Abstract
Symbiotic nitrogen fixation (SNF) involves global changes in gene expression and metabolite accumulation in both rhizobia and the host plant. In order to study the metabolic changes mediated by leaf–root interaction, photosynthesis was limited in leaves by exposure of plants to darkness, and subsequently gene expression was profiled by real-time reverse transcription–PCR (RT–PCR) and metabolite levels by gas chromatography–mass spectrometry in the nodules of the model legume Lotus japonicus. Photosynthetic carbon deficiency caused by prolonged darkness affected many metabolic processes in L. japonicus nodules. Most of the metabolic genes analysed were down-regulated during the extended dark period. In addition to that, the levels of most metabolites decreased or remained unaltered, although accumulation of amino acids was observed. Reduced glycolysis and carbon fixation resulted in lower organic acid levels, especially of malate, the primary source of carbon for bacteroid metabolism and SNF. The high amino acid concentrations together with a reduction in total protein concentration indicate possible protein degradation in nodules under these conditions. Interestingly, comparisons between amino acid and protein content in various organs indicated systemic changes in response to prolonged darkness between nodulated and non-nodulated plants, rendering the nodule a source organ for both C and N under these conditions.
Key words: Carbon starvation, Lotus japonicus, metabolomic analysis, nodule, symbiosis, transcript profile.
Introduction
Symbiotic nitrogen fixation (SNF) by rhizobia in legumes is a beneficial interaction that provides the plant with the limiting macronutrient nitrogen. SNF takes place in specialized plant organs called nodules. During symbiosis, the plant provides its endosymbiont with photosynthate in exchange for reduced nitrogen, in the form of ammonium and amino acids (Udvardi and Day, 1997; White et al., 2007).
SNF creates substantial demand for fixed carbon, rendering the nodule a strong sink for carbohydrates. The carbohydrate supply is derived from plant photosynthates, transported to the nodules via the phloem (Gordon et al., 1999). Sucrose is the principal source of carbohydrate and energy for nodule metabolism. Mutants of Pisum sativum which lack sucrose synthase activity are not able to produce nitrogen-fixing nodules, indicating that the enzyme is essential for nitrogen fixation (Gordon et al., 1999). However, in Lotus japonicus, mutants lacking the major nodule-induced isoform of sucrose synthase (LjSUS3) retained some capacity for nitrogen fixation, although they exhibited N stress symptoms (Horst et al., 2007). In addition to sucrose synthase, alkaline/neutral invertase is also important for sucrose degradation, since it releases hexoses. The activity of both enzymes is elevated in L. japonicus nodules (Flemetakis et al., 2006). The products of sucrose metabolism can be further metabolized by glycolytic enzymes, producing phosphoenolpyryvate (PEP). In L. japonicus, >20 genes involved in sugar breakdown have been found to be more highly expressed in nodules than in roots. These highly expressed genes include genes for the majority of the enzymatic steps between sucrose and PEP (Colebatch et al., 2004). It is widely accepted that dicarboxylic acids and not sugars are supplied to bacteroids, and transporters that deliver dicarboxylates to the bacteroids have been characterized (Udvardi et al., 1988; Jeong et al., 2004). PEP can be carboxylated to oxaloacetate and then reduced to malate, the primary source of carbon for bacteroid metabolism and SNF (Day and Copeland, 1991; Streeter, 1995). Both PEP carboxylase (PEPC) and malate dehydrogenase genes have been found to be nodule induced in L. japonicus (Colebatch et al., 2002; Nakagawa et al., 2003; Colebatch et al., 2004).
Carbon skeletons generated by carbohydrate breakdown are required to convert ammonium into amino acids. In nodules, ammonium is assimilated into glutamine by the combined activity of the enzymes glutamine synthetase (GS; EC 6.3.1.2) and glutamate synthase (GOGAT; EC 1.4.1.13) (GS/GOGAT pathway) (Patriarca et al., 2002; Barsch et al., 2006). Asparagine can be produced from the assimilated glutamine via the concerted activity of glutamine-dependent asparagine synthetase (AS; EC 6.3.5.4) and aspartate aminotransferase (ASAT; EC 2.6.1.1). Temperate legumes export asparagine from the nodules to the shoot, but tropical legumes export ureides (Temple et al., 1998; Goggin et al., 2003). A number of genes involved in ammonium assimilation and asparagine synthesis (including GS and AS) were induced in L. japonicus nodules (Colebatch et al., 2004).
SNF relies on photosynthate supply (Hardy and Havelka, 1976); consequently, SNF should vary together with photosynthesis. Experimental conditions that enhance photosynthesis, for example an increase in light intensity (Lawn and Brun, 1974; Hardy and Havelka, 1976; Bethlenfalvay and Phillips, 1977), are usually associated with an increase in SNF, whereas experimental conditions that limit photosynthesis, for example shading (Tricot, 1993) and limited light intensity (Feigenbaum and Mengel, 1979), decrease SNF. Extended darkness has been used to study the significance of photosynthetic carbon allocation for nodule metabolism, although a global view of transcriptomic and metabolomic changes in nodules upon carbohydrate limitation is lacking. Responses of nodules to photosynthate limitation vary amongst legume species. For example, the observed decline of nitrogenase activity after prolonged darkness was much less dramatic in soybean (Sarath et al., 1986) and cluster bean (Swaraj et al., 1994) than it was in common bean (Gogorcena et al., 1997) and pea (Matamoros et al., 1999). Exposure of nodulated soybeans to a short period of darkness was sufficient to cause a significant decline in the levels of gene transcripts for the key nodule proteins, leghaemoglobin, sucrose synthase, and glutamine synthase, and in the activity of sucrose synthase in nodules (Gordon et al., 1993). Furthermore, during dark-induced stress in soybean, a decline in sugars and some key glycolytic intermediates in nodules coincided with depletion of glutamine, asparagine, and alanine, and with accumulation of ureides, which reflected a large reduction of N2 fixation (Vauclare et al., 2010). Moreover, dark exposure decreased respiration and nitrogenase activity of soybean and lupine nodules (Layzell et al., 1990; Iannetta et al., 1993), indicating that depletion of sucrose in the nodules limits the supply of respiratory substrates to bacteroids and affects SNF. In common bean plants, nitrogenase activity was almost zero after 1 d of darkness and the activities of enzymes involved in carbon and nitrogen metabolism were reduced after 2 d of dark stress (Gorgocena et al., 1997).
Previous work on the effect of reduced photosynthesis on nodule metabolism focused on just a few genes, enzymes, and/or metabolites. The aim of this study was to assess the global effects of photosynthetic carbon limitation on metabolism in nodules of the model legume, L. japonicus. For this purpose, transcriptomic analysis was carried out, using quantitative real-time PCR, and metabolite profiling, using gas chromatography–mass spectrometry (GC-MS), of nodules subjected to photosynthetic carbon deficiency caused by prolonged darkness. In parallel, in order to study the carbon allocation under these conditions, the systemic effects of prolonged darkness on total amino acid and protein content in different organs of nodulated versus non-nodulated plants were measured. The results indicate that changes in nodule metabolism during photosynthate limitation are controlled largely at the transcriptional level. It is also shown, for the first time, that nodules can become a net source of carbon, as well as of nitrogen, for leaves in the absence of photosynthesis.
Materials and methods
Plant material and growth conditions
Lotus japonicus (Gifu B-129) seeds were kindly provided by Dr Jens Stougaard (University of Aarhus, Denmark). The plants were grown in a controlled environment with a 16h day/8h night cycle, a 22 °C day/18 °C night temperature regime, and 70% humidity (Handberg and Stougaard, 1992). Prior to germination, seeds were scarified for 5min with H2SO4 and then sterilized for 20min in a solution containing 2% NaOCl–0.02% Tween-20. Seeds were pre-germinated at 18 °C in the dark for 72h and the small plants were grown in Hoagland nutrient solution. For the infection with rhizobia, 72h seedlings were transferred into pots containing sand, and spot-infected with a 0.1 OD600 suspension culture of Mesorhizobium loti (strain R7A).
After a 4 week growth period, the plants were divided into groups subjected to the following different light regimes: (i) a normal photoperiod; (ii) a 24h dark period; (iii) a 24h dark period followed by 48h recovery to the normal photoperiod; (iv) a 72h dark period; and (v) a 72h dark period followed by 48h recovery to the normal photoperiod. Nodule harvest was performed between 2h and 4h after the beginning of the light period. Nodules of plants exposed to continuous darkness were also harvested at the same time of the day, under green light.
Starch staining
Whole plants of L. japonicus were subjected to starch staining. The plants were submerged in boiling water for 1min and then were transferred to boiling ethanol for the removal of chlorophyll. As soon as the chlorophyll was removed, the plants were rinsed in water and the starch was stained by using a potassium iodide (KI) solution.
For nodule starch staining, nodules were dipped into Jung tissue freezing medium (Leica Microsystems) and cut into thin sections (20 μm) by the use of a Leica CM 1850 cryotome at –15 °C. The sections were transferred onto slides and rinsed in water. The starch was stained by the addition of a few drops of KI solution to the slide. Starch staining visualization was carried out using an Olympus Bx40 microscope equipped with an Olympus DP71 camera.
Nitrogenase activity
Total nitrogenase activity was estimated by the acetylene reduction assay in whole plants at 26 days post-inoculation incubated at 25 °C in 28ml rubber cap tubes containing 1/10 (v/v) acetylene. The ethylene produced (Supplementary Fig. S2 available at JXB online) at different time points (after 1, 2, and 3h incubation) was quantified with a Perkin-Elmer 8500 gas chromatograph (Perkin-Elmer Life and Analytical Science Inc., Wellesley, MA, USA) equipped with a 2 m Porapak R column and a flame ionization detector. Acetylene reduction activity was calculated from production of ethylene (based on peak area) per mg of nodule dry weight (Hardy et al., 1973).
Metabolite extraction, derivatization, and GC-MS analysis
Nodules were harvested, immediately frozen in liquid nitrogen, and lyophilized. Approximately 10mg dry weight of tissue were ground in liquid nitrogen. Ground samples were extracted with 380 μl of methanol and 20 μl of ribitol in methanol (0.2mg ml–1). Samples were incubated at 70 °C for 15min with continuous shaking. After the addition of 200 μl of chloroform, the samples were further incubated at 37 °C for 5min under continuous shaking. Following the addition of 400 μl of ddH2O, the samples were vortexed and then centrifuged at 18 000 g for 5min at room temperature. The aqueous phase containing the polar metabolite fraction was transferred into new Eppendorf tubes and dried by nitrogen gas. For derivatization, dried samples were re-suspended in 25 μl of methoxyamine-HCl (20mg ml–1 in pyridine), and incubated at 30 °C for 90min with continuous gentle agitation. This was followed by addition of 50 μl of N-methyl-N-(trimethylsilyl)-trifluoroacetamide and incubated at 37 °C for 30min with continuous gentle agitation. Finally, 10 μl of an n-alkane mix were added for determination of retention indexes (RIs). Gas chromatography coupled to mass spectrometry (GC-MS) measurements were performed in a Agilent Technologies 7890A GC system coupled to an Agilent Technologies 5975C MS (Agilent Technologies, Frankfurt, Germany). For all treatments tested, five biological replications were conducted. The chromatograms were evaluated automatically using the AMDIS software, and metabolites were identified using the Golm metabolome (Kopka et al., 2005; Schauer et al., 2005), and Agilent Fienh metabolite and NIST05 metabolite databases. Results are expressed as a response that corresponds to the ratio between the area of the target metabolite divided by the area of the reference metabolite (ribitol, m/z 319) and reported relative to the dry weight.
Determination of transcript levels using quantitative real-time reverse transcription–PCR
Lotus japonicus nodules and uninfected roots were harvested and ground in liquid nitrogen. Total RNA was isolated and treated with DNase I (Promega, Madison, WI, USA) at 37 °C for 45min, in order to eliminate any traces of genomic DNA. Successful removal of genomic DNA was tested by PCR. First-strand cDNA was reverse transcribed from 2 μg of DNase-treated total RNA. All samples were denatured at 65 °C for 5min followed by quick cooling on ice in a 12 μl reaction mixture containing 500ng of oligo(dT)12–18mer and 1 μl of 10mM dNTPs. After the addition of 4 μl of 5× First-Strand buffer (Invitrogen, Paisley, UK), 1 μl (40U) of RNaseOUT (Invitrogen) RNase inhibitor and 2 μl of 0.1M dithiothreitol (DTT), the reaction was pre-heated at 42 °C for 2min before the addition of 1 μl (200U) of SuperScript II reverse transcriptase (Invitrogen). The reaction mixture was incubated at 42 °C for 50min, followed by heat inactivation at 70 °C for 15min. Target cDNAs were amplified using gene-specific primers (Supplementary Table S1 at JXB online) designed by the Primer Express 1.5 software (Applied Biosystems, Darmstadt, Germany). Quantitative RT–PCRs were performed on the Stratagene MX3005P using Power SYBR Green master mix (Applied Biosystems), gene-specific primers at a final concentration of 0.2 μM each, and 1 μl of the cDNA as template. PCR cycling started with the initial polymerase activation at 95 °C for 10min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1min. Primer specificity and formation of primer dimers were monitored by dissociation curve analysis. The expression levels of an L. japonicus ubiquitin gene were used as internal standards to normalize small differences in cDNA template amounts. Relative transcript levels of the gene of interest (X) were calculated as a ratio to the ubiquitin gene transcripts (U), as (1+E)–ΔCt, where ΔCt was calculated as (CtX–CtU). PCR efficiency (E) for each amplicon was calculated employing the linear regression method on the Log (Fluorescence) per cycle number data, using the LinRegPCR software (Ramakers et al., 2003). All real-time qPCRs were performed on three biological repeats.
13C labelling
The 13C labelling was conducted through the root system of 30-day-old inoculated L. japonicus plants, as previously described (Fotelli et al., 2011). In total, 108 plants were used for the labelling experiment, subjected to the previously described normal photoperiod and extended dark conditions. All plants were placed in vials (100ml volume) for hydroponics which were connected to a CO2-free air supply system, during a 2h acclimation period. CO2 was removed from the air supply with soda lime CO2 traps. For dark treatment, the hypergeous part of the plants was covered. Two-thirds of the plants were transferred from hydroponics to air-tight glass vials containing 10mM Na213CO3 solutions (99% enriched in 13C; pH=6). Na213CO3 dissociates in water and produces 13CO2 that is taken up by the plants. Labelling was performed by a 2h pulse. After labelling, one-third of the plants were kept in hydroponics for a 1h chase period. Nodules were harvested after each phase (2h acclimation period, 2h labelling, and 1h chase period), ground in liquid nitrogen, and stored at –80 °C.
δ13C composition
Nodules were oven-dried (3 d, 65 °C) and samples of 0.5mg were transferred into tin capsules (IVA Analysentechnik, Meerbusch, Germany). Subsequently, the samples were injected into an isotope ratio mass spectrometer (Delta Plus; Finnigan MAT GmbH, Bremen, Germany) for δ13C analysis. δ13C values are expressed relative to the VPDB (Vienna Pee Dee Belemnite) standard.
Soluble protein
To quantify total soluble protein concentrations, ~0.05g of frozen tissue were extracted in 1ml of buffer (0.1M K2HPO4/KH2PO4 pH 7.7, 5mM Na-EDTA, 10mM DTT, 1% Triton X-100, 2% PVP). The protein extracts were passed through cotton gauze. For quantification of soluble protein, 1ml of Bradford reagent plus 90 μl of 0.15M NaCl were added to 10 μl aliquots of the extracts. The optical density was measured in a Hitachi U-2800 Spectrophotometer (Tokyo, Japan) at 595nm after a 20min incubation at room temperature in the dark (Bradford, 1976). Bovine serum albumin was used as a standard.
Total amino acid content
For quantification of total soluble amino acids, aliquots of 0.05g fresh weight of tissue were homogenized in 1ml of methanol:chloroform (3.5:1.5, v:v) and 0.2ml of buffer (pH 7.0) containing 20mM HEPES, 5mM EGTA, and 10mM NaF, according to the method of Winter et al. (1992). After incubating for 30min, 600 μl of distilled water were added to the samples, mixed, and centrifuged for 5min at 14 000 g and 4 °C. This step was repeated once. Total amino acid content was determined from the combined supernatants according to the method of Liu et al. (2005). Aliquots of the extract (100 μl) and 100 μl of ninhydrin reagent [50/50 mixture of solution one containing 19.21g of citrate in 200ml of 1 N NaOH (pH 5.0), made up to 500ml with distilled water and 0.8g SnCl2·H2O, and solution two containing 20g of ninhydrin in 500ml of ethylene glycol monomethyl ether] were prepared. Isopropanol (800 μl, 50%) was added to the samples, followed by a 15min incubation at 100 °C. The optical density was measured in a Hitachi U-2800 Spectrophotometer (Tokyo, Japan) at 570nm. Glutamine was used as a standard.
Statistical analysis
Transcriptomic and metabolomic data statistical analysis was performed by one-way analysis of variance (ANOVA) at a 95% level of significance. Principal components analysis (PCA) was performed using the Unscrambler 9.5 software (CAMO Software Inc., NJ, USA). Missing values were defined as 0, assuming that missing values were unchanged when compared with the co-analysed standard samples. Statistical analysis of data of nodule δ13C abundance was carried out using SPSS 12.0 (SPSS Inc., Chicago, IL, USA). Comparisons were performed by independent-sample t-tests at a 95% level of significance. To detect differences in total protein and total amino acid content between the examined tissues, independent-sample t-tests were used at a 95% level of significance. Statistical tests were conducted using Sigmaplot 10.0 statistical software (Systat Software Inc.. San Jose, CA, USA).
Results
Extended dark reduces starch levels and nitrogenase activity
To identify global changes in nodule metabolism under carbohydrate limitation, L. japonicus plants were subjected to prolonged darkness. The length of the dark period applied to the plants was determined based on the rate of starch breakdown in nodules. For this reason, starch accumulation was visualized by direct staining in nodules after 12, 24, 36, 60, and 72h of continuous darkness. After 24h of continuous darkness, starch deposits were still evident in nodule sections via staining, but after 72h of darkness starch reserves were completely exhausted (Supplementary Fig. S1 at JXB online). Thus, these two time points were selected for further analysis.
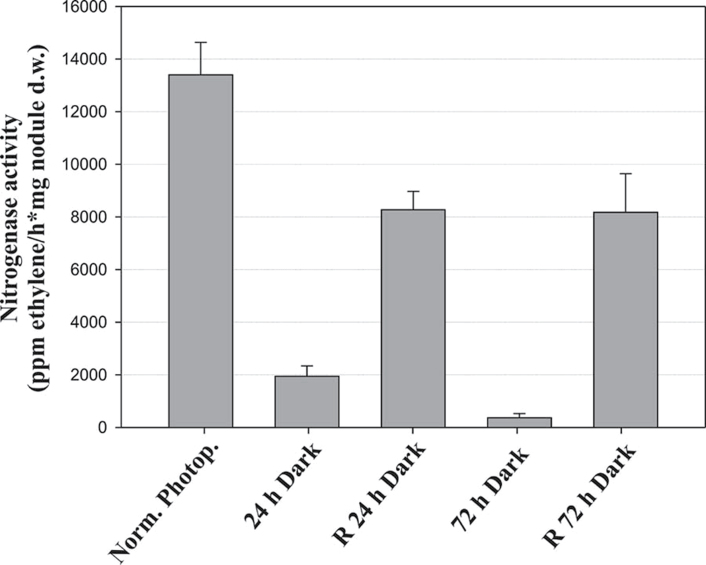
The effect of photosynthetic carbon limitation on nitrogenase activity in intact L. japonicus nodules was estimated by the acetylene reduction assay, using whole plants. After 24h of continuous darkness, nitrogenase activity was almost 7-fold reduced compared with control plants exposed to normal photoperiod conditions, while after 72h of darkness nitrogenase activity was >30-fold reduced compared with the control (Fig. 1). To test whether the reduced nitrogenase activity observed was a direct effect of carbohydrate limitation caused by prolonged darkness, nitrogenase activity was measured after 48h recovery by transferring the darkened plants back to the normal photoperiod conditions. Nitrogenase activity partially recovered, to almost 60% of its original level, following restoration of the normal photoperiod (Fig. 1).
Fig. 1.
Nitrogenase activity, assayed as acetylene reduction, in L. japonicus nodules under a normal photoperiod, after the plants have been subjected to extended dark periods for 24h and 72h, and after 48h recovery (R) under the normal photoperiod, respectively. Bars represent means (±SE) of six biological replications.
Nodule metabolite profiling under carbon-limiting conditions
For this experiment, 30-day-old nodulated L. japonicus plants were subjected to either 24h or 72h of continuous darkness, while control plants of the same age were kept under normal photoperiod conditions. GC-MS analysis in nodules identified metabolites belonging to different classes, including amino acids, organic acids, other nitrogenous compounds, sugars, and polyols.
Changes in specific metabolites were calculated by the response ratios for dark-treated versus control plants exposed to normal photoperiod conditions. Although the levels of many metabolites were not significantly different following extended darkness, ANOVA detected 39 metabolites that changed significantly (P < 0.05, Table 1). Moreover, seven metabolites were found only in dark-treated plants.
Table 1.
Identified metabolites of L. japonicus nodules harvested from plants grown under normal photoperiod (N. Ph.), or subjected to 24h and 72h continuous darkness.
| m\z | RI | PC1 (49%) loadings | PC2 (27%) loadings | N. Ph. average response | 24h darkness average response | 72h darkness average response | 24h/N. Ph. response ratio | 72h/N. Ph. response ratio | P-value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sugars | ||||||||||||||||||||
| Sucrose | 361 | 2385 | –0.5010 | 0.3580 | 8.679 | 7.866 | 3.276 | 0.91 a | 0.38 b | <0.001 | ||||||||||
| Raffinose | 87 | 3056 | –0.0197 | –0.0103 | 0.268 | 0.060 | 0.026 | 0.22 | 0.10 | 0.010 | ||||||||||
| Maltose | 361 | 2495 | 0.0005 | –0.0023 | 0.059 | 0.051 | 0.068 | 0.86 | 1.14 | 0.833 | ||||||||||
| Trehalose | 361 | 2505 | 0.0144 | –0.0178 | 0.329 | 0.413 | 0.492 | 1.25 | 1.49 | 0.310 | ||||||||||
| d-Melezitose | 361 | 3153 | –0.0380 | –0.0010 | 0.558 | 0.231 | 0.069 | 0.41 a | 0.12 b | <0.001 | ||||||||||
| Melibiose | 361 | 2512 | –0.0046 | 0.0043 | 0.216 | 0.146 | 0.155 | 0.67 | 0.72 | 0.208 | ||||||||||
| Cellobiose | 361 | 2252 | –0.0002 | –0.0006 | 0.020 | 0.016 | 0.015 | 0.79 | 0.77 | 0.870 | ||||||||||
| Maltotriose | 361 | 3034 | –0.0031 | 0.0008 | 0.039 | 0.015 | 0.002 | 0.40 | 0.06 | 0.012 | ||||||||||
| d-Lyxose | 217 | 1389 | –0.0012 | 0.0001 | 0.024 | 0.019 | 0.011 | 0.80 | 0.48 | 0.393 | ||||||||||
| d-Glucose | 319 | 1614 | –0.0187 | 0.0033 | 0.237 | 0.088 | 0.000 | 0.37 a | 0.00 b | 0.047 | ||||||||||
| Amino acids | ||||||||||||||||||||
| l-Cysteine | 220 | 1250 | 0.0047 | –0.0030 | 0.000 | 0.042 | 0.047 | Dark sp. | Dark sp. | 0.529 | ||||||||||
| l-Ornithine | 142 | 1310 | 0.0150 | –0.0091 | 0.000 | 0.000 | 0.185 | ND | Dark sp. | – | ||||||||||
| l-Homoserine | 218 | 1145 | 0.0103 | 0.0018 | 0.017 | 0.062 | 0.129 | 3.70 | 7.74 | 0.028 | ||||||||||
| l-Proline | 142 | 983 | –0.0048 | 0.0025 | 0.106 | 0.074 | 0.042 | 0.69 | 0.40 | 0.603 | ||||||||||
| l-Asparagine | 231 | 1376 | 0.4410 | 0.1160 | 11.782 | 14.955 | 17.094 | 1.27 a | 1.45 b | <0.001 | ||||||||||
| β-Alanine | 248 | 1116 | 0.0057 | –0.0005 | 0.052 | 0.060 | 0.119 | 1.16 a | 2.30 b | 0.009 | ||||||||||
| l-Glutamic acid | 246 | 1319 | 0.1840 | 0.8760 | 2.066 | 5.274 | 4.310 | 1.28 | 1.15 | 0.211 | ||||||||||
| β-Cyano-l-alanine | 141 | 1064 | 0.0022 | 0.0020 | 0.190 | 0.201 | 0.187 | 1.06 | 0.98 | 0.959 | ||||||||||
| l-Alanine | 116 | 798 | –0.0252 | 0.0160 | 0.400 | 0.397 | 0.119 | 0.99 a | 0.30 b | 0.004 | ||||||||||
| l-Serine | 204 | 1055 | 0.0262 | –0.0019 | 0.330 | 0.430 | 0.673 | 1.31 a | 2.04 b | 0.018 | ||||||||||
| l-Aspartic acid | 232 | 1216 | 0.3470 | –0.0640 | 0.873 | 2.162 | 4.931 | 2.48 a | 5.65 b | <0.001 | ||||||||||
| l-Lysine | 317 | 1618 | 0.0722 | –0.0294 | 0.212 | 0.334 | 1.066 | 1.57 a | 5.02 b | <0.001 | ||||||||||
| l-Tyrosine | 218 | 1635 | 0.0674 | 0.0217 | 0.000 | 0.682 | 0.778 | Dark sp. | Dark sp. | 0.059 | ||||||||||
| l-Methionine | 176 | 1208 | 0.0066 | –0.0036 | 0.000 | 0.000 | 0.070 | ND. | Dark sp. | – | ||||||||||
| l-Histidine | 154 | 1618 | 0.1240 | –0.0340 | 0.065 | 0.186 | 1.450 | 2.88 a | 22.45 b | <0.001 | ||||||||||
| l-Valine | 144 | 903 | 0.0888 | –0.0378 | 0.106 | 0.280 | 1.185 | 2.65 a | 11.21 b | <0.001 | ||||||||||
| l-Threonine | 218 | 1083 | 0.0211 | –0.0002 | 0.166 | 0.343 | 0.434 | 2.07 | 2.62 | <0.001 | ||||||||||
| l-Tryptophan | 202 | 1918 | 0.0878 | –0.0262 | 0.046 | 0.350 | 1.120 | 7.64 a | 24.41 b | <0.001 | ||||||||||
| dl-Isoleucine | 158 | 982 | 0.0768 | 0.0058 | 0.034 | 0.297 | 0.875 | 8.70 a | 25.58 b | <0.001 | ||||||||||
| l-Glutamine | 156 | 1470 | –0.0184 | –0.0036 | 0.411 | 0.129 | 0.130 | 0.31 | 0.32 | <0.001 | ||||||||||
| Glycine | 174 | 993 | 0.0103 | 0.0021 | 0.027 | 0.097 | 0.146 | 3.55 | 5.35 | 0.028 | ||||||||||
| Organic acids | ||||||||||||||||||||
| Dehydroascorbic acid | 173 | 1553 | –0.0025 | –0.0019 | 0.036 | 0.000 | 0.000 | 0.00 | 0.00 | – | ||||||||||
| Shikimic acid | 204 | 1517 | –0.0019 | 0.0003 | 0.032 | 0.000 | 0.000 | 0.00 | 0.00 | – | ||||||||||
| 6-Hydroxycaproic acid | 147 | 1010 | 0.0017 | –0.0011 | 0.018 | 0.031 | 0.033 | 1.68 | 1.81 | 0.626 | ||||||||||
| l-Ascorbic acid | 332 | 1659 | –0.0107 | –0.0086 | 0.173 | 0.041 | 0.000 | 0.24 | 0.00 | 0.039 | ||||||||||
| l-Lactic acid | 147 | 765 | –0.0097 | –0.0093 | 0.301 | 0.306 | 0.206 | 1.02 | 0.69 | 0.519 | ||||||||||
| Succinic acid | 148 | 999 | –0.0241 | 0.0232 | 0.541 | 0.677 | 0.250 | 1.25 a | 0.46 b | 0.006 | ||||||||||
| Pyruvic acid | 174 | 756 | –0.0010 | –0.0000 | 0.020 | 0.012 | 0.005 | 0.61 a | 0.27 b | <0.001 | ||||||||||
| Glyceric acid | 189 | 1024 | 0.0025 | 0.0005 | 0.036 | 0.038 | 0.067 | 1.05 | 1.87 | 0.165 | ||||||||||
| Pipecolic acid | 156 | 1051 | 0.0139 | 0.0068 | 0.144 | 0.171 | 0.275 | 1.18 | 1.90 | 0.263 | ||||||||||
| Mucic acid | 333 | 1763 | 0.0003 | 0.0009 | 0.317 | 0.348 | 0.317 | 1.10 | 1.00 | 0.387 | ||||||||||
| Citric acid | 347 | 1529 | –0.1140 | 0.0781 | 8.026 | 7.309 | 6.357 | 0.91 | 0.79 | 0.069 | ||||||||||
| Isocitric acid | 273 | 1531 | 0.0023 | 0.0029 | 0.000 | 0.079 | 0.056 | Dark sp. | Dark sp. | 0.537 | ||||||||||
| Citramalic acid | 147 | 1169 | 0.1520 | 0.1640 | 5.783 | 8.406 | 7.776 | 1.45 | 1.34 | 0.005 | ||||||||||
| Maleic acid | 147 | 989 | –0.0018 | –0.0019 | 0.048 | 0.055 | 0.025 | 1.15 | 0.52 | 0.502 | ||||||||||
| Fumaric acid | 245 | 1033 | –0.0020 | 0.0002 | 0.159 | 0.150 | 0.119 | 0.94 | 0.75 | 0.187 | ||||||||||
| Glucaric acid | 333 | 1732 | –0.0017 | 0.0010 | 0.480 | 0.439 | 0.434 | 0.92 | 0.90 | 0.843 | ||||||||||
| Gluconic acid | 333 | 1698 | –0.0079 | 0.0038 | 0.459 | 0.213 | 0.255 | 0.46 | 0.56 | 0.034 | ||||||||||
| 2-Hydroxycinnamic acid | 293 | 1397 | 0.0112 | –0.0026 | 0.668 | 0.884 | 0.777 | 1.32 | 1.16 | 0.298 | ||||||||||
| 4-Hydroxycinnamic acid | 293 | 1624 | –0.0011 | –0.0023 | 0.038 | 0.042 | 0.015 | 1.09 | 0.40 | 0.464 | ||||||||||
| Citraconic acid | 147 | 1040 | –0.0000 | 0.0001 | 0.002 | 0.004 | 0.001 | 2.32 | 0.77 | 0.437 | ||||||||||
| Glycolic acid | 147 | 775 | –0.0008 | 0.0002 | 0.023 | 0.012 | 0.008 | 0.53 | 0.35 | 0.021 | ||||||||||
| Malonic acid | 147 | 890 | –0.0094 | 0.0005 | 0.736 | 0.732 | 0.671 | 1.00 | 0.91 | 0.926 | ||||||||||
| d-Malic acid | 147 | 1187 | –0.3360 | 0.1160 | 5.901 | 5.240 | 1.798 | 0.89 a | 0.30 b | <0.001 | ||||||||||
| Oxalic acid | 147 | 824 | –0.0130 | 0.0027 | 0.325 | 0.272 | 0.177 | 0.84 a | 0.54 b | 0.013 | ||||||||||
| Polyols | ||||||||||||||||||||
| Galactinol | 204 | 2708 | –0.0070 | –0.0037 | 0.092 | 0.031 | 0.010 | 0.33 | 0.11 | 0.101 | ||||||||||
| Palatinitol | 361 | 2555 | 0.0011 | 0.0009 | 0.034 | 0.036 | 0.053 | 1.06 | 1.56 | 0.417 | ||||||||||
| d-Mannitol | 319 | 1648 | 0.0471 | 0.1800 | 0.477 | 1.643 | 1.161 | 3.45 | 2.43 | 0.020 | ||||||||||
| Pinitol | 260 | 1550 | –0.4390 | –0.0313 | 22.215 | 22.108 | 18.608 | 1.00 | 0.84 | 0.223 | ||||||||||
| allo-Inositol | 318 | 1745 | –0.0008 | –0.0010 | 0.216 | 0.202 | 0.201 | 0.94 | 0.93 | 0.984 | ||||||||||
| Phosphates | ||||||||||||||||||||
| O-Phospho-l-threonine | 370 | 1534 | –0.0026 | –0.0008 | 0.037 | 0.000 | 0.000 | 0.00 | 0.00 | – | ||||||||||
| Phosphoric acid | 299 | 967 | –0.0072 | 0.0056 | 3.422 | 2.796 | 3.168 | 0.82 | 0.93 | 0.245 | ||||||||||
| Phosphoenolpyruvic acid | 369 | 1302 | 0.0014 | –0.0000 | 0.008 | 0.012 | 0.028 | 1.57 a | 3.68 b | 0.007 | ||||||||||
| O-Phosphocolamine | 299 | 1485 | 0.0161 | –0.0036 | 0.010 | 0.089 | 0.203 | 8.79 a | 19.97 b | <0.001 | ||||||||||
| Pyrophosphate | 451 | 1377 | –0.0013 | 0.0001 | 0.021 | 0.009 | 0.003 | 0.41 | 0.16 | <0.001 | ||||||||||
| Adenosine-5-monophosphate | 315 | 2751 | –0.0188 | –0.0008 | 0.366 | 0.161 | 0.111 | 0.44 a | 0.30 b | <0.001 | ||||||||||
| 3-Phosphoglycerate | 357 | 1519 | 0.0015 | –0.0044 | 0.116 | 0.018 | 0.134 | 0.16 | 1.16 | 0.291 | ||||||||||
| 3-Phosphoglyceric acid | 387 | 1519 | 0.0028 | 0.0034 | 0.086 | 0.096 | 0.132 | 1.12 | 1.53 | 0.232 | ||||||||||
| d-Glucose-6-phosphate | 387 | 2052 | –0.0221 | –0.0051 | 0.348 | 0.144 | 0.098 | 0.41 | 0.28 | 0.019 | ||||||||||
| Uridine 5’-monophosphate | 315 | 2563 | –0.0029 | –0.0003 | 0.044 | 0.074 | 0.030 | 1.65 | 0.69 | 0.181 | ||||||||||
| Glycerol 1-phosphate | 357 | 1471 | –0.0011 | –0.0011 | 0.155 | 0.151 | 0.126 | 0.97 | 0.82 | 0.556 | ||||||||||
| Other metabolites | ||||||||||||||||||||
| Putrescine | 174 | 1427 | –0.0014 | –0.0003 | 0.063 | 0.045 | 0.037 | 0.73 | 0.59 | 0.009 | ||||||||||
| Spermidine | 174 | 1952 | –0.0010 | –0.0004 | 0.043 | 0.043 | 0.024 | 1.01 | 0.56 | 0.320 | ||||||||||
| Uracil | 241 | 1021 | 0.0001 | –0.0004 | 0.021 | 0.009 | 0.015 | 0.45 | 0.73 | 0.619 | ||||||||||
| Urea | 147 | 922 | 0.0009 | –0.0023 | 0.013 | 0.034 | 0.035 | 2.63 | 2.75 | 0.488 | ||||||||||
| Adenine | 264 | 1555 | –0.0014 | 0.0010 | 0.019 | 0.025 | 0.013 | 1.32 | 0.65 | 0.440 | ||||||||||
| Adenosine | 236 | 2342 | 0.0004 | 0.0066 | 0.028 | 0.111 | 0.053 | 3.96 a | 1.88 b | 0.007 | ||||||||||
| Norvaline | 72 | 783 | 0.0004 | 0.0008 | 0.000 | 0.006 | 0.007 | Dark sp. | Dark sp. | 0.246 | ||||||||||
| Loganin | 361 | 2539 | 0.0032 | 0.0032 | 0.254 | 0.290 | 0.287 | 1.14 | 1.13 | 0.557 | ||||||||||
| Thymine | 255 | 1057 | 0.0003 | 0.0004 | 0.011 | 0.024 | 0.015 | 2.14 | 1.35 | 0.235 | ||||||||||
| l-Norleucine | 158 | 960 | 0.0206 | 0.0045 | 0.014 | 0.163 | 0.226 | 11.39 | 15.79 | 0.040 | ||||||||||
| Porphine | 285 | 1020 | 0.0013 | 0.0012 | 0.008 | 0.024 | 0.028 | 2.91 | 3.38 | 0.146 | ||||||||||
| Trans-3-hydroxy-l-proline | 230 | 1221 | 0.0002 | –0.0001 | 0.000 | 0.001 | 0.002 | Dark sp. | Dark sp. | 0.186 | ||||||||||
| Methyl-β-d-galactopyranoside | 204 | 1699 | –0.0177 | –0.0082 | 0.482 | 0.258 | 0.229 | 0.54 | 0.48 | <0.001 | ||||||||||
| N-methyl-d-glutamic acid | 260 | 1382 | –0.0086 | –0.0006 | 0.110 | 0.197 | 0.033 | 1.78 a | 0.30 b | 0.009 | ||||||||||
| 1-Methyl-nicotinamide | 179 | 1155 | –0.0009 | –0.0004 | 0.015 | 0.005 | 0.002 | 0.31 | 0.11 | 0.011 | ||||||||||
For each identified metabolite, GC-MS characteristics (fragment mass, RI, retention index), loadings in principal components of Fig. 3, average response (resulting from five biological replications), and response ratios of each dark period to the normal photoperiod are presented.
Response ratios representing significant differences (P < 0.05) are indicated in bold, while significant differences between the relative responses measured after 24h and 72h darkness are indicated by different letters. The P-value represents the level of significance between the relative responses of each compound (ANOVA), when at least two average responses are >0.
Sucrose levels declined substantially during extended darkness, with a 60% reduction after 72h of darkness. Raffinose, d-melezitose, maltotriose, and d-glucose concentrations also decreased significantly with prolonged darkness (Table 1). In the organic acid group, significantly reduced concentrations of l-ascorbic acid, succinic acid, pyruvic acid, gluconic acid, glycolic acid, d-malic acid, and oxalic acid were found in nodules of dark-treated plants. Reduced concentrations of phosphates were also measured. O-Phospho-l-threonine, pyrophosphate, adenosine-S-monophosphate, and d-glucose-6-phosphate were significantly reduced in nodules of dark-treated plants; however, O-phosphocolamine accumulation was remarkably high (almost 20-fold). Putrescine, methyl-β-d-galactopyranoside, N-methyl-dl-glutamic acid, and 1-methyl-nicotinamide levels also declined significantly following extended darkness.
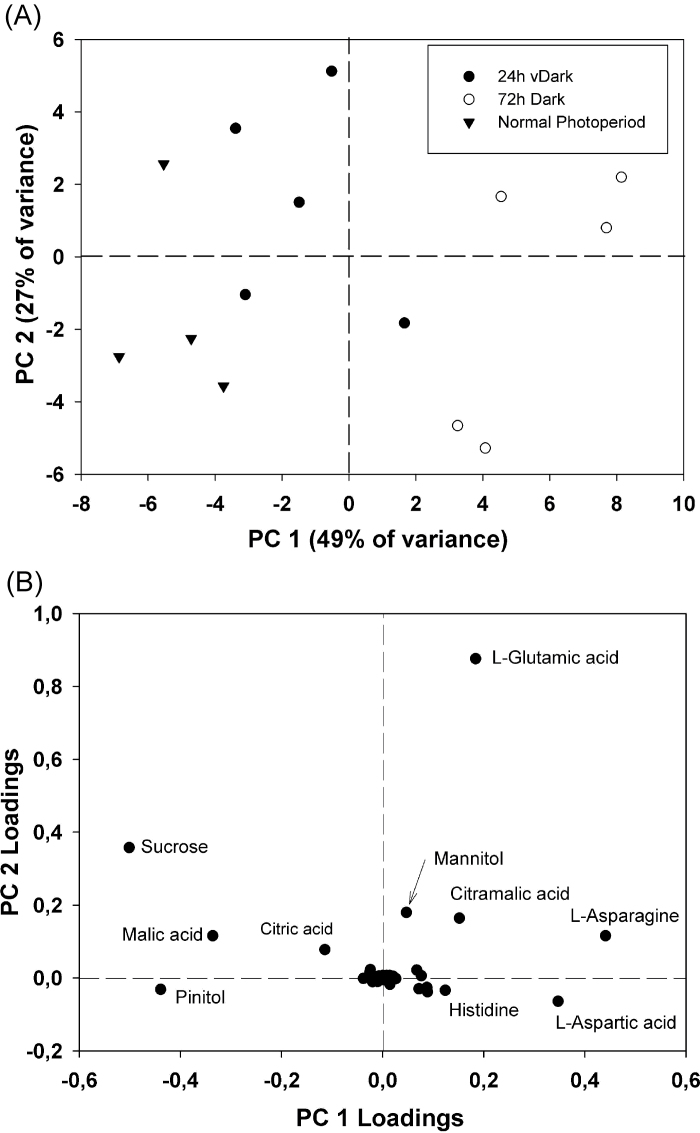
In most metabolite groups, significantly dark-affected compounds declined under darkness, with the highest reductions being observed after 72h of continuous darkness. PCA, which allows clustering of samples into groups, revealed strong differentiation in metabolite contents after 72h of continuous darkness. The first PCA component accounted for 49% of the variance in metabolite levels and separated samples of 72h darkness from those of 24h darkness and the normal photoperiod. According to the first component, 24h of darkness exhibited similarity to the normal photoperiod; however, the second component, which accounted for 27% of the variance, allowed distinction between samples of 24h darkness and the normal photoperiod (Fig. 2A).
Fig. 2.
Principal component analysis of GC-MS profiles of L. japonicus nodules harvested from plants grown under a normal photoperiod (n=4), and after the plants have been subjected to 24h and 72h of a continuous dark period (n=5). (A) Bi-plot of principal components 1 and 2 representing 76% of the total variance observed in the GC-MS profiles. (B) Loadings of individual indentified components showing the highest variability under the studied experimental conditions.
A remarkable exception to the trend of reduced metabolite levels following prolonged darkness was the high accumulation of certain amino acids. In this compound group, most of the identified metabolites were found at significantly higher concentrations under continuous darkness. These included dl-isoleucine, l-tryptophan, and l-histidine, which exhibited 25-, 24-, and 22-fold increased levels after 72h of darkness, respectively. Nine other amino acids were also found to accumulate under these conditions, with l-valine and l-homoserine presenting the highest accumulation. In addition, four amino acids (l-cysteine, l-ornithine, l-tyrosine, and l-methionine) were detected exclusively in nodules of dark-treated plants (Table 1). In PCA analysis, the first component was influenced most by three amino acids: l-asparagine, l-aspartic acid, and l-glutamic acid (Fig. 2B), which is indicative of the strong differentiation of amino acids under these conditions.
Transcriptional regulation of plant genes in nodules under carbon-limiting conditions
Gene transcript profiling was studied in nodules of plants exposed to the normal photoperiod, and extended dark for 24h and 72h. Gene transcripts were analysed by quantitative real-time PCR (qRT-PCR) using a platform containing 174 gene-specific primer pairs (Supplementary Table S1 at JXB online) for genes involved in various aspects of nodule metabolism (Ott et al., 2009). Most genes represented on the qRT-PCR platform code for enzymes involved in primary metabolic processes in nodules such as starch metabolism, carbohydrate metabolism, pentose phosphate cycle, tricarboxylic acid (TCA) cycle, glycolysis, CO2 fixation, ammonium assimilation, and amino acid metabolism.
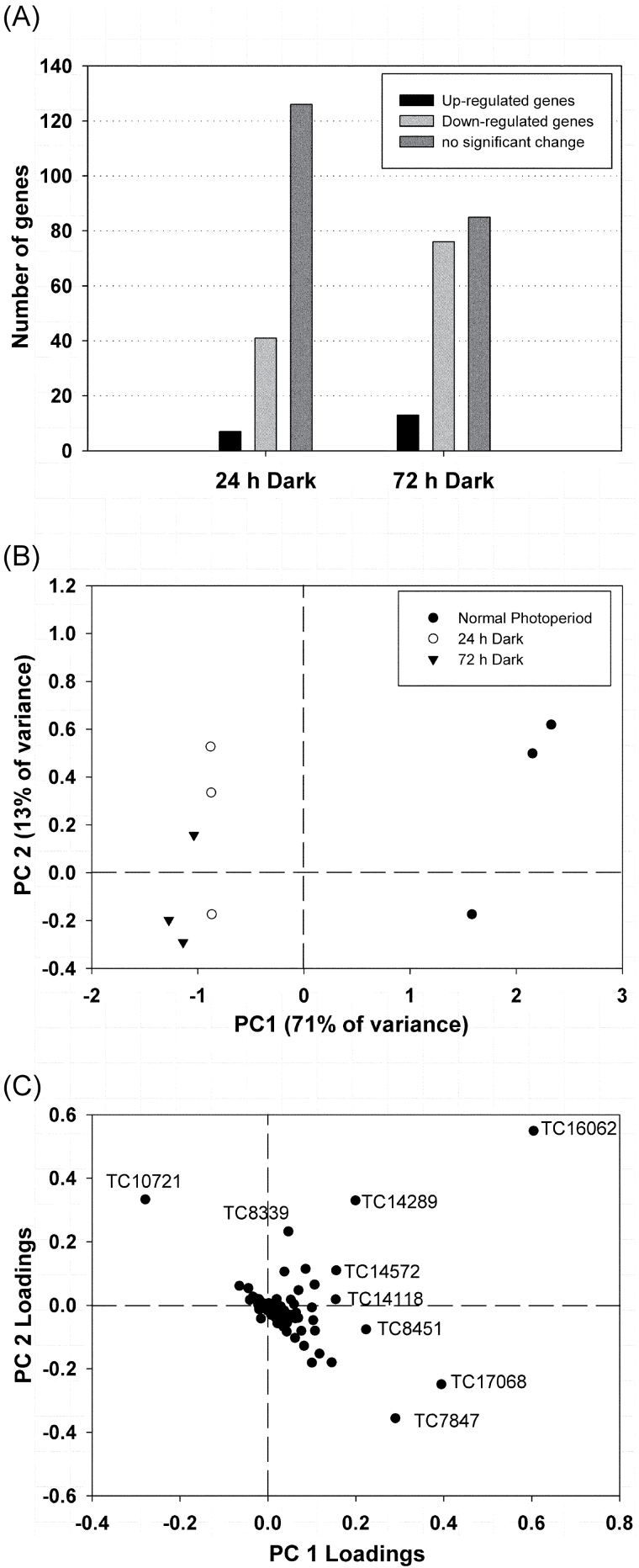
To identify changes in gene expression under photosynthetic carbon-limiting conditions, transcript levels were measured after 24h and 72h of continuous darkness and compared with transcript levels under normal photoperiod conditions. Although transcript levels of many genes represented on the qRT-PCR platform were not significantly different after the extended dark period compared with the control, ANOVA revealed 54 genes that were significantly up- or down-regulated after 24h of darkness. After 72h of extended darkness, a total of 89 genes showed significant differences in expression (P < 0.05) (Supplementary Table S2 at JXB online). The majority of the genes analysed exhibited reduced transcript levels under prolonged darkness, while only a few genes were found to be up-regulated (Fig. 3A).
Fig. 3.
Analysis of the gene expression profiling results of L. japonicus nodules harvested from plants grown under a normal photoperiod, and after the plants have been subjected to 24h and 72h of a continuous dark period (n=3). (A) Number of genes showing increased expression levels (ratio dark/normal photoperiod >2, Ρ < 0.05), decreased expression levels (ratio dark/normal photoperiod <0.5, Ρ < 0.05), or unaffected expression levels (Ρ > 0.05). (B) Bi-plot of principal components 1 and 2 representing 84% of the total variance observed in gene expression levels. (C) Loadings of individual gene transcripts showing the highest variability under the studied experimental conditions.
PCA analysis revealed great differentiation in gene expression after 24h and 72h of continuous darkness when compared with gene expression under the normal photoperiod. The first component accounted for 71% of the variance and allowed distinction of the normal photoperiod from extended darkness. According to the first component, 24h of darkness exhibited similarity to 72h of darkness; however, the second component, which accounted for 13% of the variance, allowed distinction of 24h and 72h of darkness (Fig. 3B).
Many gene transcripts were found to be >5-fold down-regulated in nodules after an extended dark period of 72h (dark/normal photoperiod ratio <0.2, P <0.05, Table 2). The majority of these genes are involved in starch and sugar metabolism, including two genes encoding ADP-glucose pyrophosphorylases, two genes encoding phosphoglucomutases, and one gene encoding sucrose synthase, which presented a 12.5-fold reduction in expression levels (Table 2). The great differentiation in the expression levels of this gene (TC7847) under extended darkness was also seen by the PCA analysis because it was among the genes that most influenced the first component (Fig. 3C). The expression of genes encoding glycolytic enzymes was also highly reduced after 72h of darkness. These included genes for two enolases and a pyruvate decarboxylase.
Table 2.
Genes highly down-regulated in nodules after extended dark periods.
| Gene | Lotus gene ID | Expression ratio versus control | P-value | |
|---|---|---|---|---|
| 24h/N. Ph. | 72h/N. Ph. | |||
| Starch metabolism | ||||
| Soluble starch synthase {Arabidopsis thaliana} | TC15963 | 0.34 | 0.17 | 0.024 |
| ADP-glucose pyrophosphorylase small subunit PvAGPS1 | TC8396 5 end | 0.08 | 0.02 | 0.004 |
| ADP-glucose pyrophosphorylase small subunit CagpS1 | TC8756 3 end | 0.06 | 0.03 | 0.011 |
| Other carbohydrate metabolism | ||||
| Trehalose-6-phosphate synthase homologue | TC14996 | 0.12 | 0.03 | 0.004 |
| Phosphoglucomutase, cytoplasmic | TC14676 | 0.14 | 0.09 | 0.025 |
| Phosphoglucomutase, cytoplasmic | TC14675 | 0.40 | 0.07 | 0.036 |
| Glucose-6-phosphate isomerase | TC17137 | 0.28 | 0.20 | 0.033 |
| Sucrose synthase 1 | TC7847 | 0.16 | 0.08 | 0.004 |
| Glycolysis | ||||
| Enolase | TC14118 | 0.36 | 0.13 | 0.004 |
| Enolase | TC16760 | 0.73 | 0.17 | 0.004 |
| Pyruvate decarboxylase II | TC7922 | 0.27 | 0.17 | <0.001 |
| Pentose phosphate pathway | ||||
| Glucose-6-phosphate 1-dehydrogenase | TC8851 | 0.29 | 0.06 | 0.004 |
| Transaldolase | TC11589 | 0.02 | 0.01 | 0.011 |
| Transaldolase ToTAL2 | TC10369 | 0.18 | 0.08 | 0.011 |
| TCA cycle | ||||
| Cytosolic malate dehydrogenase (1.1.1.37) | TC7834 | 0.28 | 0.15 | 0.011 |
| Aconitate hydratase, cytoplasmic | TC14371 | 0.37 | 0.16 | 0.004 |
| CO2 metabolism | ||||
| Carbonic anhydrase LjCA1 | TC14306 | 0.01 | 0.00 | 0.004 |
| CA α-type | Ljnest13e5 | 0.33 | 0.13 | <0.001 |
| CA α-type 2 | Ljnest20a7rc | 0.20 | 0.08 | 0.004 |
| Phosphoenolpyruvate carboxylase | TC7830 | 0.60 | 0.11 | 0.009 |
| Phosphoenolpyruvate carboxylase kinase | TC14602 | 0.19 | 0.07 | 0.004 |
| Ribulose bisphosphate carboxylase/oxygenase activase | TC14081 | 0.17 | 0.03 | 0.012 |
| Ammonium assimilation | ||||
| Glutamine synthetase, cytosolic isozyme | TC8035 | 0.39 | 0.13 | 0.007 |
| Glutamate dehydrogenase | TC10844 | 0.20 | 0.16 | 0.008 |
| Other aminoacid metabolism | ||||
| Alanine:glyoxylate aminotransferase 2 homologUE | TC9107 | 0.48 | 0.10 | 0.016 |
| Histidinol dehydrogenase | TC10151 | 0.17 | 0.06 | 0.004 |
| Serine acetyltransferase | TC17048 | 0.19 | 0.05 | 0.004 |
| Cysteine synthase | TC14692 | 0.15 | 0.08 | 0.011 |
| Plastidic cysteine synthase 1 | TC17110 | 0.03 | 0.07 | <0.001 |
| Threonine dehydratase/deaminase | TC18952 | 0.20 | 0.17 | <0.001 |
| Aspartokinase-homoserine dehydrogenase (HDH) | TC9572 | 0.07 | 0.01 | 0.004 |
| Threonine synthase | TC9527 | 0.20 | 0.07 | 0.004 |
| Pantoate-beta-alanine ligase | TC12836 | 0.19 | 0.14 | <0.001 |
| Similarity to ornithine cyclodeaminase | TC11292 | 0.23 | 0.08 | 0.001 |
| Prolyl 4-hydroxylase | TC15896 | 0.29 | 0.11 | <0.001 |
| Prolyl 4-hydroxylase, alpha subunit-like protein | TC11925 | 0.32 | 0.20 | 0.005 |
| Branched-chain amino acid aminotransferase-like protein | TC16062 | 0.13 | 0.01 | 0.004 |
| Other metabolism | ||||
| Chalcone reductase | TC8351 | 0.01 | 0.00 | 0.011 |
| Putative flavanone 3-hydroxylase | TC7931 | 0.40 | 0.14 | 0.004 |
| Acidic endochitinase | TC14167 | 0.16 | 0.04 | <0.001 |
| Thiazole biosynthetic enzyme precursor | TC14068 | 0.06 | 0.02 | 0.004 |
| Cytochrome P450 | TC15466 | 0.33 | 0.04 | 0.004 |
| Cytochrome P450 | TC8343 | 0.05 | 0.01 | <0.001 |
| N-Hydroxycinnamoyl/benzoyltransferase-like protein | TC8570 | 0.23 | 0.08 | 0.004 |
| Isoliquiritigenin 2’-O-methyltransferase | TC14525 | 0.35 | 0.17 | <0.001 |
| 4-Coumarate:CoA ligase 2 | TC7977 | 0.47 | 0.16 | 0.003 |
| Laccase | TC17617 | 0.04 | 0.02 | 0.025 |
| Pyrroline-5-carboxylate reductase | TC15604 | 0.25 | 0.17 | <0.001 |
Gene expression in L. japonicus nodules harvested from plants grown under normal photoperiod (N. Ph.), or subjected to 24h and 72h continuous darkness, respectively. Transcript levels of these genes decreased significantly (<5-fold, P < 0.05) under extended dark conditions and are marked in bold. The numbers indicate ratios of gene expression after 24h and 72h continuous darkness versus expression in the normal photoperiod (control). The P-value represents the level of significance between the relative gene expression levels (ANOVA).
Regarding CO2 fixation, six genes were found to be >5-fold down-regulated, including three genes encoding carbonic anhydrases and one gene encoding phosphoenolpyruvate carboxylase, which together fix CO2 into oxaloacetate. Moreover, a 6.5-fold reduction in expression levels was observed in one gene encoding malate dehydrogenase, which uses oxaloacetate as a substrate to produce malate, the main carbon source for bacteroid metabolism and nitrogen fixation.
In the ammonium assimilation metabolic pathway, two genes encoding glutamine synthetase and glutamate dehydrogenase presented a remarkable reduction in their expression levels after both 24h and 72h of continuous darkness (Table 2). However, one gene encoding an NADH glutamate dehydrogenase and one gene encoding an asparagine synthase were almost 40- and 20-fold up-regulated, respectively, after 72h of darkness (Table 3).
Table 3.
Genes highly induced in nodules after extended dark periods.
| Gene | Lotus gene ID | Expression ratio versus control | P-value | |
|---|---|---|---|---|
| 24h/N. Ph. | 72h/N. Ph. | |||
| Other carbohydrate metabolism | ||||
| Trehalase 1 | TC18454 | 1.88 | 2.97 | 0.007 |
| Beta-fructofuranosidase | TC16805 | 1.76 | 3.56 | 0.039 |
| Pentose phosphate pathway | ||||
| Ribulose-phosphate 3-epimerase, chloroplast precursor | TC7925 | 3.27 | 2.09 | 0.004 |
| TCA cycle | ||||
| Aconitase (aconitate hydratase) (citrate hydrolyase) | TC14176 | 8.80 | 3.80 | 0.002 |
| Cytosolic aconitase | TC15109 | 3.09 | 2.06 | 0.011 |
| Ammonium assimilation | ||||
| NADH glutamate dehydrogenase | TC10856 | 6.97 | 40.99 | 0.004 |
| Asparagine synthase | TC14104 | 12.16 | 19.19 | 0.031 |
| Other amino acid metabolism | ||||
| Ornithine aminotransferase | TC8388 | 6.31 | 7.78 | 0.007 |
| Proline dehydrogenase | TC7863 | 4.51 | 10.84 | <0.001 |
| Other metabolism | ||||
| 4-Hydroxyphenylpyruvate dioxygenase (4HPPD) | TC8682 | 3.51 | 4.93 | 0.037 |
| Cytochrome P450, isoflavone synthase | TC14262 | 1.76 | 3.16 | 0.013 |
| Chalcone isomerase | NP591666 | 4.05 | 4.47 | <0.001 |
| Isoflavone reductase | TC7899 | 1.89 | 5.52 | <0.001 |
| Trans-cinnamate 4-monooxygenase | TC14933 | 1.57 | 2.83 | 0.046 |
| Putative oxidoreductase | TC14685 | 0.81 | 4.10 | 0.026 |
| NAD-dependent sorbitol dehydrogenase | TC17752 | 8.46 | 3.33 | 0.004 |
Gene expression in L. japonicus nodules harvested from plants grown under normal photoperiod (N. Ph.), or subjected to 24h and 72h continuous darkness, respectively. Transcript levels of these genes increased significantly (>2-fold, P < 0.05) under extended dark conditions and are marked in bold. The numbers indicate ratios of gene expression after 24h and 72h continuous darkness versus expression in the normal photoperiod (control). The P-value represents the level of significance between the relative gene expression levels (ANOVA).
Several genes involved in amino acid metabolism were down-regulated after prolonged darkness. Thirteen genes exhibited >5-fold reduction in expression levels, while eight of these had transcript levels that were >10-fold lower than in the control. These included a gene encoding serine acetyltransferase and two genes encoding cysteine synthase, which are involved in cysteine biosynthesis, two genes encoding aspartokinase-homoserine dehydrogenase and threonine synthase, which are involved in threonine biosynthesis, and one gene encoding ornithine cyclodeaminase and two genes encoding prolyl 4-hydroxylase, which are involved in proline metabolism (Table 2). However, in proline metabolism, a remarkable increase was detected in the expression levels of a gene encoding an ornithine aminotransferase and another gene encoding a proline dehydrogenase (Table 3). Significant reduction was observed in the expression levels of many genes involved in flavonoid biosynthesis, lignin biosynthesis, and other aspects of secondary metabolism, including genes encoding chalcone reductase, flavanone 3-hydroxylase, isoliquiritigenin 2’-O-methyltransferase, laccase, 4-coumarate, and cytochrome P450 (Table 2). However, there are a number of gene transcripts involved in secondary metabolism which were found to be highly up-regulated (Table 3). The fact that some genes encoding enzymes involved in flavonoid biosynthesis have been detected to be up-regulated and some others down-regulated raises questions about the flavonoid changes in nodules under such conditions.
Dark CO2 fixation is limited under continuous darkness
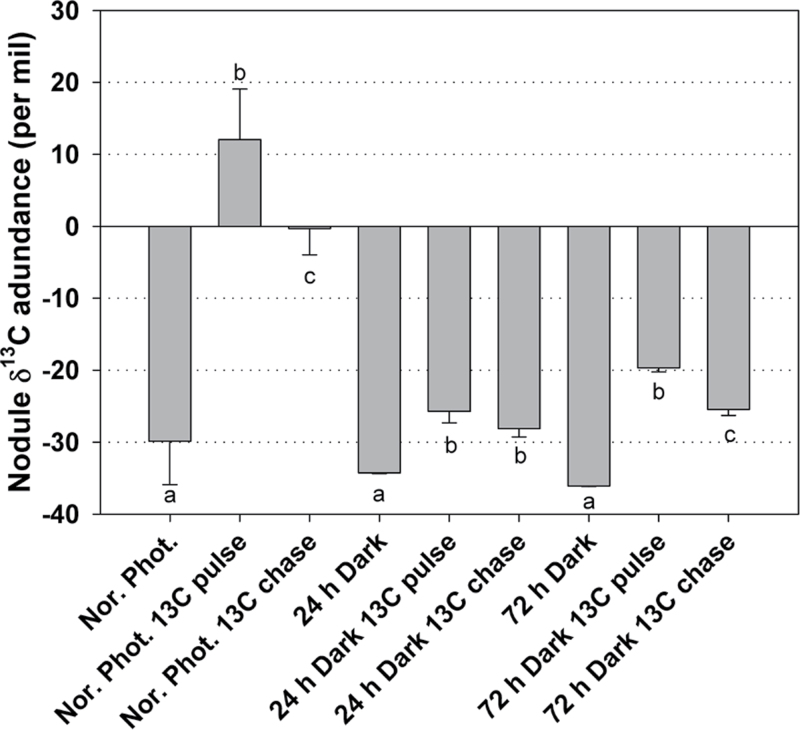
The observation that the organic acid concentration and the levels of several transcripts involved in dark CO2 fixation decreased significantly in nodules during photosynthetic carbon deficiency prompted the study of CO2 fixation using direct Na213CO2 labelling of the nodulated root system. Two hours of pulse 13C labelling under the normal photoperiod resulted in a substantial increase in the 13C composition of nodules (Fig. 4), as expected (Fotelli et al., 2011). However, such an increase in 13C composition of nodules was not observed in plants subjected to prolonged darkness. Nodule δ13C abundance of plants subjected to 24h and 72h of continuous darkness was only marginally increased after 2h of 13C labelling and a 1h chase period (Fig. 4).
Fig. 4.
δ13C abundance in nodules of L. japonicus plants grown under a normal photoperiod, and after 24h and 72h of continuous dark periods, subjected to 2h of 13C pulse labelling, and 1h of chase after the completion of the 2h 13C pulse labelling. Bars represent means (±SE) of six biological replications (three plants each). Within each light/dark condition, the statistical analysis refers to the comparison between the different labelling treatments. Significant differences (P < 0.05) are indicated by different letters.
Changes in total protein and amino acid content in symbiotic and non-symbiotic organs under extended dark
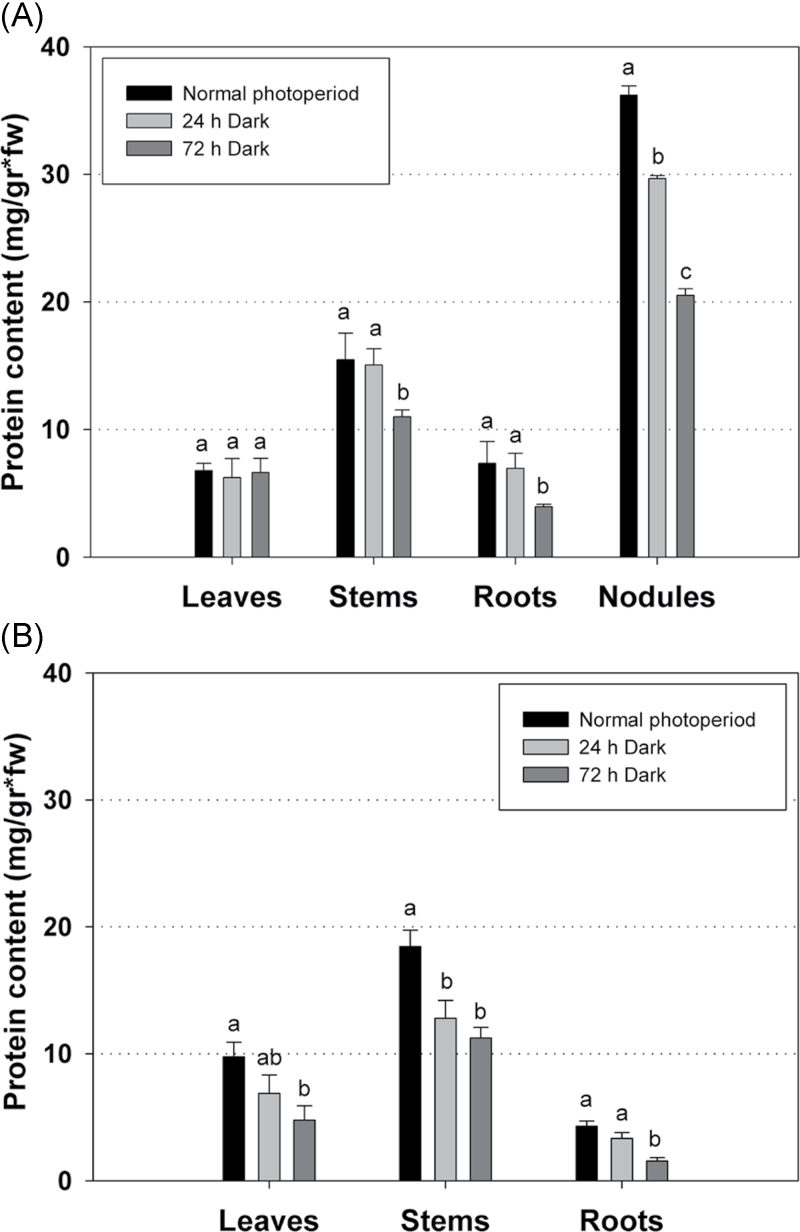
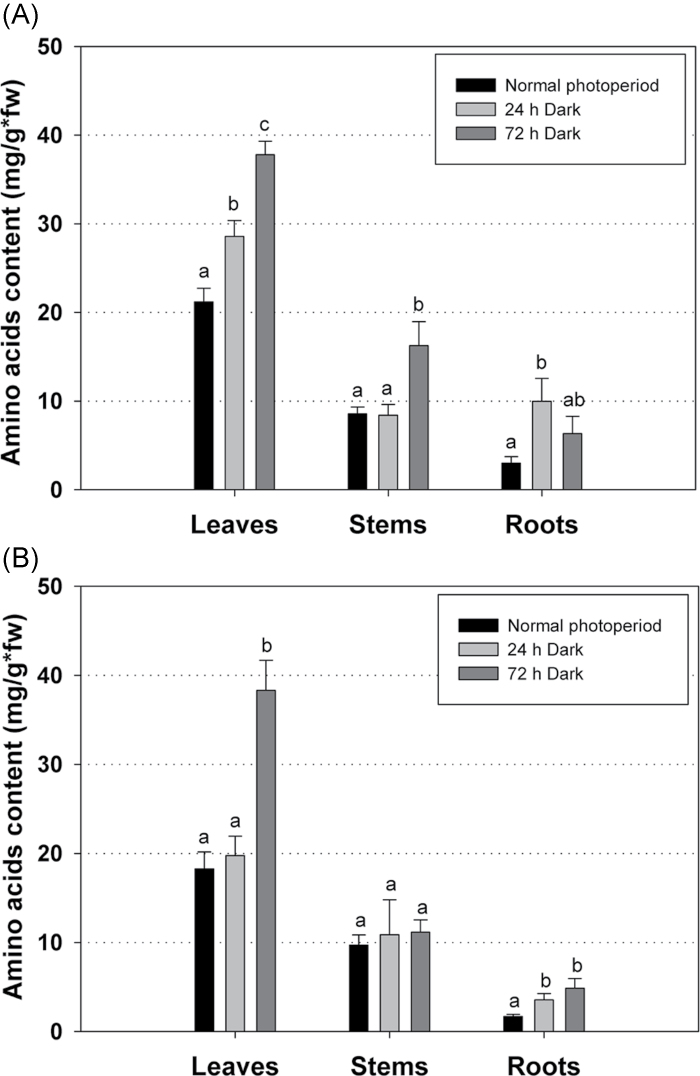
To test the hypothesis that increased amino acid content of nodules under photosynthetic carbon-limiting conditions was due to protein degradation, the total soluble protein content was measured in both symbiotic and non-symbiotic organs of L. japonicus plants subjected to prolonged darkness. Furthermore, in order to test whether the presence of nodules systemically affects the remobilization of proteins under these conditions, both 30-day-old inoculated plants harbouring nitrogen-fixing nodules and uninoculated plants, grown under the normal photoperiod or subjected to either 24h or 72h of extended dark, were tested. Dark treatment resulted in significantly reduced total protein levels in nodules after both 24h and 72h of darkness, confirming the hypothesis of protein degradation under such conditions (Fig. 5A). Reduced protein levels were also observed in stem and root of inoculated plants after 72h of darkness. However, the total protein levels in leaves of inoculated plants remained unaltered, indicating that net protein degradation did not occur under these conditions in this organ (Fig. 5A). In contrast to inoculated plants, a reduced protein content was observed in all organs (leaves, stem, and root) of uninoculated plants after both 24h and 72h of extended darkness (Fig. 5B). These results indicate that nodules provided amino acids to other plant organs and thereby prevented protein degradation in leaves during darkness. To confirm this result, the amino acid concentration was quantified in leaves, stems, and roots of inoculated and uninoculated 30-day-old L. japonicus plants that had been subjected to extended dark periods. Amino acids accumulated in all organs under these conditions, including leaves of inoculated (Fig. 6A) and uninoculated plants (Fig. 6B) after 72h of dark treatment. Under carbon limitation, the elevated levels of amino acids in leaves of nodulated plants, where net protein degradation did not take place, may be attributed to amino acid transfer from nodules (Fig. 7).
Fig. 5.
Total protein content in various organs of (A) inoculated and (B) uninoculated L. japonicus plants during growth under a normal photoperiod and after 24h and 72h of continuous darkness. Bars represent means (±SE) of four biological replications. Within each graph, the statistical analysis refers to the comparison between the protein content of each organ. Significant differences (P < 0.05) are indicated by different letters.
Fig. 6.
Total amino acid content in leaves, stems, and roots of (A) inoculated and (B) uninoculated L. japonicus plants during growth under a normal photoperiod and after 24h and 72h of continuous darkness. Bars represent means (±SE) of four biological replications. Within each graph, the statistical analysis refers to the comparison between the total amino acid content of each organ. Significant differences (P < 0.05) are indicated by different letters.
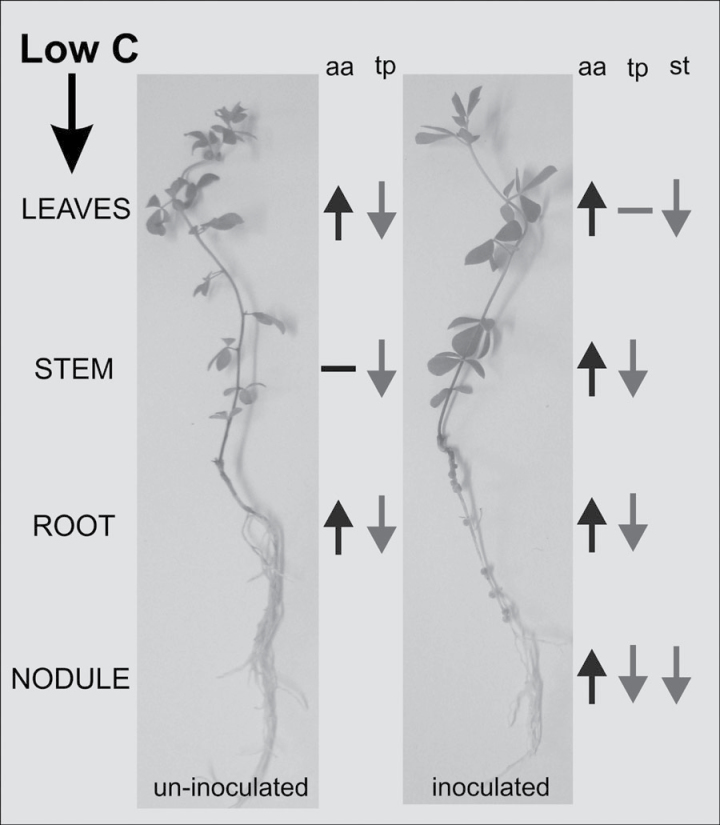
Fig. 7.
Diagram of the systemic changes observed in uninfected versus infected L. japonicus plants under carbon-limited conditions. The high total amino acid (aa) concentration observed in all plant organs is accompanied by a reduction in total protein (tp) concentration, denoting protein degradation. In infected plants, the amino acids mobilized by protein degradation in nodules and other organs may have been a source of fixed carbon for leaves when photosynthesis was blocked, preventing protein degradation in these leaves. These results arise from the analysis of plants subjected to 72h of continuous darkness, when starch (st) reserves were completely exhausted.
Discussion
Photosynthetic carbon deficiency affects several aspects of nodule metabolism
Nodulation results in dramatic differentiation and reprogramming of multiple aspects of primary and secondary metabolism in legumes. Among the metabolic pathways up-regulated during development in L. japonicus nodules, several were involved in C metabolism, including starch and sucrose metabolism, glycolysis, and dark CO2 fixation (Colebatch et al., 2004; Fotelli et al., 2011), demonstrating the strong need for carbon and energy for nodule organogenesis and function.
In an attempt to study the effects of photosynthetic carbon deficiency on nodule metabolism, transcriptomic and metabolomic analyses were carried out in L. japonicus plants exposed to prolonged darkness. The length of the dark periods applied to the plants was chosen according to nodule starch content and nitrogenase activity measurements. Lotus japonicus exhibited a decline in nitrogenase activity of 85% after 24h of continuous darkness (Fig. 1) but starch was still detectable in nodules, while after 72h of darkness, nodule starch content was exhausted completely (Supplementary Fig. S1 at JXB online) and nitrogenase activity was decreased by 97% (Fig. 1). Although limited nitrogenase activity can be explained by different hypotheses (including interruption of photosynthesis and N feedback regulation), this dramatic decline in nitrogenase activity was attributed primarily to sugar deprivation, as it was partially restored after transferring the plants to the normal photoperiod for 48h (Fig. 1). Consequently, even after 72h of continuous darkness the effect of carbon starvation is still reversible.
Metabolite and transcript profiling in the present study revealed that limited photosynthate supply resulted in substantial changes, both in gene expression and in metabolite content, in nodules of dark-treated L. japonicus plants (Fig. 7). Interestingly, the results show that just 24h of extended darkness was sufficient to trigger most of the variation observed in transcript levels (Fig. 3B). Changes in metabolite levels occurred more gradually during extended darkness (Fig. 2A), indicating that changes in nodule metabolism due to photosynthate limitation are mainly under transcriptional regulation, although the observed latency in metabolite changes may, in part, be accounted for post-transcriptional regulation as well.
Lotus japonicus nodules showed a remarkable decline in sucrose content (Table 1), accompanied by a significant reduction of sucrose synthase transcript levels under continuous darkness (Table 2). Similarly, sucrose synthase enzymatic activity decreased to a great extent in nodules of common bean plants subjected to 2 d of darkness (Gogorcena et al., 1997). Although transcript levels of several invertase genes were unaltered (Supplementary Table S2 at JXB online), significant induction of a gene encoding a β-fructofuranosidase (Table 3) indicates that specific members of the invertase family could play a role in the hydrolysis of sucrose under dark-stress conditions.
After 72h of continuous darkness, the observation that the starch content was exhausted (Supplementary Fig. S1 at JXB online) is in accordance with the observation that three genes involved in starch synthesis (one gene coding for starch synthase and two for ADP-glucose pyrophosphorylase) were substantially down-regulated (Table 2). The other genes of starch metabolism tested did not show a significant differentiation between treatments, neither did the genes involved in starch biosynthesis nor the genes involved in starch degradation (Supplementary Table S2).
Like sucrose, the levels of a number of metabolites in the group of sugars decreased in nodules of dark-treated plants (Table 1). Glucose became undetectable after 72h of continuous darkness, and in addition to that a down-regulation was observed in genes involved in glucose breakdown in nodules (see below).
Interestingly, during photosynthate limitation, trehalose content in nodules remained unaffected, although a plant trehalase isoform, which is likely to be involved in trehalose degradation, was found to be significantly up-regulated during prolonged darkness (Table 3). Moreover, transcripts of two plant genes coding for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase, which are involved in trehalose synthesis, were down-regulated during extended darkness. Taken together, these results indicate that most of the measured trehalose may have been in the bacteroids. Trehalose is a well-known osmo-protectant and its biosynthesis has been linked to competitiveness of rhizobia during nodulation (Domínguez-Ferreras et al., 2009). The present data indicate that trehalose, a relatively abundant disaccharide in L. japonicus nodules (Desbrosses et al., 2005), cannot be utilized as an alternative carbon source during photosynthate limitation. This result attributes different roles to trehalose in nodules, probably involving adaptation to osmotic stress.
Induction of a sorbitol dehydrogenase gene in nodules is consistent with greater polyol biosynthesis in this plant organ (Desbrosses et al., 2005). With the exception of mannitol, the levels of polyols were not significantly affected by photosynthate limitation. Interestingly, mannitol content was found to be >3-fold higher after 24h of prolonged darkness. In contrast, the expression of an NADPH-dependent mannose-6-phosphate reductase, involved in mannitol biosynthesis, was found to be significantly down-regulated, whereas transcripts of mannitol and sorbitol dehydrogenase isoforms, which are involved in mannitol catabolism, were found to be significantly up-regulated. This discrepancy again may point to major participation of bacteroids in compatible solute biosynthesis and storage. Moreover, pinitol, a C-rich metabolite which is abundant in nodules (Fougère et al., 1991; Desbrosses et al., 2005), could also not be mobilized during carbon starvation in nodules, indicating that its main role is not in carbon and energy metabolism. These results suggest that polyol biosynthesis in nodules plays an alternative role, possibly in osmoregulation.
Concomitant with the depletion of sugars, a considerable number of genes involved in carbohydrate metabolism and glycolysis were found to be strongly down-regulated following dark treatment, including genes coding for phosphoglucomutase, glucose-6P isomerase, fructokinase, enolase, triosephosphate isomerase, and pyruvate decarboxylase (Table 2; Supplementary Table S2 at JXB online).
Sugar depletion was followed by depletion of a number of organic acids: dehydroascorbic acid, shikimic acid, ascorbic acid, succinic acid, pyruvic acid, gluconic acid, glycolic acid, d-malic acid, and oxalic acid (Table 1). The accumulation of isocitric acid (Table 1), which was found to be dark specific, can be attributed to the up-regulation of two genes coding for aconitate hydratase (Table 3).
The observation that three genes coding for both α- and β-type carbonic anhydrases and one gene coding for phosphoenolpyruvate carboxylase, which together fix CO2 to oxaloacetate, are highly down-regulated in dark treatment (Table 2) correlates well with the observed reduction in 13C incorporation in nodules under extended dark conditions (Fig. 4). Similarly, when L. japonicus plants were subjected to prolonged darkness and subsequent illumination, the mRNA levels and the activity of PEPC decreased and then recovered in nodules, suggesting that PEPC transcripts and activity are regulated by the amount of photosynthate transported from leaves to nodules (Nakagawa et al., 2003). The limited activity of this enzyme under extended darkness probably explains the high PEP accumulation observed after both 24h and 72h of darkness (Table 1). Limited dark CO2 fixation also reflects on TCA accumulation, which supports N2 assimilation in bacteroids (for a review, see Lodwig and Poole, 2003). As suspected, reduced amounts of malate were detected in the present metabolomic analysis (Table 1), coinciding with a significant decline in the expression levels of a gene coding for cytosolic malate dehydrogenase, TC7834 (Table 2). Similarly, in soybean, extended darkness was accompanied by a collapse of the malate level (Vauclare et al., 2010).
l-Glutamine, one of the two main amino acids assimilating ammonium, and the transcript levels of a gene encoding glutamine synthetase (TC8035) were significantly lower in nodules of dark-treated plants compared with the control (Tables 1, 2). In bean nodules, both glutamine synthetase and glutamate synthase activities decreased after plants were subjected to 1 d of darkness (Gogorcena et al., 1997). Moreover, the accumulation of l-alanine was substantially decreased in prolonged darkness (Table 1). Alanine is a possible alternative product of N2 fixation in the bacteroid, under some conditions (for a review, see Day et al., 2001). Although ammonia is the major product of N2 fixation when nitrogenase activity is optimized (Li et al., 2002), soybean bacteroids were shown to synthesize and secrete alanine under conditions of oxygen deprivation in a closed system (Waters et al., 1998). As in L. japonicus, nodules of dark-treated soybean plants showed depletion of glutamine and asparagine, and disappearance of alanine (Vauclare et al., 2010).
The above results are indicative of a limited carbon supply to nodules of L. japonicus plants under extended dark periods, resulting in dramatic nitrogenase activity reduction and limited nitrogen assimilation (Fig. 7).
Nodules can act as source organs when photosynthesis is abolished
Plants, like other organisms, have developed sophisticated mechanisms for recycling intracellular constituents under nutrient-limiting conditions. Autophagy, for example, has been suggested to play a central role in several stress responses including nutrient recycling during starvation (for a review, see Han et al., 2011). In the present work, photosynthetic carbon starvation resulted in significant decreases in total protein content in both symbiotic and non-symbiotic organs of L. japonicus plants (Fig. 5). Journet et al. (1986) reported that protein degradation occurs as a consequence of sucrose starvation and proposed that this protein degradation is due to autophagy. In dark-stressed bean nodules, total soluble protein decreased by 13% and 54% after 2 d and 4 d of darkness, respectively (Gogorcena et al., 1997). Similarly, in Arabidopsis, carbon starvation triggered a rapid degradation of cellular proteins and the up-regulation of autophagy-related genes (Rose et al., 2006). In addition, disruption of ATG (autophagy-related genes) proteins resulted in Arabidopsis plants that were hypersensitive to C starvation induced by extended darkness (Thompson et al., 2005).
Thus, the accumulation of amino acids observed in L. japonicus nodules after both 24h and 72h of extended darkness (Table 1) could be attributed to protein degradation (Fig. 5). Taylor et al. (2004) suggested that catabolism of branched-chain amino acids is important in providing an alternative carbon sources for oxidative phosphorylation and energy production during carbon starvation. The 40-fold increase in transcript level for NADH-dependent glutamate dehydrogenase (GDH) (TC10856) found after 72h of darkness (Table 3) may reflect the enzyme’s role in the breakdown of amino acids to provide alternative carbon skeletons for energy production. It has been hypothesized that glutamate catabolized by GDH enters the TCA cycle as an alternative C source during C starvation (Terce-Laforgue et al., 2004a, b; Skopelitis et al., 2006). In Arabidopsis, NADH-dependent glutamate dehydrogenase was suggested to be a key enzyme in the breakdown of several amino acids under carbon deficiency (Miyashita and Good, 2008). Total protein declined in most organs of nodulated and non-nodulated plants following dark treatment (Fig. 5), while amino acid levels increased (Fig. 6), which indicated that protein degradation in these organs is probably the main source of the accumulating amino acids. Leaves of nodulated plants represented an exception: elevated amino acids levels (Fig. 6) were not accompanied by reduced total protein (Fig. 5). Thus, amino acids mobilized by protein degradation in nodules and other organs may have been a source of fixed carbon for leaves when photosynthesis was blocked, preventing protein degradation in these leaves (Fig. 7).
In conclusion, extended darkness was used as a tool to study the metabolic responses of nodules under photosynthetic C limitation, combining for the first time both transcript profiling and metabolomic analysis. From the data obtained, it is proposed that nodulation of the model legume L. japonicus results in systemic changes in the responses of the plant to photosynthate limitation triggered by the extended dark period, as nodules appeared to act as a source of both C and N, limiting protein degradation in the leaves under non-photosynthetic conditions. Although the xylem sap of L. japonicus could not be directly measured due to technical limitations, amino acids accumulating in nodules as a result of localized protein degradation are likely to have been transported to the shoot where they served as an alternative carbon source. Future work will aim to quantify the contribution of specific compounds exported from nodules to the total plant C budget under photosynthate limitation, using alternative legume species that allow the direct analysis of xylem sap under these conditions.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Starch accumulation in leaves and nodules of L. japonicus plants.
Figure S2. Ethylene production during the acetylene reduction assay.
Table S1. Primer pairs used in quantitative real-time PCRs.
Table S2. Gene expression in nodules after prolonged darkness.
Acknowledgements
This work was partially funded by a joint DAAD and Greek State Scholarship Foundation project and by The Samuel Roberts Noble Foundation.
References
- Barsch A, Carvalho HG, Cullimore JV, Niehaus K. 2006. GC-MS based metabolite profiling implies three interdependent ways of ammonium assimilation in Medicago truncatula root nodules. Journal of Biotechnology 127, 79–83 [DOI] [PubMed] [Google Scholar]
- Bethlenfalvay GJ, Phillips DA. 1977. Effect of light intensity on efficiency of carbon dioxide and nitrogen reduction in Pisum sativum L. Plant Physiology 60, 868–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72, 248–254 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK. 2004. Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus . The Plant Journal 39, 487–512 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK. 2002. Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Molecular Plant-Microbe Interactions 15, 411–420 [DOI] [PubMed] [Google Scholar]
- Day DA, Copeland L. 1991. Carbon metabolism and compartmentation in nitrogen-fixing legume nodules. Plant Physiology and Biochemistry 29, 185–201 [Google Scholar]
- Day DA, Poole PS, Tyerman SD, Rosendahl L. 2001. Ammonia and amino acid transport across symbiotic membranes in nitrogen-fixing legume nodules. Cellular and Molecular Life Sciences 58, 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses GG, Kopka J, Udvardi MK. 2005. Lotus japonicus metabolic profiling. Development of gas chromatography–mass spectrometry resources for the study of plant–microbe interactions. Plant Physiology 137, 1302–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Ferreras A, Soto MJ, Pérez-Arnedo R, Olivares J, Sanjuán J. 2009. Importance of trehalose biosynthesis for Sinorhizobium meliloti osmotolerance and nodulation of alfalfa roots. Journal of Bacteriology 191, 7490–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenbaum S, Mengel K. 1979. The effect of reduced light intensity and sub-optimal potassium supply on N2 fixation and N turnover in Rhizobium-infected lucerne. Physiologia Plantarum 45, 245–249 [Google Scholar]
- Flemetakis E, Efrose RC, Ott T, Stedel C, Aivalakis G, Udvardi MK, Katinakis P. 2006. Spatial and temporal organization of sucrose metabolism in Lotus japonicus nitrogen-fixing nodules suggests a role for the elusive alkaline/neutral invertase. Plant Molecular Biology 62, 53–69 [DOI] [PubMed] [Google Scholar]
- Fotelli MN, Tsikou D, Kolliopoulou A, Aivalakis G, Katinakis P, Udvardi MK, Rennenberg H, Flemetakis E. 2011. Nodulation enhances dark CO2 fixation and recycling in the model legume Lotus japonicus . Journal of Experimental Botany 62, 2959–2971 [DOI] [PubMed] [Google Scholar]
- Fougère F, Le Rudulier D, Streeter JG. 1991. Effects of salt stress on amino acid, organic acid, and carbohydrate composition of roots, bacteroids, and cytosol of alfalfa (Medicago sativa L). Plant Physiology 96, 1228–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin DE, Lipscombe R, Fedorova E, Millar AH, Mann A, Atkins CA, Smith PMC. 2003. Dual intracellular localization and targeting of aminoimidazole ribonucleotide synthetase in cowpea. Plant Physiology 131, 1033–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogorcena Y, Gordon AJ, Escuredo PR, Minchin FR, Witty JF, Moran JF, Becana M. 1997. N2 fixation, carbon metabolism, and oxidative damage in nodules of dark-stressed common bean plants. Plant Physiology 113, 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJ, Minchin FR, James CL, Komina O. 1999. Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiology 120, 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJ, Ougham HJ, James CL. 1993. Changes in the levels of gene transcripts and their corresponding proteins in nodules of soybean plants subjected to dark-induced stress. Journal of Experimental Botany 44, 1453–1460 [Google Scholar]
- Han S, Yu B, Wang Y, Liu Y. 2011. Role of plant autophagy in stress response. Protein and Cell 2, 784–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. 1992. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. The Plant Journal 2, 487–496 [Google Scholar]
- Hardy RWF, Burns RC, Holsten RD. 1973. Application of acetylene–ethylene assay for measurement of nitrogen fixation. Soil Biology and Biochemistry 5, 47–81 [Google Scholar]
- Hardy RWF, Havelka UD. 1976. Photosynthate as a major factor limiting nitrogen fixation by field-grown legumes with emphasis on soybeans. In: Nutman PS, ed. Symbiotic nitrogen fixation. Cambridge: Cambridge University Press; 421–439 [Google Scholar]
- Horst I, Welham T, Kelly S, Kaneko T, Sato S, Tabata S, Parniske M, Wang TL. 2007. TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiology 144, 806–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannetta PPM, de Lorenzo C, James EK, Fernandez-Pascual M, Sprent JI, Lucas MM, Witty JF, de Felipe MR, Minchin FR. 1993. Oxygen diffusion in lupin nodules I. Visualization of diffusion barrier operation. Journal of Experimental Botany 44, 1461–1467 [Google Scholar]
- Jeong J, Suh S, Guan C, Tsay YF, Moran N, Oh CJ, An CS, Demchenko KN, Pawlowski K, Lee Y. 2004. A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiology 134, 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet E, Bligny R, Douce R. 1986. Biochemical changes during sucrose deprivation in higher plant cells. Journal of Biological Chemistry 261, 3193–3199 [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, et al. 2005. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21, 1635–1638 [DOI] [PubMed] [Google Scholar]
- Lawn RJ, Brun WA. 1974. Symbiotic nitrogen fixation in soybeans. I. Effect of photosynthate source–sink manipulation. Crop Science 14, 11–16 [Google Scholar]
- Layzell DB, Hunt S, Palmer GR. 1990. Mechanisms of nitrogenase inhibition in soybean nodules: pulse-modulated spectroscopy indicates that nitrogenase activity is limited by O2. Plant Physiology 92, 1101–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Parsons R, Day DA, Bergersen FJ. 2002. Reassessment of major products of N2 fixation by bacteroids from soybean root nodules. Microbiology 148, 1959–1966 [DOI] [PubMed] [Google Scholar]
- Liu X, Grams TEE, Matyssek R, Rennenberg H. 2005. Effects of elevated pCO2 and ⁄or pO3 on C-, N-, and S-metabolites in the leaves of juvenile beech and spruce differ between trees grown in monoculture and mixed culture. Plant Physiology and Biochemistry 43, 147–154 [DOI] [PubMed] [Google Scholar]
- Lodwig E, Poole P. 2003. Metabolism of Rhizobium bacteroids. Critical Reviews in Plant Sciences 22, 37–78 [Google Scholar]
- Matamoros MA, Baird LM, Escuredo PR, Dalton DA, Minchin FR, Iturbe-Ormaetxe I, Rubio MC, Moran JF, Gordon AJ, Becana M. 1999. Stress-induced legume root nodule senescence. Physiological, biochemical, and structural alterations. Plant Physiology 121, 97–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y, Good AG. 2008. NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. Journal of Experimental Botany 59, 667–680 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Izumi T, Banba M, Umehara Y, Kouchi H, Izui K, Hata S. 2003. Characterization and expression analysis of genes encoding phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxylase kinase of Lotus japonicus, a model legume. Molecular Plant-Microbe Interactions 16, 281–288 [DOI] [PubMed] [Google Scholar]
- Ott T, Sullivan J, James EK, Flemetakis E, Günther C, Gibon Y, Ronson C, Udvardi M. 2009. Absence of symbiotic leghemoglobins alters bacteroid and plant cell differentiation during development of Lotus japonicus root nodules. Molecular Plant-Microbe Interactions 22, 800–808 [DOI] [PubMed] [Google Scholar]
- Patriarca EJ, Tate R, Iaccarino M. 2002. Key role of bacterial NH4+ metabolism in Rhizobium–plant symbiosis. Microbiology and Molecular Biology Reviews 66, 203–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66 [DOI] [PubMed] [Google Scholar]
- Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. 2006. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biology of the Cell 98, 53–67 [DOI] [PubMed] [Google Scholar]
- Sarath G, Pfeiffer NE, Sodhi CS, Wagner FW. 1986. Bacteroids are stable during dark-induced senescence of soybean root nodules. Plant Physiology 82, 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, et al. 2005. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Letters 579, 1332–1337 [DOI] [PubMed] [Google Scholar]
- Skopelitis DS, Paranychianakis NV, Paschalidis KA, et al. 2006. Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. The Plant Cell 18, 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter JG. 1995. Recent developments in carbon transport and metabolism in symbiotic systems. Symbiosis 19, 175–196 [Google Scholar]
- Swaraj K, Laura JS, Bishnoi NR. 1994. Dark treatment effects on nitrogen fixation and enzymes associated with scavenging hydrogen peroxide in clusterbean nodules. Plant Physiology and Biochemistry 32, 115–119 [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Miller AH. 2004. Lipoic acid-dependent oxidative catabolism of α-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiology 134, 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SJ, Vance CP, Gantt JS. 1998. Glutamate synthase and nitrogen assimilation. Trends in Plant Science 3, 51–56 [Google Scholar]
- Tercé-Laforgue T, Dubois F, Ferrario-Méry S, Pou de Crecenzo MA, Sangwan R, Hirel B. 2004a. Glutamate dehydrogenase of tobacco is mainly induced in the cytosol of phloem companion cells when ammonia is provided either externally or released during photorespiration. Plant Physiology 136, 4308–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercé-Laforgue T, Mäck G, Hirel B. 2004b. New insights towards the function of glutamate dehydrogenase revealed during source–sink transition of tobacco (Nicotiana tabacum) plants grown under different nitrogen regimes. Physiologia Plantarum 120, 220–228 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. 2005. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiology 138, 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricot F. 1993. Mise en place des nodosites du pois proteagineux de printemps (Pisum sativum L.). Influence de la nutrition carbonee. PhD thesis, Universite Paris-Sud Orsay; [Google Scholar]
- Udvardi MK, Day DA. 1997. Metabolite transport across symbiotic membranes of legume nodules. Annual Review of Plant Physiology 48, 493–523 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Price GD, Gresshoff PM, Day DA. 1988. A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Letters 231, 36–40 [Google Scholar]
- Vauclare P, Bligny R, Gout E, De Meuron V, Widmer F. 2010. Metabolic and structural rearrangement during dark-induced autophagy in soybean (Glycine max L.) nodules: an electron microscopy and 31P and 13C nuclear magnetic resonance study. Planta 231, 1495–1504 [DOI] [PubMed] [Google Scholar]
- