Abstract
Because rapamycin, an inhibitor of the nutrient sensor mammalian target of rapamycin, and dietary restriction both increase life span of mice, it has been hypothesized that they act through similar mechanisms. To test this hypothesis, we compared various biological parameters in dietary restriction mice (40% food restriction) and mice fed rapamycin (14 ppm). Both treatments led to a significant reduction in mammalian target of rapamycin signaling and a corresponding increase in autophagy. However, we observed striking differences in fat mass, insulin sensitivity, and expression of cell cycle and sirtuin genes in mice fed rapamycin compared with dietary restriction. Thus, although both treatments lead to significant downregulation of mammalian target of rapamycin signaling, these two manipulations have quite different effects on other physiological functions suggesting that they might increase life span through a common pathway as well as pathways that are altered differently by dietary restriction and rapamycin.
Keywords: Rapamycin, Dietary restriction, mTOR, Autophagy, Gene expression
Recently, it has been shown that rapamycin (Rapa) results in a significant extension of both median and maximal life span in mice (1,2). Rapa is an antifungal drug that was first discovered in the soil of Easter Island. Brown and coworkers (3) found that rapamycin inhibited mammalian cell cycle progression and interfered with the activation of S6K1. The signaling protein found to be directly inhibited by Rapa was thus named mammalian target of rapamycin (mTOR). mTOR forms two major complexes: mTORC1, which has been widely studied in regards to Rapa and is Rapa sensitive (4), and mTORC2, which has been shown to be Rapa insensitive in short-term Rapa treatment, but recent data suggest that it may be affected by long-term Rapa treatment (5). mTORC1, which consists of mTOR, Raptor, mLST8, FKBP38, PRAS40, and Deptor, is a downstream effector for many inputs, such as nutrients, growth factors, and cytokines (4). Activation of mTORC1 results in increased protein synthesis (through activation of S6 kinase [S6K] and eIF4-BP), a reduction in autophagy, and an increase cell growth. Through specific binding of Rapa to the immunophilin, FKBP12, Rapa-FKBP12 complex inhibits the activity of mTOR by directly binding to the FKBP12-rapamycin–binding domain in mTOR, which is adjacent to the kinase domain of mTOR and disrupting the interaction of mTOR and Raptor (6). Inhibition of mTORC1 by Rapa in turn decreases protein synthesis, increases autophagy, and promotes inhibition of cell growth (7).
Because mTOR is involved in nutrient sensing, it has been argued that Rapa and dietary restriction (DR) act through similar mechanisms (8). Using epistasis analysis, several laboratories have studied whether Rapa and DR extend life span through a similar mechanism(s) in invertebrates. Kaeberlein's laboratory showed that in Saccharomyces cerevisiae, mutation of TOR (tor1Δ) results in an extension of replicative life span similar to what is observed in wild-type S. cerevisiae on a DR diet (low sugar). Interestingly, placing the tor1Δ mutants on a DR diet produces no further extension in replicative life span, suggesting that DR and tor1Δ share a common mechanism(s) to extend yeast life span (9). Kenyon's laboratory found that reduction of TOR by RNAi increased both the mean and maximum life span of Caenorhabditis elegans worms over the N2 background (10) and that TOR RNAi does not further extend the life span of eat-2 worms, one of the DR models in C. elegans. In contrast to the observations in yeast and C. elegans, Partridge’s laboratory (11) reported that Drosophila on DR-fed Rapa had a further life-span extension beyond that observed with flies on a DR diet alone (protein restriction by decrease of yeast concentration). These results suggest that in Drosophila, DR and Rapa extend life span at least partially through different mechanisms.
As a first step to determine whether Rapa and DR increase life span in mice through similar or different pathways, we compared the effect of short-term DR and Rapa feeding on several pathways that have been proposed to be involved in enhanced longevity in mice. Our data showed that although DR and Rapa have a similar effect on mTOR signaling and autophagy, other processes such as insulin signaling, glutathione (GSH) redox status, and messenger RNA (mRNA) levels of cell cycling and sirtuin genes differ significantly in these two experimental manipulations.
METHODS
Animals and Feeding Regiment
Male C57BL/6 mice used in this study were purchased from The Jackson Labs (Bar Harbor, ME). The mice were fed a mouse chow, 7012 Teklad LM-450 (Harlan Laboratories, Madison, WI) until 2 months of age, after which, they were divided into three dietary regimens: ad libitum (AL), where mice were maintained on a commercial mouse chow, Purina Mills Test Diet Control #1810306 (Purina Mills, St Louis, MO); 40% diet restriction (DR), where the mice were fed 60% of the amount of Purina Mills chow eaten by the ad-libitum fed mice at 3 PM each day. Mice given Rapa were fed ad libitum the Purina Mills chow containing 14 ppm of encapsulated rapamycin in the diet as described by Harrison and coworkers (1). Mice were maintained on the three diets for 6 months, until they were 8 months of age. All procedures followed the guidelines approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio. The body weights and body composition (determined by quantitative magnetic resonance [Echo Medial Systems, Houston, TX]) were measured in the groups of mice at 2 (before dietary regimens) and 8 months of age. Mice were euthanized by carbon dioxide and liver tissues were harvested, snap frozen in liquid nitrogen, and stored at −80°C until used.
