen ED, Klymkowsky MW. The β-catenin/VegT-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Development. 2003;130:5609–5624. doi: 10.1242/dev.00798. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Hernandez-Lagunas L, Simpson P, Stemple DL, Artinger KB, Klymkowsky MW. Repression of nodal expression by maternal B1-type SOXs regulates germ layer formation in Xenopus and zebrafish. Dev Biol. 2004;273:23–37. doi: 10.1016/j.ydbio.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Dee CT, Hirst CS, Shih Y-H, Tripathi VB, Patient RK, Scotting PJ. Sox3 regulates both neural fate and differentiation in the zebrafish ectoderm. Dev Biol. 2008;320:289–301. doi: 10.1016/j.ydbio.2008.05.542. [DOI] [PubMed] [Google Scholar]
- Okuda Y, Ogura E, Kondoh H, Kamachi Y. B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo. PloS Genet. 2010;6:e1000936. doi: 10.1371/journal.pgen.1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CS, März M, Strähle U. gfap and nestin reporter lines reveal characteristics of neural progenitors in the adult zebrafish brain. Dev Dyn. 2009;238:475–486. doi: 10.1002/dvdy.21853. [DOI] [PubMed] [Google Scholar]
- Reim G, Brand M. Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development. 2006;133:2757–2770. doi: 10.1242/dev.02391. [DOI] [PubMed] [Google Scholar]
- Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, Smith B, Ribecco-Lutkiewicz M, Kennedy J, Walker PR, Sikorska M. Role of Sox2 in the development of the mouse neocortex. Dev Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Valotta M, Ferri ALM, Latorre E, Mariani J, Giachino C, Lancini C, Tosetti V, Ottolenghi S, Taylor V, Nicolis SK. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12:1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Buckley CE, Ren X, Ward LC, Girdler GC, Araya C, Green MJ, Clark BS, Link BA, Clarke JDW. Mirror-symmetric microtubule assembly and cell interactions drive lumen formation in the zebrafish neural rod. EMBO J. 2012;32:30–44. doi: 10.1038/emboj.2012.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. Role of polarized cell divisions in zebrafish neural tube formation. Curr Opin Neurobiol. 2009;19:134–138. doi: 10.1016/j.conb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk M, Araya C, Lyons DA, Reugels AM, Girdler GC, Bayley PR, Hyde DR, Tada M, Clarke JDW. A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature. 2007;446:797–800. doi: 10.1038/nature05722. [DOI] [PubMed] [Google Scholar]
- Yang X, Zou J, Hyde DR, Davidson LA, Wei X. Stepwise maturation of apicobasal polarity of the neuroepithelium is essential for vertebrate neurulation. J Neurosci. 2009;29:11426–11440. doi: 10.1523/JNEUROSCI.1880-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papan C, Campos-Ortega JA. Region-specific cell clones in the developing spinal cord of the zebrafish. Dev Genes Evol. 1999;209:135–144. doi: 10.1007/s004270050237. [DOI] [PubMed] [Google Scholar]
- Alexandre P, Reugels AM, Barker D, Blanc E, Clarke JDW. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat Neurosci. 2010;13:673–679. doi: 10.1038/nn.2547. [DOI] [PubMed] [Google Scholar]
- Dong Z, Yang N, Yeo S-Y, Chitnis A, Guo S. Intralineage directional Notch signaling regulates self-renewal and differentiation of asymmetrically dividing radial glia. Neuron. 2012;74:65–78. doi: 10.1016/j.neuron.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye LM, Link BA. Nuclear migration during retinal development. Brain Res. 2008;1192:29–36. doi: 10.1016/j.brainres.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel B, Chitnis A. Neurogenesis and specification of neuronal identity. Results Probl Cell Differ. 2002;40:237–251. doi: 10.1007/978-3-540-46041-1_12. [DOI] [PubMed] [Google Scholar]
- Blader P, Fischer N, Gradwohl G, Guillemot F, Strähle U. The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development. 1997;124:4557–4569. doi: 10.1242/dev.124.22.4557. [DOI] [PubMed] [Google Scholar]
- Korzh V, Strähle U. Proneural, prosensory, antiglial: the many faces of neurogenins. Trends Neurosci. 2002;25:603–605. doi: 10.1016/S0166-2236(02)02275-0. [DOI] [PubMed] [Google Scholar]
