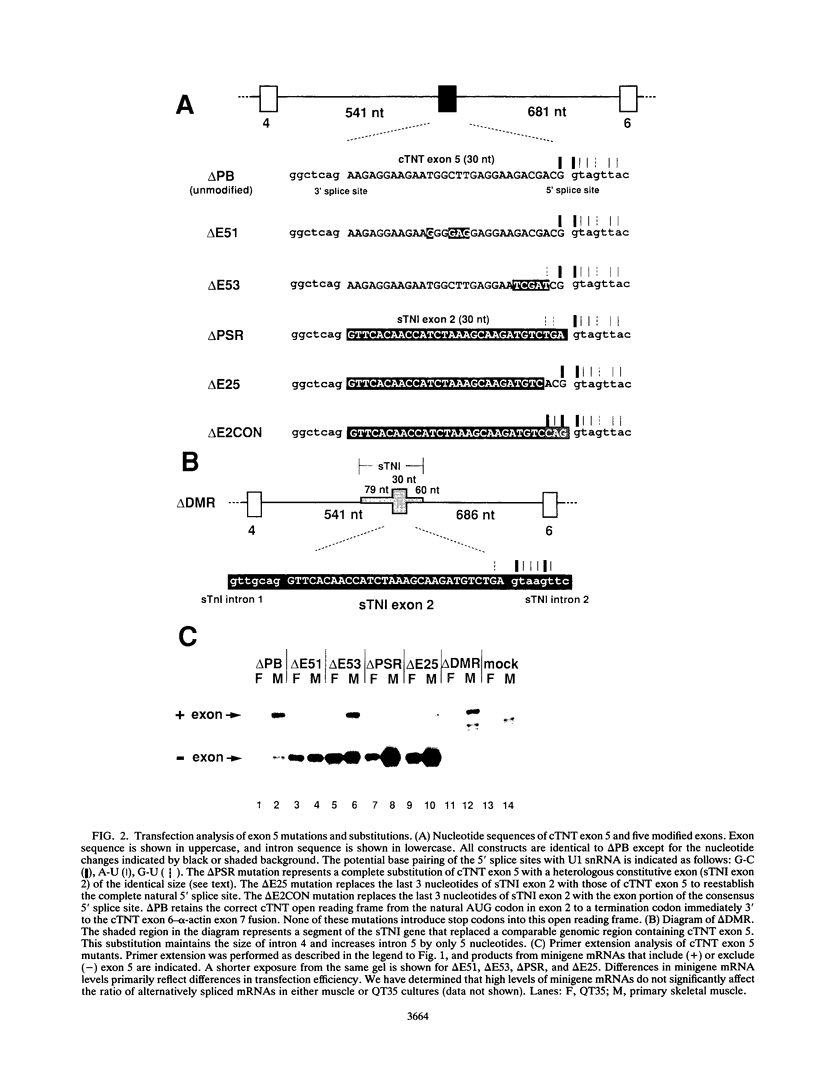
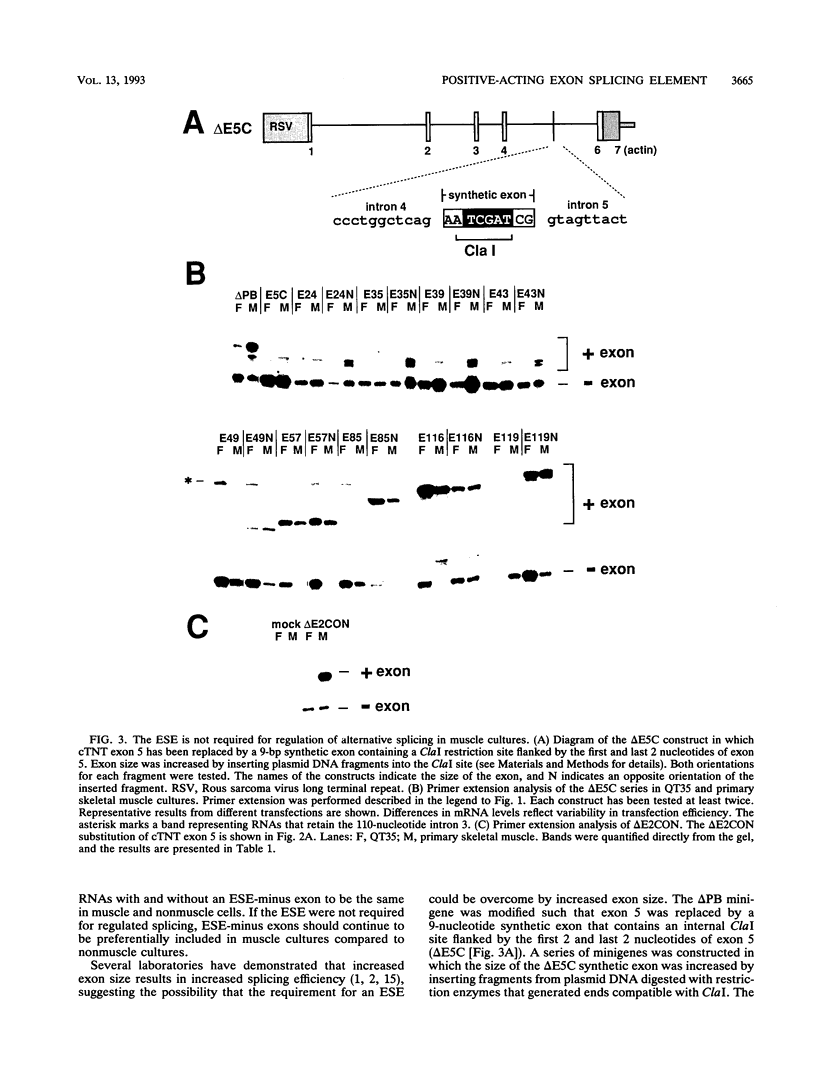
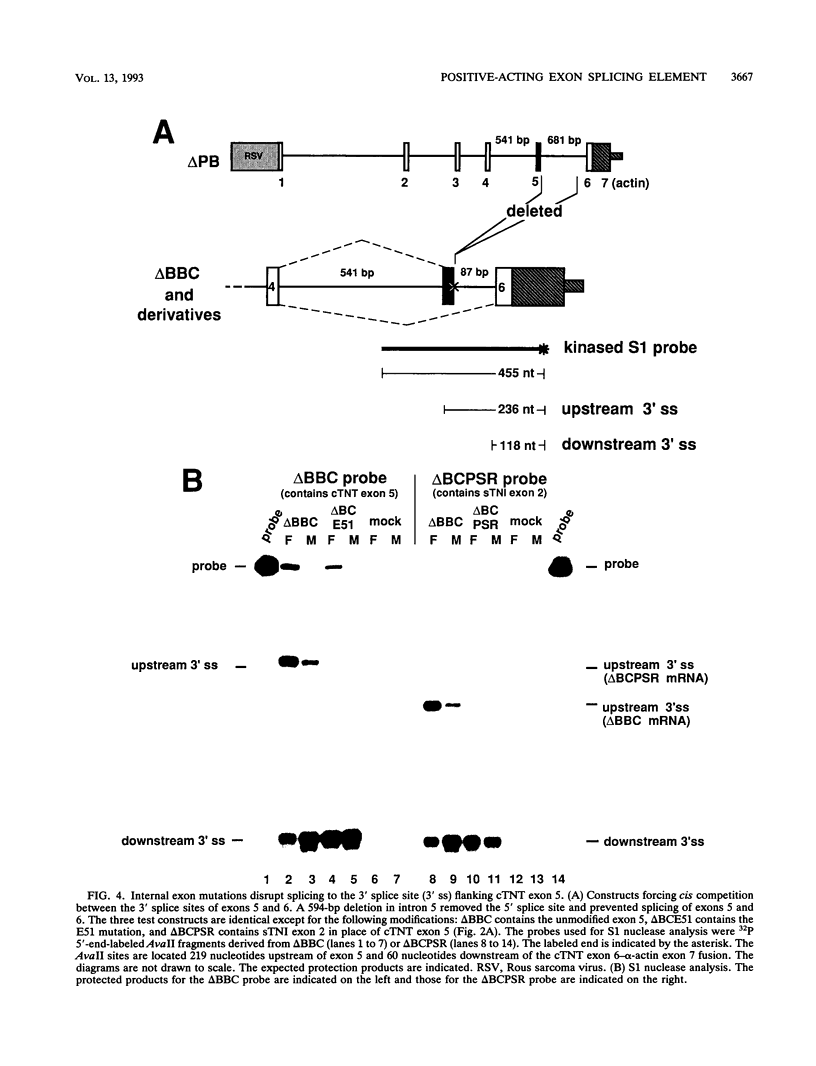
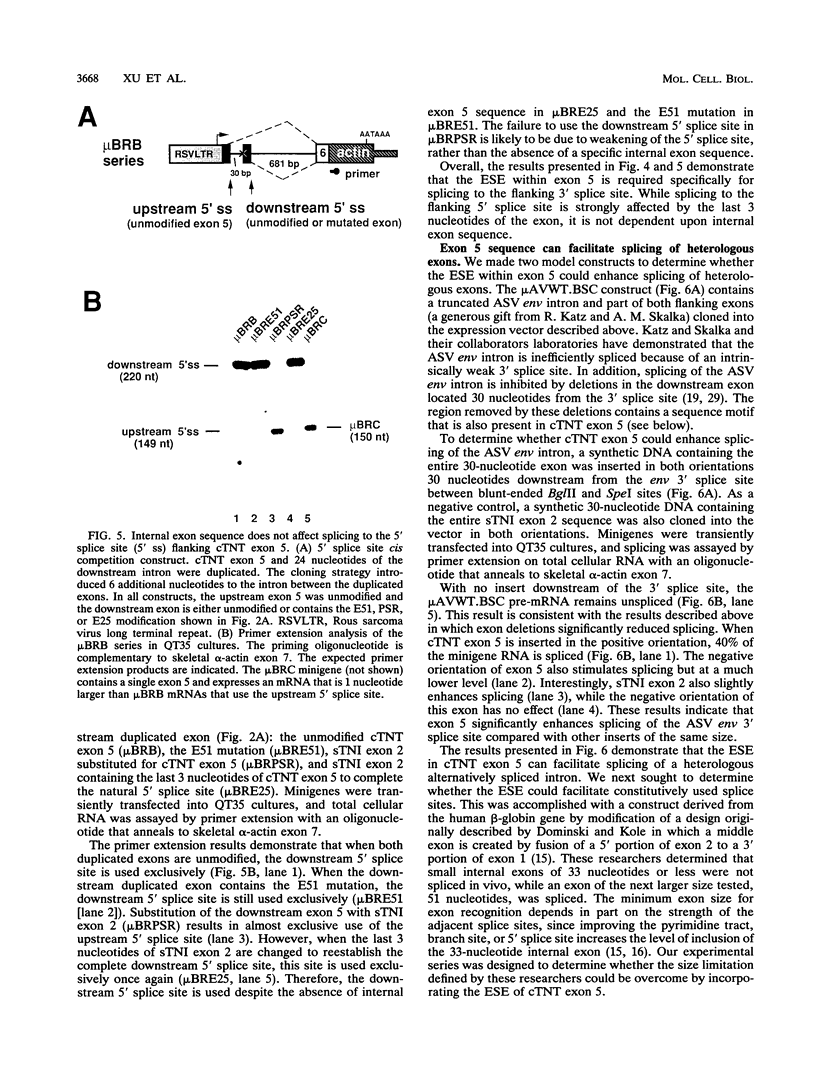
Abstract
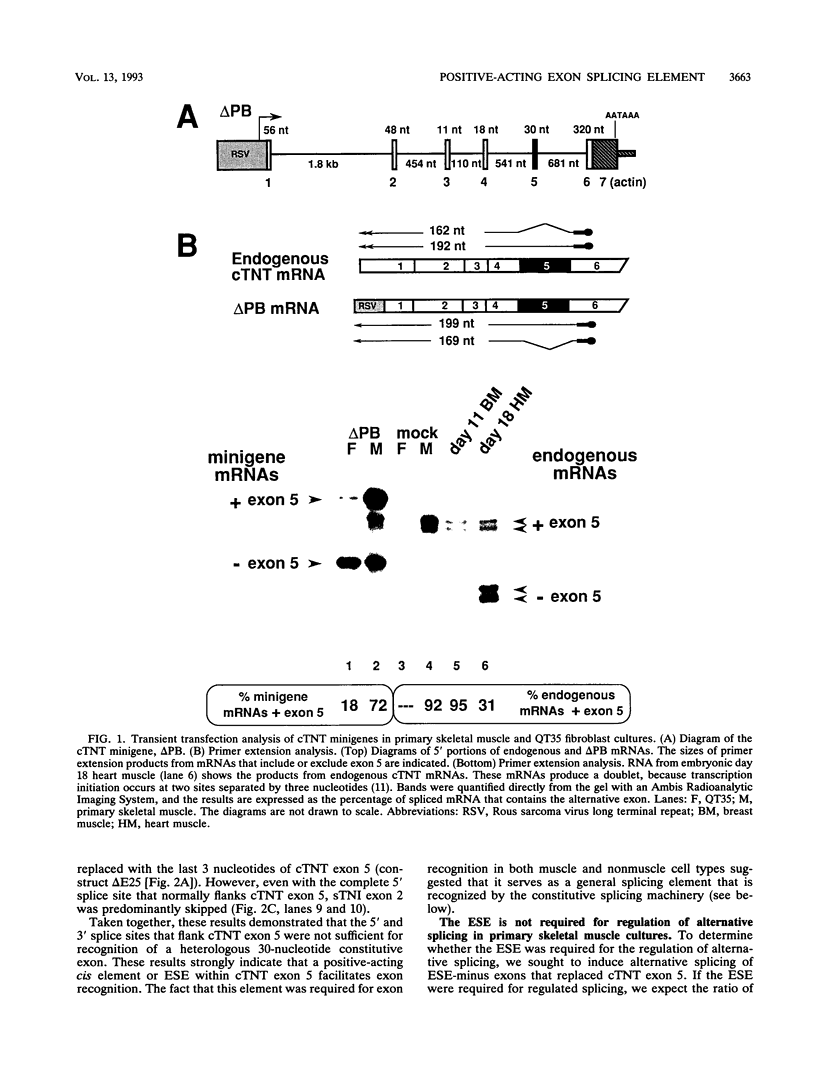
We have characterized a novel positive-acting splicing element within the developmentally regulated alternative exon (exon 5) of the cardiac troponin T (cTNT) gene. The exon splicing element (ESE) is internal to the exon portions of the splice sites and is required for splicing to the 3' splice site but not the 5' splice site flanking the exon. Sequence comparisons between cTNT exon 5 and other exons that contain regions required for splicing reveal a common purine-rich motif. Sequence within cTNT exon 5 or a synthetic purine-rich motif facilitates splicing of heterologous alternative and constitutive splice sites in vivo. Interestingly, the ESE is not required for the preferential inclusion of cTNT exon 5 observed in primary skeletal muscle cultures. Our results strongly suggest that the purine-rich ESE serves as a general splicing element that is recognized by the constitutive splicing machinery.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G. R., Carmichael G. G. The length but not the sequence of the polyoma virus late leader exon is important for both late RNA splicing and stability. Nucleic Acids Res. 1987 Mar 25;15(6):2593–2610. doi: 10.1093/nar/15.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 1991 Mar;5(3):389–402. doi: 10.1101/gad.5.3.389. [DOI] [PubMed] [Google Scholar]
- Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991 Jul 5;220(1):49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Chain A. C., Zollman S., Tseng J. C., Laski F. A. Identification of a cis-acting sequence required for germ line-specific splicing of the P element ORF2-ORF3 intron. Mol Cell Biol. 1991 Mar;11(3):1538–1546. doi: 10.1128/mcb.11.3.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Fogel-Petrovic M., Maquat L. E. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol Cell Biol. 1990 Oct;10(10):5215–5225. doi: 10.1128/mcb.10.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Cardone M. H., Ordahl C. P. Cis requirements for alternative splicing of the cardiac troponin T pre-mRNA. Nucleic Acids Res. 1988 Sep 12;16(17):8443–8465. doi: 10.1093/nar/16.17.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. A. In vitro splicing of cardiac troponin T precursors. Exon mutations disrupt splicing of the upstream intron. J Biol Chem. 1992 Mar 15;267(8):5330–5338. [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J Biol Chem. 1985 Sep 15;260(20):11140–11148. [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. A single troponin T gene regulated by different programs in cardiac and skeletal muscle development. Science. 1984 Nov 23;226(4677):979–982. doi: 10.1126/science.6095446. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989 Oct 11;17(19):7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote G. J., Stolow D. T., Peleg S., Berget S. M., Gagel R. F. Identification of exon sequences and an exon binding protein involved in alternative RNA splicing of calcitonin/CGRP. Nucleic Acids Res. 1992 May 11;20(9):2361–2366. doi: 10.1093/nar/20.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenjoud L., Gallinaro H., Kister L., Meyer S., Jacob M. Identification of a specific exon sequence that is a major determinant in the selection between a natural and a cryptic 5' splice site. Mol Cell Biol. 1991 Sep;11(9):4581–4590. doi: 10.1128/mcb.11.9.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Cooperation of pre-mRNA sequence elements in splice site selection. Mol Cell Biol. 1992 May;12(5):2108–2114. doi: 10.1128/mcb.12.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991 Dec;11(12):6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Fornwald J. A., Kuncio G., Peng I., Ordahl C. P. The complete nucleotide sequence of the chick a-actin gene and its evolutionary relationship to the actin gene family. Nucleic Acids Res. 1982 Jul 10;10(13):3861–3876. doi: 10.1093/nar/10.13.