Glucose Tolerance Tests and Insulin Tolerance Tests
Both the glucose tolerance test (GTT) and insulin tolerance test (ITT) assays were performed following 16 hours of food deprivation in 8-month-old mice. Food was removed from the AL and Rapa mice immediately after the DR mice had eaten their daily allotment. GTTs were performed with an intraperitoneal injection of glucose (1.5 g/kg body weight) solution in saline into the mice, and blood glucose was measured 0, 15, 30, 60, and 120 minutes after glucose injection. ITTs were performed with an intraperitoneal injection of human recombinant insulin in saline (Novalin; Novo Nordisk) at a dose of 0.75 U/kg body weight, and blood glucose was measured 0, 15, 30, and 60 minutes after insulin injection. Blood glucose concentrations were measured using a One-Touch Ultra glucometer. Area under the curve (4) for GTT and ITT was estimated by summing the numerical integration values of successive linear segments of the glucose disposal curve at each set of time points. The data were expressed as mean ± standard error of the mean and analyzed using one-way analysis of variance with the Holm–Sidak post hoc test.
mTOR Signaling and Autophagy Activity
Frozen liver tissue was homogenized with a Dounce homogenizer in ice-cold RadioImmunoPrecipitation Assay buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) supplemented with inhibitors of protease and phosphatase (Roche, Indiana) on ice. The supernatant was collected after centrifugation at 4°C, 12,000g for 10 minutes. Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel followed by transfer to nitrocellulose membrane. Target proteins were detected with the following specific monoclonal or polyclonal antibodies: actin, S6, phospho-S6 (Ser235/236), and LC3 (Cell Signaling, Danvers, MA). mTOR signaling was assessed using the ratio of phosopho-s6 levels to total S6 levels. Autophagy activity was measured using the ratio of LC3II/LC3I, and actin was used as a loading control for both. Data were expressed as means ± standard deviation (SD).
GSH and Thioredoxin Redox State
The GSH redox state was determined by measuring the levels of reduced and oxidized GSH in liver collected from the mice. Fifty milligrams of frozen liver was homogenized in 0.5 mL of 5% PCA/0.2 M Boric Acid/10 μM r-EE solution provided by Clinical Biomarkers Lab (Jones Lab, Emory University). Samples were sonicated to breakdown any aggregates and were then spun down and 300 μL of supernatant were transferred to a new tube. Samples were stored at −80°C and until assayed. The levels of reduced and oxidized GSH were determined using N-dansyl derivatives and high-performance liquid chromatography with fluorescence detection as described by Jones and coworkers (12).
The Thioredoxin1 (Trx1) redox state status was determined in liver tissue by measuring the levels of reduced and oxidized Trx1 with modifications to the methods described in (13,14). Briefly, frozen liver tissue was homogenized in 20 mM Tris pH 8.0, containing 15 mM 4-acetoamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS; Molecular Probes, Eugene, OR), supplemented with protease cocktail inhibitor III (Calbiochem, La Jolla, CA). Cytosolic fractions were obtained from homogenates by centrifugation at 16,000g for 15 minutes at 4°C. The cytosolic fractions (1 mg/mL) were then incubated in the dark with 15 mM of AMS in 20 mM Tris, pH.8.0 for 3 hours at room temperature. Excess AMS was removed using Microcon YM-3 (Millipore Corporation, Billerica, MA). Reduced and oxidized Trx1 were then separated on a sodium dodecyl sulfate/15% polyacrylamide gel (Bio-Rad, Hercules, CA) under nonreducing conditions. The gel was transferred onto a polyvinylidene fluoride membrane, and proteins were then detected with a specific Trx1 polyclonal antibody obtained from LabFrontier (Seoul, Korea). The intensities of the bands corresponding to the reduced Trx1 (top band) and the oxidized Trx1 (bottom band) were calculated using ImageQuant 5.1 (Molecular Dynamics, Amersham) software.
Quantitative Real-Time PCR
Total RNA was extracted from frozen liver tissues (25 mg) using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions, and DNA contamination was removed by Turbo free DNase (Ambion Foster City, CA). The RNA yield of each sample was determined spectrophotometrically, assuming that 1 optical density at 260 nm (OD260) unit = 40 mg/L. The quality of total RNA extracted from each sample was monitored by A260:A280 ratio and 1.0% agarose formaldehyde gel electrophoresis.
One microgram of RNA was used to generate complementary DNA using the Retroscript kit (Ambion). All complementary DNAs were diluted to 1, 1:10, 1:100 before being used as a PCR template. Primers were designed using Primer Express (Applied Biosystem, Foster City, CA) and Primer-BLAST (NCBI). Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green PCR Master Mix (Applied Biosystem) in a 96-well plate using Gapdh as a housekeeping control with detection by a 7500 Real-Time PCR Detection System (Applied Biosystem). The primer pairs used are described in Supplementary Table 3. Analysis of the qRT-PCR results was done using the ΔΔCT method, and gene products were assayed using agarose gels and dissociation curves.
Statistical Analysis
Unless specified, all data were expressed as mean ± standard error of the mean and were analyzed by one-way analysis of variance with pair-wise comparisons using Turkey’s post hoc test. Statistical significance is indicated by p less than .05.
RESULTS
We first compared the effect of Rapa and DR on body weight/composition because it has been shown that DR results in reduced body size and fat content in rats and mice (15) and that Rapa has an adverse effect on body weight in young rats (16). As shown in Figure 1A, mice fed AL showed approximately 50% increase in body weight between 2 and 8 months of age. Mice fed Rapa showed comparable increase in body weight to the AL mice; however, DR mice showed less than a 5% increase in body weight from 2 to 8 months of age. Thus, at 8 months of age, the DR mice had a 34% lower gain in body weight compared with AL and 29.5% decrease compared with Rapa-fed mice. Using quantitative magnetic resonance imaging, we observed a major decrease in fat content when measured relative to body weight in the DR mice and no change between the AL- and Rapa-fed mice in fat content (Figure 1B). This decrease in fat content is correlated to a decrease in the eipdidymal, perirenal, and mesenteric fat depots in the DR mice (Supplementary Figure 1A); no difference was observed in the fat depots of AL and Rapa mice. No significant differences were observed in overall lean mass (everything except fat) relative to body weight among AL-, DR-, and Rapa-treated mice (Figure 1C). Besides fat, we observed differences in liver, heart, brain, lungs, and hindlimb skeletal muscles relative to body weight among AL, DR, and Rapa mice (Supplementary Figure 1B and C). DR mice showed a decrease in the liver weight relative to body weight compared with AL and Rapa but an increase in the brain weight relative to body weight compared with AL and Rapa. DR mice also showed an increase in the heart weight relative to body weight compared with Rapa and an increase in the lungs weight relative to body weight compared with AL (Supplementary Figure 1B). In the hindlimb skeletal muscles, DR showed an increase in gastrocnemius, tibialis, and quadriceps weights relative to body weight compared with AL and Rapa.