- Allende ML, Weinberg ES. The expression pattern of two zebrafish achaete-scute homolog (ash) genes is altered in the embryonic brain of the cyclops mutant. Dev Biol. 1994;166:509–530. doi: 10.1006/dbio.1994.1334. [DOI] [PubMed] [Google Scholar]
- Wilson SW, Ross LS, Parrett T, Easter SS. The development of a simple scaffold of axon tracts in the brain of the embryonic zebrafish Brachydanio rerio. Development. 1990;108:121–145. doi: 10.1242/dev.108.1.121. [DOI] [PubMed] [Google Scholar]
- Stigloher C, Chapouton P, Adolf B, Bally-Cuif L. Identification of neural progenitor pools by E(Spl) factors in the embryonic and adult brain. Brain Res Bull. 2008;75:266–273. doi: 10.1016/j.brainresbull.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Tallafuss A, Adolf B, Bally-Cuif L. Selective control of neuronal cluster size at the forebrain/midbrain boundary by signaling from the prechordal plate. Dev Dyn. 2003;227:524–535. doi: 10.1002/dvdy.10329. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Delogu A, Gilthorpe J, Peukert D, Schindler S, Lumsden A. Her6 regulates the neurogenetic gradient and neuronal identity in the thalamus. Proc Natl Acad Sci USA. 2009;106:19895–19900. doi: 10.1073/pnas.0910894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapouton P, Godinho L. Neurogenesis. Methods Cell Biol. 2010;100:73–126. doi: 10.1016/B978-0-12-384892-5.00004-9. [DOI] [PubMed] [Google Scholar]
- Geling A, Itoh M, Tallafuss A, Chapouton P, Tannhäuser B, Kuwada JY, Chitnis AB, Bally-Cuif L. bHLH transcription factor Her5 links patterning to regional inhibition of neurogenesis at the midbrain-hindbrain boundary. Development. 2003;130:1591–1604. doi: 10.1242/dev.00375. [DOI] [PubMed] [Google Scholar]
- Geling A, Plessy C, Rastegar S, Strähle U, Bally-Cuif L. Her5 acts as a prepattern factor that blocks neurogenin1 and coe2 expression upstream of Notch to inhibit neurogenesis at the midbrain-hindbrain boundary. Development. 2004;131:1993–2006. doi: 10.1242/dev.01093. [DOI] [PubMed] [Google Scholar]
- Ninkovic J, Tallafuss A, Leucht C, Topczewski J, Tannhäuser B, Solnica-Krezel L, Bally-Cuif L. Inhibition of neurogenesis at the zebrafish midbrain-hindbrain boundary by the combined and dose-dependent activity of a new hairy/E(spl) gene pair. Development. 2005;132:75–88. doi: 10.1242/dev.01525. [DOI] [PubMed] [Google Scholar]
- Chapouton P, Adolf B, Leucht C, Tannhäuser B, Ryu S, Driever W, Bally-Cuif L. Her5 expression reveals a pool of neural stem cells in the adult zebrafish midbrain. Development. 2006;133:4293–4303. doi: 10.1242/dev.02573. [DOI] [PubMed] [Google Scholar]
- Hans S, Scheer N, Riedl I, Weizsäcker VE, Blader P, Campos-Ortega JA. her3, a zebrafish member of the hairy-E(spl) family, is repressed by Notch signalling. Development. 2004;131:2957–2969. doi: 10.1242/dev.01167. [DOI] [PubMed] [Google Scholar]
- Blader P, Plessy C, Strähle U. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech Dev. 2003;120:211–218. doi: 10.1016/S0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- Haddon C, Smithers L, Schneider-Maunoury S, Coche T, Henrique D, Lewis J. Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development. 1998;125:359–370. doi: 10.1242/dev.125.3.359. [DOI] [PubMed] [Google Scholar]
- Appel B, Givan LA, Eisen JS. Delta-Notch signaling and lateral inhibition in zebrafish spinal cord development. BMC Dev Biol. 2001;1:13. doi: 10.1186/1471-213X-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takke C, Dornseifer P, Weizsäcker VE, Campos-Ortega JA. her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development. 1999;126:1811–1821. doi: 10.1242/dev.126.9.1811. [DOI] [PubMed] [Google Scholar]
- Kroehne V, Freudenreich D, Hans S, Kaslin J, Brand M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development. 2011;138:4831–4841. doi: 10.1242/dev.072587. [DOI] [PubMed] [Google Scholar]
