Abstract
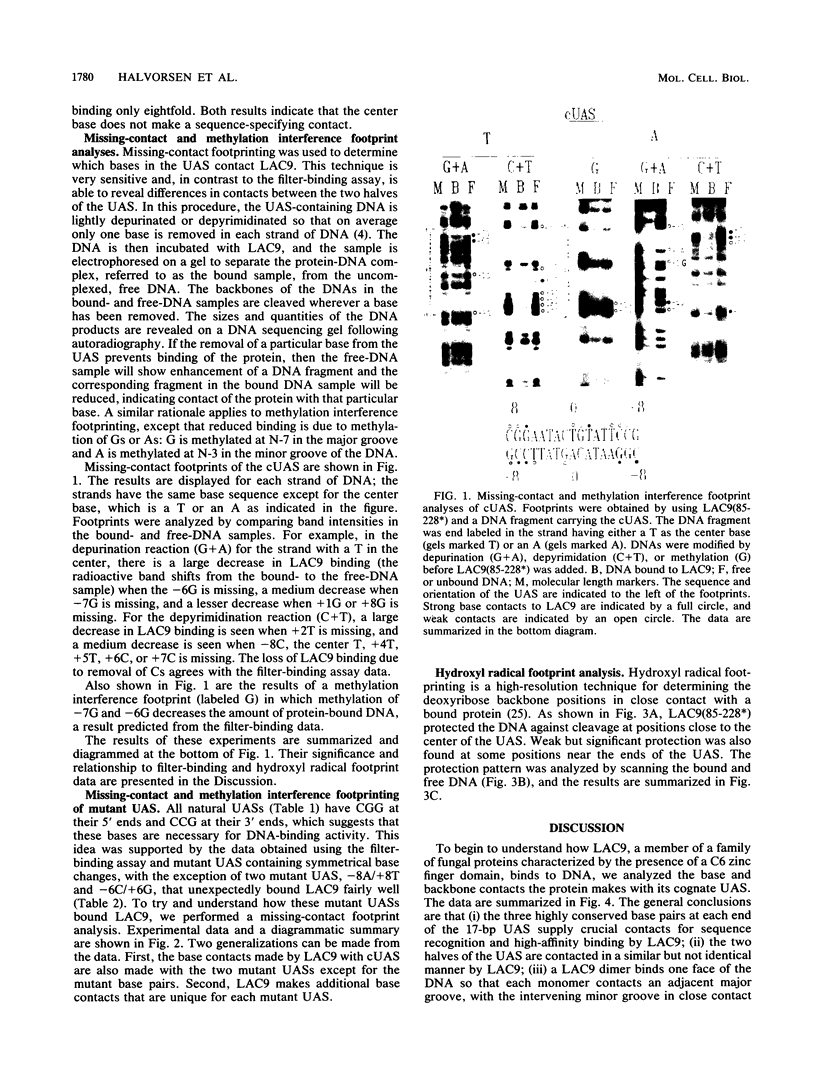
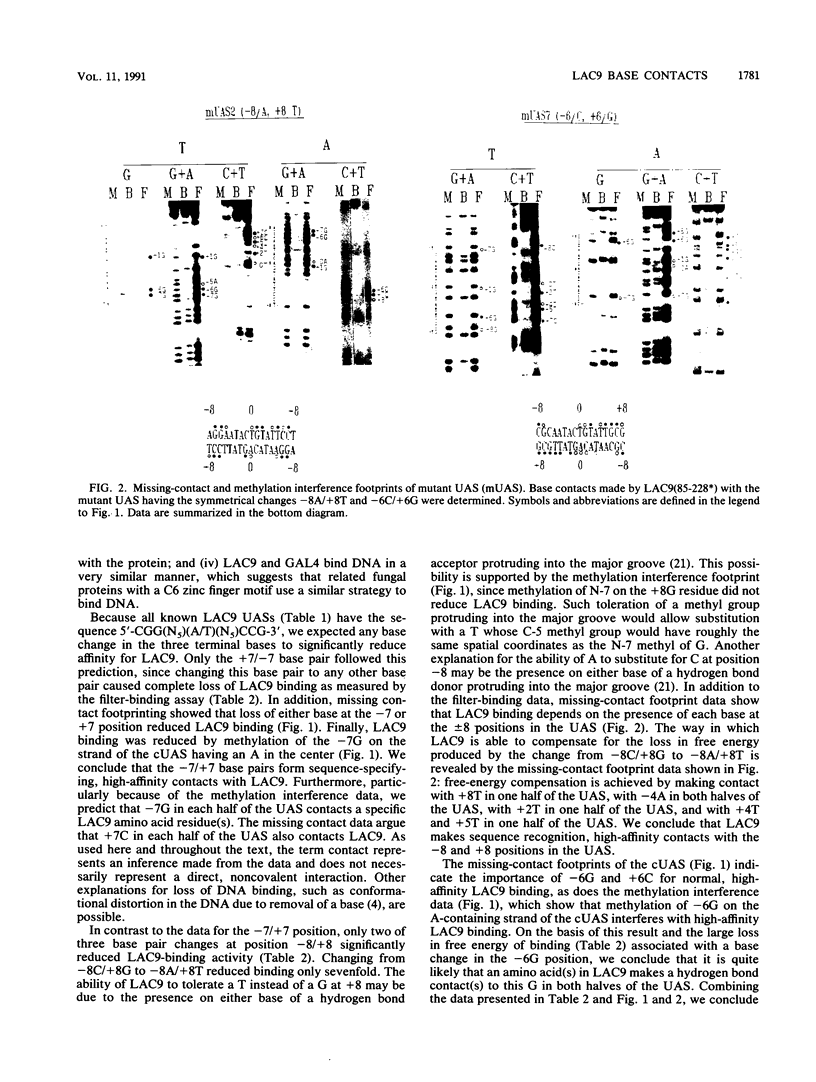
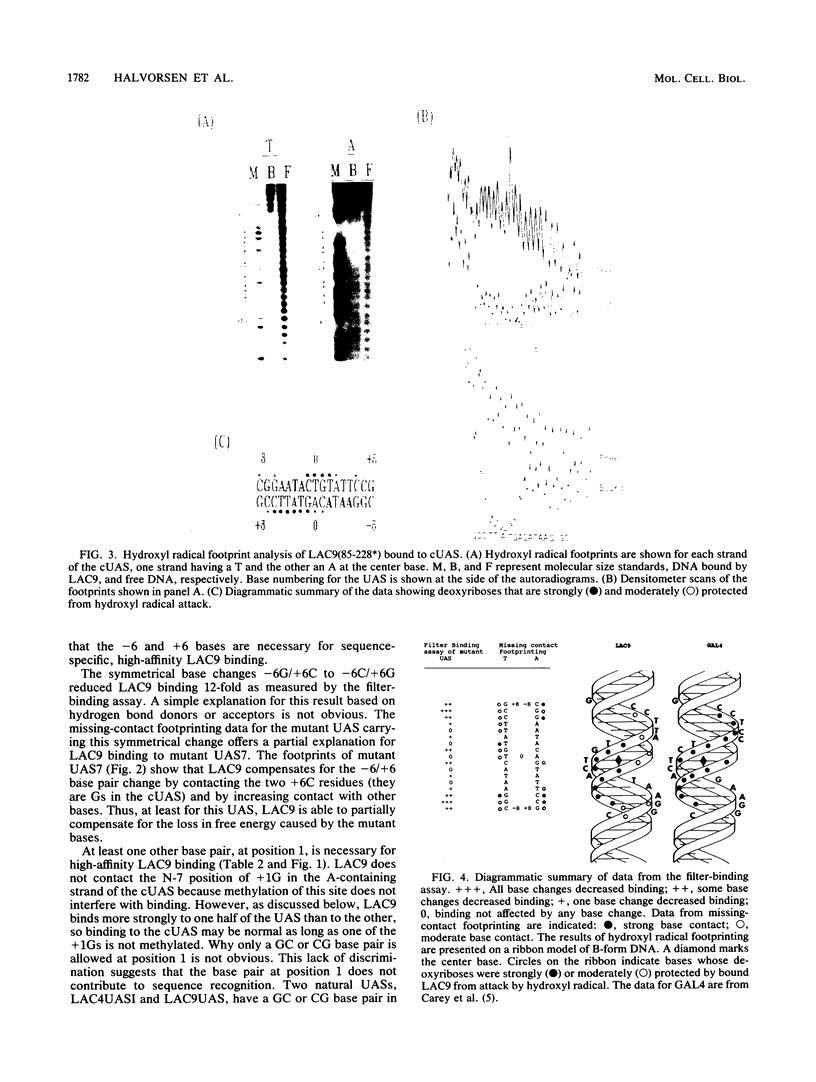
The LAC9 protein of Kluyveromyces lactis is a transcriptional regulator of genes in the lactose-galactose regulon. To regulate transcription, LAC9 must bind to 17-bp upstream activator sequences (UASs) located in front of each target gene. LAC9 is homologous to the GAL4 protein of Saccharomyces cerevisiae, and the two proteins must bind DNA in a very similar manner. In this paper we show that high-affinity, sequence-specific binding by LAC9 dimers is mediated primarily by 3 bp at each end of the UAS: [Formula: see text]. In addition, at least one half of the UAS must have a GC or CG base pair at position 1 for high-affinity binding; LAC9 binds preferentially to the half containing the GC base pair. Bases at positions 2, 3, and 4 in each half of the UAS make little if any contribution to binding. The center base pair is not essential for high-affinity LAC9 binding when DNA-binding activity measured in vitro. However, the center base pair must play an essential role in vivo, since all natural UASs have 17, not 16, bp. Hydroxyl radical footprinting shows that a LAC9 dimer binds an unusually broad region on one face of the DNA helix. Because of the data, we suggest that LAC9 contacts positions 6, 7, and 8, both plus and minus, of the UAS, which are separated by more than one turn of the DNA helix, and twists part way around the DNA, thus protecting the broad region of the minor groove between the major-groove contacts.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- André B. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Mol Gen Genet. 1990 Jan;220(2):269–276. doi: 10.1007/BF00260493. [DOI] [PubMed] [Google Scholar]