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. D., Katz R. A., Skalka A. M., Maniatis T. The role of branchpoint and 3'-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991 Feb;5(2):211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- Ge H., Noble J., Colgan J., Manley J. L. Polyoma virus small tumor antigen pre-mRNA splicing requires cooperation between two 3' splice sites. Proc Natl Acad Sci U S A. 1990 May;87(9):3338–3342. doi: 10.1073/pnas.87.9.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandchamp B., Picat C., de Rooij F., Beaumont C., Wilson P., Deybach J. C., Nordmann Y. A point mutation G----A in exon 12 of the porphobilinogen deaminase gene results in exon skipping and is responsible for acute intermittent porphyria. Nucleic Acids Res. 1989 Aug 25;17(16):6637–6649. doi: 10.1093/nar/17.16.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley M. L., Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell. 1991 May 17;65(4):579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M., Finn L. A. Alternative splicing of tropomyosin pre-mRNAs in vitro and in vivo. Genes Dev. 1988 Dec;2(12A):1627–1638. doi: 10.1101/gad.2.12a.1627. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991 May 10;252(5007):833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizuka A., Ingi T., Murai T., Nakanishi S. A set of U1 snRNA-complementary sequences involved in governing alternative RNA splicing of the kininogen genes. J Biol Chem. 1990 Jun 15;265(17):10102–10108. [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990 Feb;10(2):696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreivi J. P., Zerivitz K., Zefrivitz K., Akusjärvi G. A U1 snRNA binding site improves the efficiency of in vitro pre-mRNA splicing. Nucleic Acids Res. 1991 Dec 25;19(24):6956–6956. doi: 10.1093/nar/19.24.