- Chapouton P, Webb KJ, Stigloher C, Alunni A, Adolf B, Hesl B, Topp S, Kremmer E, Bally-Cuif L. Expression of hairy/enhancer of split genes in neural progenitors and neurogenesis domains of the adult zebrafish brain. J Comp Neurol. 2011;519:1748–1769. doi: 10.1002/cne.22599. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Nigtevecht GV. Neural activation and transformation in explants of competent ectoderm under the influence of fragments of anterior notochord in urodeles. J Embryol Exp Morphol. 1954;2:175–193. [Google Scholar]
- Foley AC, Skromne I, Stern CD. Reconciling different models of forebrain induction and patterning: a dual role for the hypoblast. Development. 2000;127:3839–3854. doi: 10.1242/dev.127.17.3839. [DOI] [PubMed] [Google Scholar]
- Wilson SW, Brand M, Eisen JS. Patterning the zebrafish central nervous system. Results Probl Cell Differ. 2002;40:181–215. doi: 10.1007/978-3-540-46041-1_10. [DOI] [PubMed] [Google Scholar]
- Wilson SW, Rubenstein JL. Induction and dorsoventral patterning of the telencephalon. Neuron. 2000;28:641–651. doi: 10.1016/S0896-6273(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, Chandrasekharappa SC, Driever W, Chitnis AB. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature. 2000;407:913–916. doi: 10.1038/35038097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg CP, Brand M, Jiang YJ, Warga RM, Beuchle D, Van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nusslein-Volhard C. Genes involved in forebrain development in the zebrafish Danio rerio. Development. 1996;123:191–203. doi: 10.1242/dev.123.1.191. [DOI] [PubMed] [Google Scholar]
- Heisenberg CP, Houart C, Take-Uchi M, Rauch GJ, Young N, Coutinho P, Masai I, Caneparo L, Concha ML, Geisler R, Dale TC, Wilson SW, Stemple DL. A mutation in the Gsk3-binding domain of zebrafish Masterblind/Axin1 leads to a fate transformation of telencephalon and eyes to diencephalon. Genes Dev. 2001;15:1427–1434. doi: 10.1101/gad.194301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M, Lun K, Luz M, Werner M, Brand M. Positioning of the midbrain-hindbrain boundary organizer through global posteriorization of the neuroectoderm mediated by Wnt8 signaling. Development. 2005;132:1261–1272. doi: 10.1242/dev.01685. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Lun K, Ahrendt R, Geffarth M, Brand M. Zebrafish gbx1 refines the midbrain-hindbrain boundary border and mediates the Wnt8 posteriorization signal. Neural Dev. 2009;4:12. doi: 10.1186/1749-8104-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houart C, Westerfield M, Wilson SW. A small population of anterior cells patterns the forebrain during zebrafish gastrulation. Nature. 1998;391:788–792. doi: 10.1038/35853. [DOI] [PubMed] [Google Scholar]
- Houart C, Caneparo L, Heisenberg C, Barth K, Take-Uchi M, Wilson S. Establishment of the telencephalon during gastrulation by local antagonism of Wnt signaling. Neuron. 2002;35:255–265. doi: 10.1016/S0896-6273(02)00751-1. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Lohs C, Brand M. Engrailed and Fgf8 act synergistically to maintain the boundary between diencephalon and mesencephalon. Development. 2003;130:4881–4893. doi: 10.1242/dev.00683. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Oates AC, Brand M. Boundary formation and maintenance in tissue development. Nat Rev Genet. 2011;12:43–55. doi: 10.1038/nrg2902. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Lumsden A. Building a bridal chamber: development of the thalamus. Trends Neurosci. 2010;33:373–380. doi: 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D, Kobayashi M, Matsumoto K, Ogura T, Nakafuku M, Shimamura K. Early subdivisions in the neural plate define distinct competence for inductive signals. Development. 2002;129:83–93. doi: 10.1242/dev.129.1.83. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat Neurosci. 2004;7:1242–1249. doi: 10.1038/nn1338. [DOI] [PubMed] [Google Scholar]
- Vieira C, Garda A-L, Shimamura K, Martinez S. Thalamic development induced by Shh in the chick embryo. Dev Biol. 2005;284:351–363. doi: 10.1016/j.ydbio.2005.05.031. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Wolf O, Brand M, Lumsden A. Hedgehog signalling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development. 2006;133:855–864. doi: 10.1242/dev.02248. [DOI] [PubMed] [Google Scholar]
- Hagemann AIH, Scholpp S. The tale of the three brothers – Shh, Wnt, and Fgf during development of the thalamus. Front Neurosci. 2012;6:76. doi: 10.3389/fnins.2012.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Dolson DK, Waclaw RR, Matise MP, Sussel L, Campbell K, Kaestner KH, Epstein DJ. Spatial and temporal requirements for sonic hedgehog in the regulation of thalamic interneuron identity. Development. 2011;138:531–541. doi: 10.1242/dev.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2:99–108. doi: 10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- Raible F, Brand M. Divide et impera–the midbrain-hindbrain boundary and its organizer. Trends Neurosci. 2004;27:727–734. doi: 10.1016/j.tins.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Reifers F, Böhli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Lekven AC, Buckles GR, Kostakis N, Moon RT. Wnt1 and wnt10b function redundantly at the zebrafish midbrain-hindbrain boundary. Dev Biol. 2003;254:172–187. doi: 10.1016/S0012-1606(02)00044-1. [DOI] [PubMed] [Google Scholar]
- Mattes B, Weber S, Peres J, Chen Q, Davidson G, Houart C, Scholpp S. Wnt3 and Wnt3a are required for induction of the mid-diencephalic organizer in the caudal forebrain. Neural Dev. 2012;7:12. doi: 10.1186/1749-8104-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckles GR, Thorpe CJ, Ramel M-C, Lekven AC. Combinatorial Wnt control of zebrafish midbrain-hindbrain boundary formation. Mech Dev. 2004;121:437–447. doi: 10.1016/j.mod.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Salzberg A, Bellen HJ. Invertebrate versus vertebrate neurogenesis: variations on the same theme? Dev Genet. 1996;18:1–10. doi: 10.1002/(SICI)1520-6408(1996)18:1<1::AID-DVG1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Altman J. Development of the preoptic area: time and site of origin, migratory routes, and settling patterns of its neurons. J Comp Neurol. 1987;265:65–95. doi: 10.1002/cne.902650106. [DOI] [PubMed] [Google Scholar]
- Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, Wallace VA. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184:101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Coolen M, Thieffry D, Drivenes O, Becker TS, Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev Cell. 2012;22:1052–1064. doi: 10.1016/j.devcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Guy AT, Clarke JDW. Monitoring neural progenitor fate through multiple rounds of division in an intact vertebrate brain. Development. 2003;130:3427–3436. doi: 10.1242/dev.00569. [DOI] [PubMed] [Google Scholar]
- Peukert D, Weber S, Lumsden A, Scholpp S. Lhx2 and Lhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating Wnt signaling. PloS Biol. 2011;9:e1001218. doi: 10.1371/journal.pbio.1001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullimann MF, Mueller T. Teleostean and mammalian forebrains contrasted: Evidence from genes to behavior. J Comp Neurol. 2004;475:143–162. doi: 10.1002/cne.20183. [DOI] [PubMed] [Google Scholar]
- Broglio C, Gómez A, Durán E, Ocaña FM, Jiménez-Moya F, Rodríguez F, Salas C. Hallmarks of a common forebrain vertebrate plan: specialized pallial areas for spatial, temporal and emotional memory in actinopterygian fish. Brain Res Bull. 2005;66:277–281. doi: 10.1016/j.brainresbull.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF. An evolutionary interpretation of teleostean forebrain anatomy. Brain Behav Evol. 2009;74:30–42. doi: 10.1159/000229011. [DOI] [PubMed] [Google Scholar]
- Folgueira M, Bayley P, Navratilova P, Becker TS, Wilson SW, Clarke JD. Morphogenesis underlying the development of the everted teleost telencephalon. Neural Dev. 2012;7:32. doi: 10.1186/1749-8104-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey BW, Darabie A, Tropepe V. The cellular composition of neurogenic periventricular zones in the adult zebrafish forebrain. J Comp Neurol. 2012;520:2275–2316. doi: 10.1002/cne.23065. [DOI] [PubMed] [Google Scholar]