Figure 1.
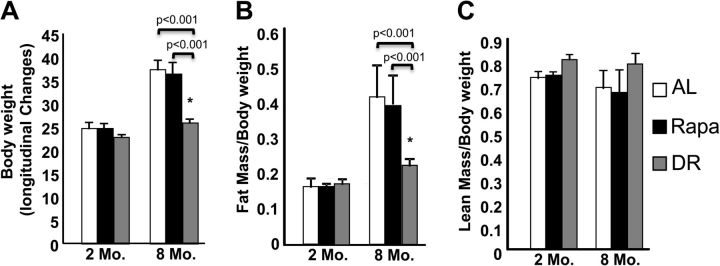
Effect of dietary restriction (DR) and rapamycin (Rapa) on body weight and body composition. Changes in body weight (A), fat mass (B), and lean mass (C) were determined in ad libitum (AL; open bars), Rapa (solid bars), and DR (gray bars) fed mice at 2 months of age (before treatment) and at 8 months of age (after 6 months of treatment). The data were obtained from 11 to 12 mice per group and expressed as mean ± standard error of the mean. Data were analyzed using one-way analysis of variance with the Turkey’s post hoc test; an asterisk denotes those values that are significantly different (p ≤ .05) from AL mice. Specific p values are denoted in the figure.
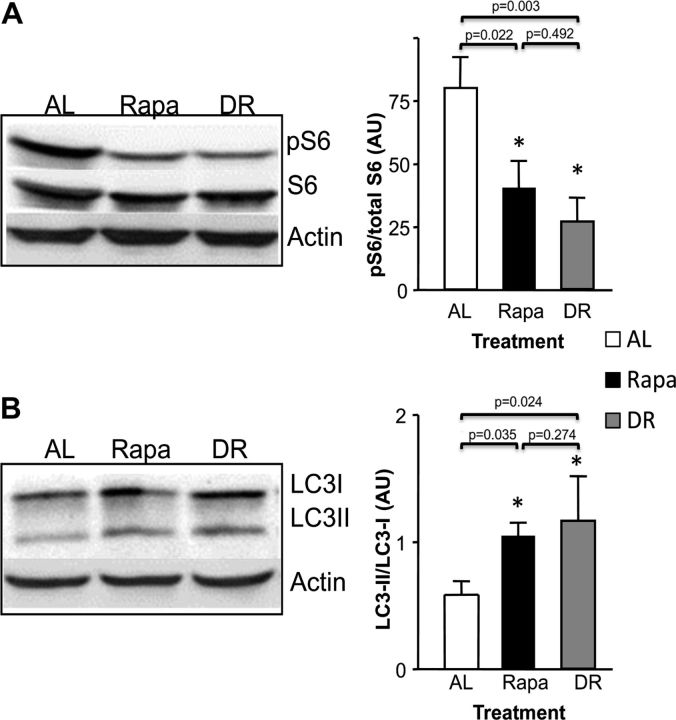
It is well known that Rapa inhibits mTOR, which is the master regulator of the nutrient sensing pathways and responds to many factors such as amino acid, glucose, growth factor, cytokines, and insulin (17). Because Rapa is known to be a specific inhibitor of mTOR signaling, we determined whether DR and Rapa had similar effects on the mTOR signaling pathway by measuring the levels of phosphorylated ribosomal protein S6 (p-S6) protein, one of the downstream targets of the mTORC1 complex. Phosphorylation of ribosomal S6 is important in the regulation of protein synthesis. The data in Figure 2A show the phosphorylation of S6 is reduced approximately 50% in mice fed Rapa. These data are consistent with previous studies showing that feeding mice a Rapa diet reduces mTOR signaling, as measured by pS6 phosphorylation (1). In addition, the data in Figure 2A show that mice on DR for 6 months also show a decrease in S6 phosphorylation that is similar in magnitude to that observed in mice fed Rapa for 6 months. Because a decrease in mTOR activity has been linked to an increase in autophagy (18), we also compared the level of autophagy in the livers of AL-, DR-, and Rapa-fed mice by measuring the ratio of LC3II to LC3I. LC3II is the cleaved form of LC3I that is present in the formation of the autophagosome; therefore, the ratio between LC3II/LC3I is a marker widely used to measure autophagy (19). The data in Figure 2B show that in both, DR- and Rapa-fed mice, autophagy is increased approximately twofold as compared with AL mice.
Figure 2.
Effect of dietary restriction (DR) and rapamycin (Rapa) on mammalian target of rapamycin (mTOR) signaling and autophagy. mTOR pathway was assessed in liver samples by measuring levels of phosphorylated S6 respect to the total S6 (A) and autophagy by measuring levels of LC3II and LC3I (LC3II/LC3I ratio; B). A representative Western blot of pooled samples for pS6/S6 and LC3II/LC3I measurements is showed next to the respective quantification. The data were obtained from 11 to 12 mice per group: ad libitum (AL; open bars), Rapa (solid bars), and DR (gray bars). The data were expressed as mean ± standard deviation and analyzed using one-way analysis of variance with the Turkey’s post hoc test; an asterisk denotes those values that are significantly different from AL mice at the p ≤ .05 level. Specific p values are denoted in the figure.
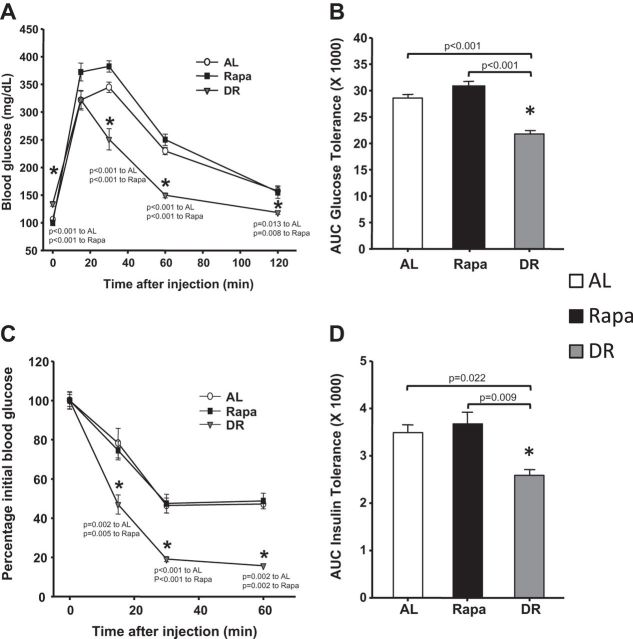
Because DR has been shown to increase insulin sensitivity in rats and mice (15), we measured glucose and insulin tolerance in mice fed with Rapa and with DR diets for 6 months. The data in Figure 3A and B show that the clearance of glucose (measured by the rate of glucose disappearance or the area under the curve) after an injection of glucose is significantly greater in DR mice compared with AL mice and that glucose clearance in mice fed Rapa is not significantly different than AL mice. We also measured insulin sensitivity by the ITT. As shown in Figure 3C and D, the decrease in blood glucose in response to an injection of insulin is significantly greater in the DR mice compared with AL mice. However, we found no significance difference in the blood glucose levels at each time point (Figure 3C) or area under the curve (AUC insulin tolerance; Figure 3D) between AL- and Rapa-fed mice. These results indicate that Rapa-fed mice have similar insulin sensitivity as the AL mice in contrast to mice on DR, which improves glucose uptake from the blood stream by increasing the insulin sensitivity.
Figure 3.
Effect of dietary restriction (DR) and rapamycin (Rapa) on glucose and insulin tolerance test. The glucose tolerance test (GTT; A) and insulin tolerance test (ITT; C) were performed as described in the Methods section. The area under the curves for GTT (B) and for ITT (D) was estimated by summing the numerical integration values of successive linear segments of each time point, respectively. The data were obtained from 10 mice per group for ad libitum (AL; open symbols/bars), Rapa (solid symbols/bars), and DR (gray symbols/bars) mice. The data were expressed as mean ± standard error of the mean and analyzed using one-way analysis of variance with the Holm–Sidak post hoc test; an asterisk denotes those values that are significantly different (p ≤ .05) from AL mice. Specific p values are denoted in the figure.
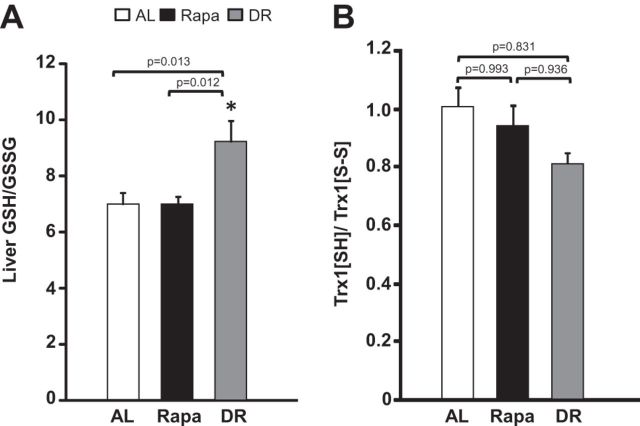
DR has been shown to increase the resistance of animals to oxidative stress (20) by altering the GSH redox state, that is the ratio of reduced to oxidized GSH (21) is increased by DR in rats (22) and mice (23,24). There have been no previous studies on the effect of Rapa in mice on GSH redox state; therefore, we measured the levels of reduced and oxidized GSH in the livers of mice on the DR and Rapa diets. The data in Figure 4A show that the ratio of reduced/oxidized GSH is significantly higher (approximately 30%) for DR mice compared with AL mice; however, mice fed Rapa showed no significant change in the reduced/oxidized GSH ratio compared with the AL mice. Because the redox state in cells is made up of two components, the GSH redox couple and the thioredoxin1 (Trx1) redox couple, we also measured the ratio of reduced/oxidized Trx1 in the livers of mice on DR and Rapa diets. We found that neither DR nor Rapa treatment significantly altered the ratio of reduced/oxidized Trx1 (Figure 4B).
Figure 4.
Effect of dietary restriction and rapamycin on the glutathione (GSH) and Thioredoxin1 (Trx1) redox states. The ratio of reduced GSH respect to oxidized GSH for liver and plasma is shown in Graph A, and the ratio of reduced and oxidized TRX1 in liver is shown in Graph B for ad libitum (AL; open bars), Rapa (solid bars), and dietary restriction (DR; gray bars) mice. The data were obtained from 10 mice per group and expressed as mean ± standard error of the mean. Data were analyzed using one-way analysis of variance with the Turkey’s post hoc test; an asterisk denotes those values that are significantly different (p ≤ .05) from AL mice. Specific p values are denoted in the figure.
Figure 5.
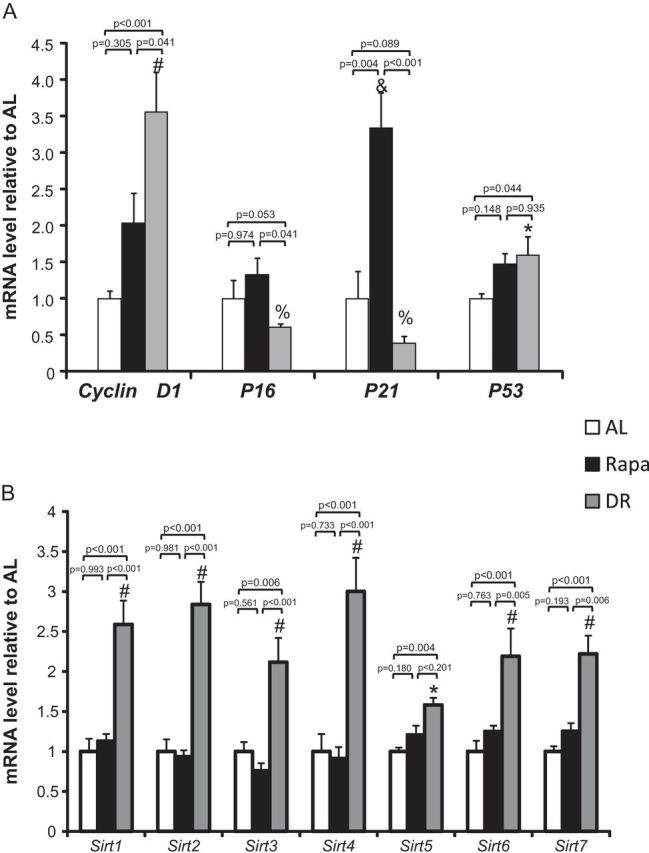
Effect of dietary restriction (DR) and rapamycin (Rapa) on gene expression. Gene expression of cell cycle genes (A) and sirtuin genes (B) were determined in the livers of ad libitum (AL; open bars), Rapa (solid bars), and DR (gray bars) mice. Gene expression was determined by quantitative RT-PCR on representative cell cycle genes: cyclin d1, p16, p21, and p53 and all seven sirtuin genes: Sirt1, Sirt2, Sirt3, Sirt4, Sirt5, Sirt6, and Sirt7. The data were obtained from 10 mice per group and expressed as mean ± standard error of the mean. Data were analyzed using one-way analysis of variance with the Turkey’s post hoc test; an asterisk denotes those values that are significantly different (p ≤ .05) from AL mice, a number sign denotes those values that are significantly different (p ≤ .05) from AL and Rapa mice, an ampersand sign denotes those values that are significantly different (p ≤ .05) from AL and DR mice, and a percent sign denotes those values that are significantly different (p ≤ .05) from Rapa mice. Specific p values are denoted in the figure.
We also compared the effect of Rapa and DR on gene expression by measuring the transcripts of genes involved in the cell cycle. The expression of genes important in the regulation of cell cycle checkpoints and initiation of the cell cycle, such as Cyclin D1, P16, P21, and P53, was measured by qRT-PCR in the liver tissue of AL, DR, and Rapa mice (Figure 5A). Cyclin D1 was found to be significantly upregulated (3.5-fold) by DR (p < .001); we also observed a twofold increase in Cyclin D1 mRNA levels in mice fed Rapa; however, this increase did not reach statistical significance (p = .305). P16 mRNA levels were reduced (50%) by DR but did not reach statistical significance (p = .053) and were not change by Rapa treatment (p = 0.974). P21 mRNA levels were decreased (60%) by DR but did not reach statistical significance (p = .089); however, liver tissue from mice fed Rapa showed a dramatic 3.5-fold increase in P21 mRNA levels compared with AL mice (p = .004). P53 levels were upregulated by DR.
The family of Sirtuin (Sirt) genes code for proteins involved of in a variety of cellular functions ranging from genome maintenance to metabolism (25), and Sir2, the homolog of Sirt1 in invertebrates, has been shown to increase life span of invertebrates (26). The levels of the transcripts for the seven Sirt genes in liver tissue from DR- and Rapa-fed mice compared with levels in AL mice are shown in Figure 5B. Sirt1, the best studied of the sirtuins, is involved in metabolism and stress response and is localized to the nucleus (27). Sirt1 was upregulated by DR but not by Rapa. Sirt2 is involved in cell cycle regulation in G2/M transition and is localized to the cytosol. Again, we observed an increase in Sirt2 by DR but not by Rapa. Sirt3, 4, and 5 are localized to the mitochondria. In mice on DR, it has been observed that the urea cycle and fatty oxidation are increased by Sirt3 (28). We observed an increase in Sirt3 in mice on DR but not in mice fed Rapa. We observed that DR, but not Rapa, upregulates Sirt4, an ADP-ribosyltransferase that is involved in insulin secretion (29). Sirt5 is also involved in the regulation of the urea cycle via interactions with carbamoyl phosphate synthease1, which catalyzes the first step in the urea cycle (30). We observed that Sirt5 was upregulated by mice on DR and not Rapa. Sirt6 is localized to the nucleus and is involved in DNA damage repair (31) and has been suggested to play a role in inflammation (32). We observed an upregulation of Sirt6 in DR but not in Rapa-fed mice. Finally, Sirt7, which is involved in ribosomal DNA transcription (33) and is localized to the nucleus, is upregulated under DR and not Rapa. Thus, DR upregulates the expression of all seven of the sirtuin genes in the liver, whereas feeding Rapa had no significant effect on the levels of these transcripts.