- Breunig K. D., Kuger P. Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol Cell Biol. 1987 Dec;7(12):4400–4406. doi: 10.1128/mcb.7.12.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M., Kakidani H., Leatherwood J., Mostashari F., Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989 Oct 5;209(3):423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- Corton J. C., Johnston S. A. Altering DNA-binding specificity of GAL4 requires sequences adjacent to the zinc finger. Nature. 1989 Aug 31;340(6236):724–727. doi: 10.1038/340724a0. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Markin J. S. Physiological studies of beta-galactosidase induction in Kluyveromyces lactis. J Bacteriol. 1980 Jun;142(3):777–785. doi: 10.1128/jb.142.3.777-785.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Riley M. I. The lactose-galactose regulon of Kluyveromyces lactis. Biotechnology. 1989;13:19–40. [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen Y. C., Nandabalan K., Dickson R. C. LAC9 DNA-binding domain coordinates two zinc atoms per monomer and contacts DNA as a dimer. J Biol Chem. 1990 Aug 5;265(22):13283–13289. [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Leonardo J. M., Bhairi S. M., Dickson R. C. Identification of upstream activator sequences that regulate induction of the beta-galactosidase gene in Kluyveromyces lactis. Mol Cell Biol. 1987 Dec;7(12):4369–4376. doi: 10.1128/mcb.7.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J. L., Erfle M., Wykes E. J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984 Dec;32(3):481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pan T., Coleman J. E. GAL4 transcription factor is not a "zinc finger" but forms a Zn(II)2Cys6 binuclear cluster. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2077–2081. doi: 10.1073/pnas.87.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. I., Hopper J. E., Johnston S. A., Dickson R. C. GAL4 of Saccharomyces cerevisiae activates the lactose-galactose regulon of Kluyveromyces lactis and creates a new phenotype: glucose repression of the regulon. Mol Cell Biol. 1987 Feb;7(2):780–786. doi: 10.1128/mcb.7.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzi M., Breunig K. D., Ficca A. G., Hollenberg C. P. Positive regulation of the beta-galactosidase gene from Kluyveromyces lactis is mediated by an upstream activation site that shows homology to the GAL upstream activation site of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):991–997. doi: 10.1128/mcb.7.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarai A., Takeda Y. Lambda repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6513–6517. doi: 10.1073/pnas.86.17.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers J. W., Vincent A. C., Struhl K. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol Cell Biol. 1990 Oct;10(10):5077–5086. doi: 10.1128/mcb.10.10.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Sarai A., Rivera V. M. Analysis of the sequence-specific interactions between Cro repressor and operator DNA by systematic base substitution experiments. Proc Natl Acad Sci U S A. 1989 Jan;86(2):439–443. doi: 10.1073/pnas.86.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte M. M., Dickson R. C. Cysteine residues in the zinc finger and amino acids adjacent to the finger are necessary for DNA binding by the LAC9 regulatory protein of Kluyveromyces lactis. Mol Cell Biol. 1988 Sep;8(9):3726–3733. doi: 10.1128/mcb.8.9.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte M. M., Dickson R. C. The C6 zinc finger and adjacent amino acids determine DNA-binding specificity and affinity in the yeast activator proteins LAC9 and PPR1. Mol Cell Biol. 1990 Oct;10(10):5128–5137. doi: 10.1128/mcb.10.10.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]