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. A., Horvath C. M. Diversity of coding strategies in influenza viruses. Trends Genet. 1991 Aug;7(8):261–266. doi: 10.1016/0168-9525(91)90326-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear A. L., Eperon L. P., Wheatley I. M., Eperon I. C. Hierarchy for 5' splice site preference determined in vivo. J Mol Biol. 1990 Jan 5;211(1):103–115. doi: 10.1016/0022-2836(90)90014-D. [DOI] [PubMed] [Google Scholar]
- Libri D., Goux-Pelletan M., Brody E., Fiszman M. Y. Exon as well as intron sequences are cis-regulating elements for the mutually exclusive alternative splicing of the beta tropomyosin gene. Mol Cell Biol. 1990 Oct;10(10):5036–5046. doi: 10.1128/mcb.10.10.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar J. H., Antin P. B., Cooper T. A., Ordahl C. P. Analysis of the upstream regions governing expression of the chicken cardiac troponin T gene in embryonic cardiac and skeletal muscle cells. J Cell Biol. 1988 Aug;107(2):573–585. doi: 10.1083/jcb.107.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M., Masumura T., Nishio H., Nakajima T., Kitoh Y., Takumi T., Koga J., Nakamura H. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy kobe. J Clin Invest. 1991 Jun;87(6):2127–2131. doi: 10.1172/JCI115244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo I., Holmes E. W. Exon recognition and nucleocytoplasmic partitioning determine AMPD1 alternative transcript production. Mol Cell Biol. 1991 Oct;11(10):5356–5363. doi: 10.1128/mcb.11.10.5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Urlaub G., Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986 Jun;6(6):1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger L. K., Schoborg R. V., Zhao Q., Tullis G. E., Pintel D. J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992 Jun;6(6):1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Nagoshi R. N., Baker B. S. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990 Jan;4(1):89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- Nasim F. H., Spears P. A., Hoffmann H. M., Kuo H. C., Grabowski P. J. A Sequential splicing mechanism promotes selection of an optimal exon by repositioning a downstream 5' splice site in preprotachykinin pre-mRNA. Genes Dev. 1990 Jul;4(7):1172–1184. doi: 10.1101/gad.4.7.1172. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988 Mar;2(3):319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- Nikovits W., Jr, Kuncio G., Ordahl C. P. The chicken fast skeletal troponin I gene: exon organization and sequence. Nucleic Acids Res. 1986 Apr 25;14(8):3377–3390. doi: 10.1093/nar/14.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Recio L., Simpson D., Cochrane J., Liber H., Skopek T. R. Molecular analysis of hprt mutants induced by 2-cyanoethylene oxide in human lymphoblastoid cells. Mutat Res. 1990 Nov;242(3):195–208. doi: 10.1016/0165-1218(90)90085-g. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Siebel C. W., Rio D. C. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990 Jun 8;248(4960):1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsdottir H., Rowley G., Dorado G., Cole J., Lehmann A. R. Mutations which alter splicing in the human hypoxanthine-guanine phosphoribosyltransferase gene. Nucleic Acids Res. 1992 Mar 25;20(6):1201–1208. doi: 10.1093/nar/20.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohman R. C., Bayne E., Spector D., Obinata T., Micou-Eastwood J., Maniotis A. Myogenesis and histogenesis of skeletal muscle on flexible membranes in vitro. In Vitro Cell Dev Biol. 1990 Feb;26(2):201–208. doi: 10.1007/BF02624113. [DOI] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992 Apr 10;256(5054):237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- Tsai A. Y., Streuli M., Saito H. Integrity of the exon 6 sequence is essential for tissue-specific alternative splicing of human leukocyte common antigen pre-mRNA. Mol Cell Biol. 1989 Oct;9(10):4550–4555. doi: 10.1128/mcb.9.10.