- Morshead CM, Reynolds BA, Craig CG, McBurney MW, Staines WA, Morassutti D, Weiss S, Van der Kooy D. Neural stem cells in the adult mammalian forebrain: a relatively quiescent subpopulation of subependymal cells. Neuron. 1994;13:1071–1082. doi: 10.1016/0896-6273(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisén J. Identification of a neural stem cell in the adult mammalian central nervous system. Cell. 1999;96:25–34. doi: 10.1016/S0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- Seaberg RM, Van der Kooy D. Adult rodent neurogenic regions: the ventricular subependyma contains neural stem cells, but the dentate gyrus contains restricted progenitors. J Neurosci. 2002;22:1784–1793. doi: 10.1523/JNEUROSCI.22-05-01784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Kang E, Liu CY, Ming G-L, Song H. Development of neural stem cell in the adult brain. Curr Opin Neurobiol. 2008;18:108–115. doi: 10.1016/j.conb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Verdugo JM, Ferrón S, Flames N, Collado L, Desfilis E, Font E. The proliferative ventricular zone in adult vertebrates: a comparative study using reptiles, birds, and mammals. Brain Res Bull. 2002;57:765–775. doi: 10.1016/S0361-9230(01)00769-9. [DOI] [PubMed] [Google Scholar]
- Ganz J, Kaslin J, Hochmann S, Freudenreich D, Brand M. Heterogeneity and Fgf dependence of adult neural progenitors in the zebrafish telencephalon. Glia. 2010;58:1345–1363. doi: 10.1002/glia.21012. [DOI] [PubMed] [Google Scholar]
- Kishimoto N, Alfaro-Cervello C, Shimizu K, Asakawa K, Urasaki A, Nonaka S, Kawakami K, Garcia-Verdugo JM, Sawamoto K. Migration of neuronal precursors from the telencephalic ventricular zone into the olfactory bulb in adult zebrafish. J Comp Neurol. 2011;519:3549–3565. doi: 10.1002/cne.22722. [DOI] [PubMed] [Google Scholar]
- Byrd CA, Brunjes PC. Addition of new cells to the olfactory bulb of adult zebrafish. Ann NY Acad Sci. 1998;855:274–276. doi: 10.1111/j.1749-6632.1998.tb10582.x. [DOI] [PubMed] [Google Scholar]
- Byrd CA, Brunjes PC. Neurogenesis in the olfactory bulb of adult zebrafish. Neuroscience. 2001;105:793–801. doi: 10.1016/S0306-4522(01)00215-9. [DOI] [PubMed] [Google Scholar]
- März M, Schmidt R, Rastegar S, Strähle U. Expression of the transcription factor Olig2 in proliferating cells in the adult zebrafish telencephalon. Dev Dyn. 2010;239:3336–3349. doi: 10.1002/dvdy.22455. [DOI] [PubMed] [Google Scholar]
- Rothenaigner I, Krecsmarik M, Hayes JA, Bahn B, Lepier A, Fortin G, Götz M, Jagasia R, Bally-Cuif L. Clonal analysis by distinct viral vectors identifies bona fide neural stem cells in the adult zebrafish telencephalon and characterizes their division properties and fate. Development. 2011;138:1459–1469. doi: 10.1242/dev.058156. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, García-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Garcia-Verdugo JM. Neurogenesis in adult subventricular zone. J Neurosci. 2002;22:629–634. doi: 10.1523/JNEUROSCI.22-03-00629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F. The glial identity of neural stem cells. Nat Neurosci. 2003;6:1127–1134. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- Zupanc GKH, Clint SC, Takimoto N, Hughes ATL, Wellbrock UM, Meissner D. Spatio-temporal distribution of microglia/macrophages during regeneration in the cerebellum of adult teleost fish, Apteronotus leptorhynchus: a quantitative analysis. Brain Behav Evol. 2003;62:31–42. doi: 10.1159/000071958. [DOI] [PubMed] [Google Scholar]
- Pellegrini E, Mouriec K, Anglade I, Menuet A, Page YLE, Gueguen M, Marmignon NE. Identification of aromatase-positive radial glial cells as progenitor cells in the ventricular layer of the forebrain in zebrafish. J Comp Neurol. 2007;167:150–167. doi: 10.1002/cne.21222. [DOI] [PubMed] [Google Scholar]
- Chapouton P, Skupien P, Hesl B, Coolen M, Moore JC, Madelaine R, Kremmer E, Faus-Kessler T, Blader P, Lawson ND, Bally-Cuif L. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J Neurosci. 2010;30:7961–7974. doi: 10.1523/JNEUROSCI.6170-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kim C-H, Palardy G, Oda T, Jiang Y-J, Maust D, Yeo S-Y, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/S1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 2001;4:143–150. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- Marcus RC, Delaney CL, Easter SS. Neurogenesis in the visual system of embryonic and adult zebrafish (Danio rerio) Vis Neurosci. 1999;16:417–424. doi: 10.1017/S095252389916303X. [DOI] [PubMed] [Google Scholar]
- Kaslin J, Ganz J, Brand M. Proliferation, neurogenesis and regeneration in the non-mammalian vertebrate brain. Philos Trans R Soc Lond B Biol Sci. 2008;363:101–122. doi: 10.1098/rstb.2006.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Tanaka H, Okamoto H, Ohshima T. Characterization of neural stem cells and their progeny in the adult zebrafish optic tectum. Dev Biol. 2010;342:26–38. doi: 10.1016/j.ydbio.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Bravo R, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri ALM, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, Nicolis SK. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci. 2000;22:139–153. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- Oteíza P, Köppen M, Concha ML, Heisenberg C-P. Origin and shaping of the laterality organ in zebrafish. Development. 2008;135:2807–2813. doi: 10.1242/dev.022228. [DOI] [PubMed] [Google Scholar]
- Higashijima S-I, Mandel G, Fetcho JR. Distribution of prospective glutamatergic, glycinergic, and GABAergic neurons in embryonic and larval zebrafish. J Comp Neurol. 2004;480:1–18. doi: 10.1002/cne.20278. [DOI] [PubMed] [Google Scholar]
- Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF. BrdU-, neuroD (nrd)- and Hu-studies reveal unusual non-ventricular neurogenesis in the postembryonic zebrafish forebrain. Mech Dev. 2002;117:123–135. doi: 10.1016/S0925-4773(02)00194-6. [DOI] [PubMed] [Google Scholar]
- Arvidsson A, Collin T, Kirik D, Kokaia Z, Lindvall O. Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat Med. 2002;8:963–970. doi: 10.1038/nm747. [DOI] [PubMed] [Google Scholar]
- Chen J, Magavi SSP, Macklis JD. Neurogenesis of corticospinal motor neurons extending spinal projections in adult mice. Proc Natl Acad Sci USA. 2004;101:16357–16362. doi: 10.1073/pnas.0406795101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep CQ, Ma D, Deo RC, Holm TM, Naylor RW, Arora N, Wingert RA, Bollig F, Djordjevic G, Lichman B, Zhu H, Ikenaga T, Ono F, Englert C, Cowan CA, Hukriede NA, Handin RI, Davidson AJ. Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature. 2011;470:95–100. doi: 10.1038/nature09669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. 2012;22:879–886. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien C-L, Harrison MR, Tuan T-L, Starnes VA. Heart repair and regeneration: recent insights from zebrafish studies. Wound Repair Regen. 2012;20:638–646. doi: 10.1111/j.1524-475X.2012.00814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscha V, Frazer SL, Dias TB, Hibi M, Becker T, Becker CG. Lesion-induced generation of interneuron cell types in specific dorsoventral domains in the spinal cord of adult zebrafish. J Comp Neurol. 2012;520:3604–3616. doi: 10.1002/cne.23115. [DOI] [PubMed] [Google Scholar]
- Becker CG, Becker T. Adult zebrafish as a model for successful central nervous system regeneration. Restor Neurol Neurosci. 2008;26:71–80. [PubMed] [Google Scholar]
- Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. J Comp Neurol. 1997;377:577–595. doi: 10.1002/(SICI)1096-9861(19970127)377:4<577::AID-CNE8>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Cameron DA. Cellular proliferation and neurogenesis in the injured retina of adult zebrafish. Vis Neurosci. 2000;17:789–797. doi: 10.1017/S0952523800175121. [DOI] [PubMed] [Google Scholar]
- Reimer MM, Sörensen I, Kuscha V, Frank RE, Liu C, Becker CG, Becker T. Motor neuron regeneration in adult zebrafish. J Neurosci. 2008;28:8510–8516. doi: 10.1523/JNEUROSCI.1189-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsche W. Regenerative processes in the brain and spinal cord. Ergeb Anat Entwicklungsgesch. 1965;38:143–194. [PubMed] [Google Scholar]
- Zupanc GK, Ott R. Cell proliferation after lesions in the cerebellum of adult teleost fish: time course, origin, and type of new cells produced. Exp Neurol. 1999;160:78–87. doi: 10.1006/exnr.1999.7182. [DOI] [PubMed] [Google Scholar]
- Clint SC, Zupanc GK. Neuronal regeneration in the cerebellum of adult teleost fish, Apteronotus leptorhynchus: guidance of migrating young cells by radial glia. Brain Res Dev Brain Res. 2001;130:15–23. doi: 10.1016/S0165-3806(01)00193-6. [DOI] [PubMed] [Google Scholar]
- Zupanc GKH. Adult neurogenesis and neuronal regeneration in the brain of teleost fish. J Physiol Paris. 2008;102:357–373. doi: 10.1016/j.jphysparis.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Ayari B, El Hachimi KH, Yanicostas C, Landoulsi A, Soussi-Yanicostas N. Prokineticin 2 expression is associated with neural repair of injured adult zebrafish telencephalon. 2010. 2010;27:959–972. doi: 10.1089/neu.2009.0972. [DOI] [PubMed] [Google Scholar]
- März M, Schmidt R, Rastegar S, Strähle U. Regenerative response following stab injury in the adult zebrafish telencephalon. Dev Dyn. 2011;240:2221–2231. doi: 10.1002/dvdy.22710. [DOI] [PubMed] [Google Scholar]
- Baumgart EV, Barbosa JS, Bally-Cuif L, Götz M, Ninkovic J. Stab wound injury of the zebrafish telencephalon: a model for comparative analysis of reactive gliosis. Glia. 2012;60:343–357. doi: 10.1002/glia.22269. [DOI] [PubMed] [Google Scholar]
- Kizil C, Dudczig S, Kyritsis N, Machate A, Blaesche J, Kroehne V, Brand M. The chemokine receptor cxcr5 regulates the regenerative neurogenesis response in the adult zebrafish brain. Neural Dev. 2012;7:27. doi: 10.1186/1749-8104-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32:638–647. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizil C, Kyritsis N, Dudczig S, Kroehne V, Freudenreich D, Kaslin J, Brand M. Regenerative neurogenesis from neural progenitor cells requires injury-induced expression of Gata3. Dev Cell. 2012;23:1230–1237. doi: 10.1016/j.devcel.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Strähle U, Schmidt R. A universal program for tissue regeneration? Dev Cell. 2012;23:1123–1124. doi: 10.1016/j.devcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Zupanc GK, Kompass KS, Horschke I, Ott R, Schwarz H. Apoptosis after injuries in the cerebellum of adult teleost fish. Exp Neurol. 1998;152:221–30. doi: 10.1006/exnr.1998.6853. [DOI] [PubMed] [Google Scholar]
- Vajda FJE. Neuroprotection and neurodegenerative disease. J Clin Neurosci. 2002;9:4–8. doi: 10.1054/jocn.2001.1027. [DOI] [PubMed] [Google Scholar]
- Liou AKF, Clark RS, Henshall DC, Yin X-M, Chen J. To die or not to die for neurons in ischemia, traumatic brain injury and epilepsy: a review on the stress-activated signaling pathways and apoptotic pathways. Prog Neurobiol. 2003;69:103–142. doi: 10.1016/S0301-0082(03)00005-4. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Krebs CJ, Guth L. Experimental analysis of progressive necrosis after spinal cord trauma in the rat: etiological role of the inflammatory response. Exp Neurol. 1997;143:141–152. doi: 10.1006/exnr.1996.6355. [DOI] [PubMed] [Google Scholar]
- Buffo A, Vosko MR, Ertürk D, Hamann GF, Jucker M, Rowitch D, Götz M. Expression pattern of the transcription factor Olig2 in response to brain injuries: implications for neuronal repair. Proc Natl Acad Sci U S A. 2005;102:18183–18188. doi: 10.1073/pnas.0506535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyritsis N, Kizil C, Zocher S, Kroehne V, Kaslin J, Freudenreich D, Iltzsche A, Brand M. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science. 2012;459:921–923. doi: 10.1126/science.1228773. [DOI] [PubMed] [Google Scholar]
- Wullimann MF, Rupp B, Reichert H. Neuroanatomy of the Zebrafish brain: a topological Atlas. Boston: Birkhauser; 1996. pp. 1–144. [Google Scholar]