DISCUSSION
As a first attempt to determine whether Rapa and DR affect similar pathways, we compared the effect of DR and Rapa on various pathways that have been proposed to play a role in aging. Starting at 2 months of age, 6 months of feeding Rapa or DR has a similar effect on the mTOR pathway in liver; mTOR signaling (measured by phosphorylation of ribosomal protein S6) was reduced approximately 50% (p = .022 Rapa compared with AL and p = .003 DR compared with AL), and autophagy (measured by the LC3II/LC3I ratio), a downstream pathway regulated by mTOR, was increased twofold (p = .035 Rapa compared with AL and p = .024 DR compared with AL). Thus, both DR and Rapa alter the major nutrient sensing system in liver to the same extent. Because the liver is the first organ to receive the nutrients absorbed by the intestine (34), these data indicate that at 14 ppm Rapa is equivalent to 40% DR with respect to the organism’s mTOR pathway and nutrient response system.
Although both DR and Rapa reduced nutrient sensing to a similar extent, we found that these two manipulations had quite different effects on the other parameters studied. First, as has consistently been observed, DR resulted in smaller body weight gains compared with AL mice; in fact, there was very little growth of the DR mice (<5%) over the 6-month study period. In addition to reduced weight gain, DR also altered body composition; it resulted in a major decrease in adiposity, which was observed in epididymal, perirenal, and mesenteric fat depots. In contrast, Rapa had no effect on either the weight of the mice or body fat relative to body weight compared with AL mice. We were somewhat surprised that the Rapa did not have any effect on growth (increase in body weight) of the mice from 2 to 8 months of age because one of the hallmark characteristics of Rapa is the inhibition of growth in cells. Rapa has inhibitory effects on cell proliferation in many cancer cell lines such as B-cell chronic lymphocytic leukemia (21) and cervical and ovarian cancer cell lines (35). Rapa has been reported to reduce bone growth and weight gain in rats over a period of 2 weeks when Rapa is given by gavage (2.5 mg/kg daily) starting at 3 weeks of age (16). It is possible that the lack of an effect of Rapa on growth in our study of mice compared with previous work is due to the age of the animals used (8 weeks vs 3 weeks for the rat study) or the route of administration, daily gavage versus feeding microencapsulated Rapa.
One of the most striking differences between DR and Rapa observed in our study was the effect on insulin sensitivity as measured by glucose and insulin tolerance. Previous studies have shown that DR increases insulin sensitivity in mice (36) and rats (37). In contrast to AL, Rapa tended to reduce insulin sensitivity (though not significantly), whereas DR significantly increased insulin sensitivity (p < .001 GTT DR compared with AL and p = .022 ITT DR compared with AL). Fraenkel and coworkers (38) previously reported that normal and diabetic rats given Rapa (0.2 mg/kg) for 2 days had reduced insulin tolerance, suggesting that Rapa might lead to insulin resistance. Because DR and other long-lived mouse models (Ames dwarf and the growth hormone receptor knockout mice) exhibit an increase in insulin sensitivity, it has been hypothesized that insulin sensitivity plays a major role in life-span extension (39). However, the observation that Rapa does not increase insulin sensitivity (and may decrease insulin sensitivity, as shown in rats) demonstrates that the increase in life span of Rapa-fed mice is achieved without an increase in insulin sensitivity.
One of the most well-studied theories of aging is the oxidative stress theory, and data from DR have been important in supporting this theory (20). DR mice and rats show reduced oxidative damage to various macromolecules (40) and have a reduced cellular environment as shown by the GSH/GSSG ratio (23). Rapa has been shown to decrease production of reactive oxygen species in hepatocytes of rats (41). After only 6 months on a DR diet, DR-fed mice increased the GSH/GSSG ratio, whereas feeding Rapa had no significant effect. The increase in GSH/GSSG ratio indicates a reduced GSH redox state produced by DR. We also observed no significant changes in the ratio or reduced Trx1 to oxidized Trx1 in either DR- or Rapa-treated mice, which indicates that there were no changes to the Trx1 redox state. These data would suggest that Rapa might not reduce oxidative damage over the life span of the mice as has been shown for DR because a more reduced GSH redox state is correlated to decrease oxidative damage (42,43).
To obtain a broader perspective on the pathways/processes altered by DR and Rapa, we studied the effect of DR and Rapa on the expression of genes involved in the cell cycle and the sirtuin family. DR has been shown to decrease cell proliferation and growth, whereas Rapa has been shown to decrease proliferation in cancer cells. The effect of DR and Rapa on cell cycle gene expression varied depending on the transcript studied, for example, they had a similar effect (upregulation) on mRNAs for cyclin D1and p53; however, only DR samples reach statistical significance. For other side, the effect on mRNAs for p16 and p21 were in the opposite direction. The differences in the effect of DR and Rapa on p21 were particularly striking; p21 mRNA levels were increased 3.5-fold by Rapa and reduced approximately threefold in DR. We also observed that the level of p21 protein was significantly increased in the liver of the mice fed Rapa compared with the DR and AL mice. Our data agree with previous reports in lymphocytes where Rapa treatment increased p21 protein levels (44), and this may explain how Rapa treatment induces specific G1 cell cycle arrest in many cells (42,43).
The sirtuins play an important role in the regulation of a variety of pathways, ranging from maintenance of genome integrity to metabolism. Of particular, importance to this study are the reports that overexpression of Sir2 extends life span in invertebrates (45). However, recent studies call these data into question (46). There are seven sirtuins in the mammals of which Sirt1 is the Sir2 homolog in yeast. The current data indicate that the protein levels of many of the sirtuins are altered by DR in tissues of rats and mice, for example, Sirt1 (47) and Sirt3 protein levels increased significantly in the liver (28), Sirt2 protein levels increased significantly in white adipose tissue and kidney but not the liver and brain (48), Sirt4 protein levels appear to be reduced but no calculation was done to show significance or magnitude of the decrease (49), Sirt5 protein levels were unchanged (30), and Sirt6 protein levels increased in the heart, brain, and white adipose tissue (50). Our data show that DR upregulates (two- to threefold for all genes except Sirt5, which was increased approximately 50%) the levels of the mRNA transcript of all seven sirtuin genes in liver. In contrast, feeding Rapa between 2 and 8 months of age had no significant effect on the levels of any of the sirtuin transcripts in the liver.
From an in vivo point of view, rapamycin and DR also have common and different effects, for example, both of them have protective effects against age-related diseases, such as cancer, Alzheimer, and Parkinson diseases, and they also shared side effects, such as wound healing problems and immunosuppressant properties [reviewed in (51,52)]. In this regards, it is important to consider that in clinical treatment rapamycin is not used alone, and therefore, some of these immunosuppressant effects may be attributed to interactions with other drugs. Recent studies performed in mouse models indicate that rapamycin acts as an immune modulator rather than an immune suppressant; for example, in mouse studies, rapamycin enhances the efficacy of vaccination and inhibits HIV infection (53,54). However, in contrast to DR, rapamycin treatment extends life span in mice even when this treatment was started late in life (52). However, whether rapamycin can be used safety in human aging due to the side effect profile of rapamycin treatment is a major question. For example, rapamycin has been associated with the development of lung toxicity in patients, which can be treated clinically (55).
In summary, although 40% DR and 14 ppm Rapa have a similar effect on mTOR signaling in C57BL/6J male mice, their effects on several pathways believed to play a role in aging are quite different. Although these data do not allow us to determine definitively whether DR and Rapa could increase life span through different pathways, these data are the first to show that these two manipulations have different effects on a variety of pathways that have been correlated to increased longevity in mice. Based on these data, we propose that it is likely that DR and Rapa affect life span at least partially through different mechanisms in mice. These data are important because they suggest that a combination of DR and Rapa could lead to an increase in life span in mice even greater that that found in DR alone.
FUNDING
Financial support was provided by The San Antonio Nathan Shock Aging Center (1P30-AG-13319 to A.R.), National Institut es of Health (NIH) RC2 Grand Opportunity grant (AG036613 to A.R.), NIH T32 Training Grant (AG021890 to W.C.F.), and The Ellison Medical Foundation (to V.I.P.).
SUPPLEMENTARY MATERIAL
Supplementary material can be found at: http://biomedgerontology.oxfordjournals.org/.
Acknowledgments
We thank Vivian Diaz and the department of Laboratory Animal Resources at University Health Science Center at San Antonio for mice husbandry and Elizabeth Fernandez.
References
- 1.Harrison DE, Strong R, Sharp ZD, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller RA, Harrison DE, Astle CM, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown EJ, Albers MW, Shin TB, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 4.Caron E, Ghosh S, Matsuoka Y, et al. A comprehensive map of the mTOR signaling network. Mol Syst Biol. 2010;6:453. doi: 10.1038/msb.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarbassov DD, Ali SM, Sengupta S, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 6.Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaeberlein M, Kennedy BK. Ageing: a midlife longevity drug? Nature. 2009;460:331–332. doi: 10.1038/460331a. [DOI] [PubMed] [Google Scholar]
- 9.Kaeberlein M, Powers RW, Steffen KK, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 10.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 11.Bjedov I, Toivonen JM, Kerr F, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones DP, Carlson JL, Samiec PS, et al. Glutathione measurement in human plasma. Evaluation of sample collection, storage and derivatization conditions for analysis of dansyl derivatives by HPLC. Clin Chim Acta. 1998;275:175–184. doi: 10.1016/s0009-8981(98)00089-8. [DOI] [PubMed] [Google Scholar]
- 13.Ritz D, Beckwith J. Redox state of cytoplasmic thioredoxin. Methods Enzymol. 2002;347:360–370. doi: 10.1016/s0076-6879(02)47036-x. [DOI] [PubMed] [Google Scholar]
- 14.Halvey PJ, Watson WH, Hansen JM, Go YM, Samali A, Jones DP. Compartmental oxidation of thiol-disulphide redox couples during epidermal growth factor signalling. Biochem J. 2005;386:215–219. doi: 10.1042/BJ20041829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das M, Gabriely I, Barzilai N. Caloric restriction, body fat and ageing in experimental models. Obes Rev. 2004;5:13–19. doi: 10.1111/j.1467-789x.2004.00115.x. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez CP, He YZ. Bone growth during rapamycin therapy in young rats. BMC Pediatr. 2009;9:3. doi: 10.1186/1471-2431-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 18.Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 19.Karim MR, Kanazawa T, Daigaku Y, Fujimura S, Miotto G, Kadowaki M. Cytosolic LC3 ratio as a sensitive index of macroautophagy in isolated rat hepatocytes and H4-II-E cells. Autophagy. 2007;3:553–560. doi: 10.4161/auto.4615. [DOI] [PubMed] [Google Scholar]
- 20.Bokov A, Chaudhuri A, Richardson A. The role of oxidative damage and stress in aging. Mech Ageing Dev. 2004;125:811–826. doi: 10.1016/j.mad.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Decker T, Hipp S, Ringshausen I, et al. Rapamycin-induced G1 arrest in cycling B-CLL cells is associated with reduced expression of cyclin D3, cyclin E, cyclin A, and survivin. Blood. 2003;101:278–285. doi: 10.1182/blood-2002-01-0189. [DOI] [PubMed] [Google Scholar]
- 22.Cho CG, Kim HJ, Chung SW, et al. Modulation of glutathione and thioredoxin systems by calorie restriction during the aging process. Exp Gerontol. 2003;38:539–548. doi: 10.1016/s0531-5565(03)00005-6. [DOI] [PubMed] [Google Scholar]
- 23.Rebrin I, Kamzalov S, Sohal RS. Effects of age and caloric restriction on glutathione redox state in mice. Free Radic Biol Med. 2003;35:626–635. doi: 10.1016/s0891-5849(03)00388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rebrin I, Forster MJ, Sohal RS. Effects of age and caloric intake on glutathione redox state in different brain regions of C57BL/6 and DBA/2 mice. Brain Res. 2007;1127:10–18. doi: 10.1016/j.brainres.2006.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen D, Guarente L. SIR2: a potential target for calorie restriction mimetics. Trends Mol Med. 2007;13:64–71. doi: 10.1016/j.molmed.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Boily G, Seifert EL, Bevilacqua L, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallows WC, Yu W, Smith BC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahuja N, Schwer B, Carobbio S, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Kawahara TL, Michishita E, Adler AS, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci. 2009;122:489–498. doi: 10.1242/jcs.042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arias IM. The Liver: Biology and Pathobiology. Hoboken, NJ: John Wiley & Sons; 2009. [Google Scholar]
- 35.Bae-Jump VL, Zhou C, Gehrig PA, Whang YE, Boggess JF. Rapamycin inhibits hTERT telomerase mRNA expression, independent of cell cycle arrest. Gynecol Oncol. 2006;100:487–494. doi: 10.1016/j.ygyno.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 36.Heilbronn LK, Ravussin E. Calorie restriction and aging: review of the literature and implications for studies in humans. Am J Clin Nutr. 2003;78:361–369. doi: 10.1093/ajcn/78.3.361. [DOI] [PubMed] [Google Scholar]
- 37.Bowman TA, Ramakrishnan SK, Kaw M, et al. Caloric restriction reverses hepatic insulin resistance and steatosis in rats with low aerobic capacity. Endocrinology. 2010;151:5157–5164. doi: 10.1210/en.2010-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraenkel M, Ketzinel-Gilad M, Ariav Y, et al. mTOR inhibition by rapamycin prevents beta-cell adaptation to hyperglycemia and exacerbates the metabolic state in type 2 diabetes. Diabetes. 2008;57:945–957. doi: 10.2337/db07-0922. [DOI] [PubMed] [Google Scholar]
- 39.Bartke A. Insulin and aging. Cell Cycle. 2008;7:3338–3343. doi: 10.4161/cc.7.21.7012. [DOI] [PubMed] [Google Scholar]
- 40.Gredilla R, Barja G. Minireview: the role of oxidative stress in relation to caloric restriction and longevity. Endocrinology. 2005;146:3713–3717. doi: 10.1210/en.2005-0378. [DOI] [PubMed] [Google Scholar]
- 41.Tunon MJ, Sanchez-Campos S, Gutierrez B, Culebras JM, Gonzalez-Gallego J. Effects of FK506 and rapamycin on generation of reactive oxygen species, nitric oxide production and nuclear factor kappa B activation in rat hepatocytes. Biochem Pharmacol. 2003;66:439–445. doi: 10.1016/s0006-2952(03)00288-0. [DOI] [PubMed] [Google Scholar]
- 42.Shen D, Dalton TP, Nebert DW, Shertzer HG. Glutathione redox state regulates mitochondrial reactive oxygen production. J Biol Chem. 2005;280:25305–25312. doi: 10.1074/jbc.M500095200. [DOI] [PubMed] [Google Scholar]
- 43.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 44.Gu L, Gao J, Li Q, et al. Rapamycin reverses NPM-ALK-induced glucocorticoid resistance in lymphoid tumor cells by inhibiting mTOR signaling pathway, enhancing G1 cell cycle arrest and apoptosis. Leukemia. 2008;22:2091–2096. doi: 10.1038/leu.2008.204. [DOI] [PubMed] [Google Scholar]
- 45.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burnett C, Valentini S, Cabreiro F, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 48.Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 49.Haigis MC, Mostoslavsky R, Haigis KM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 50.Kanfi Y, Shalman R, Peshti V, et al. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minor RK, Allard JS, Younts CM, Ward TM, de Cabo R. Dietary interventions to extend life span and health span based on calorie restriction. J Gerontol A Biol Sci Med Sci. 2010;65:695–703. doi: 10.1093/gerona/glq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharp ZD, Strong R. The role of mTOR signaling in controlling mammalian life span: what a fungicide teaches us about longevity. J Gerontol A Biol Sci Med Sci. 2010;65:580–589. doi: 10.1093/gerona/glp212. [DOI] [PubMed] [Google Scholar]
- 53.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–276. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 54.Nicoletti F, Lapenta C, Donati S, et al. Inhibition of human immunodeficiency virus (HIV-1) infection in human peripheral blood leucocytes-SCID reconstituted mice by rapamycin. Clin Exp Immunol. 2009;155:28–34. doi: 10.1111/j.1365-2249.2008.03780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Filippone EJ, Carson JM, Beckford RA, et al. Sirolimus-induced pneumonitis complicated by pentamidine-induced phospholipidosis in a renal transplant recipient: a case report. Transplant Proc. 2011;43:2792–2797. doi: 10.1016/j.transproceed.2011.06.060. [DOI] [PubMed] [Google Scholar]