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellard M., Sureau A., Soret J., Martinerie C., Perbal B. A potential splicing factor is encoded by the opposite strand of the trans-spliced c-myb exon. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2511–2515. doi: 10.1073/pnas.89.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaud M., Gattoni R., Stevenin J., Vidaud D., Amselem S., Chibani J., Rosa J., Goossens M. A 5' splice-region G----C mutation in exon 1 of the human beta-globin gene inhibits pre-mRNA splicing: a mechanism for beta+-thalassemia. Proc Natl Acad Sci U S A. 1989 Feb;86(3):1041–1045. doi: 10.1073/pnas.86.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu N., Kobayashi H., Miyatake T., Tsuji S. A novel exon mutation in the human beta-hexosaminidase beta subunit gene affects 3' splice site selection. J Biol Chem. 1992 Feb 5;267(4):2406–2413. [PubMed] [Google Scholar]
- Watakabe A., Tanaka K., Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993 Mar;7(3):407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Weil D., D'Alessio M., Ramirez F., de Wet W., Cole W. G., Chan D., Bateman J. F. A base substitution in the exon of a collagen gene causes alternative splicing and generates a structurally abnormal polypeptide in a patient with Ehlers-Danlos syndrome type VII. EMBO J. 1989 Jun;8(6):1705–1710. doi: 10.1002/j.1460-2075.1989.tb03562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. H., Vrieling H., van Zeeland A. A., Jenssen D. Spectrum of spontaneously occurring mutations in the hprt gene of V79 Chinese hamster cells. J Mol Biol. 1992 Feb 5;223(3):627–635. doi: 10.1016/0022-2836(92)90979-t. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. The conserved dinucleotide AG of the 3' splice site may be recognized twice during in vitro splicing of mammalian mRNA precursors. Gene. 1990 Jun 15;90(2):263–269. doi: 10.1016/0378-1119(90)90189-x. [DOI] [PubMed] [Google Scholar]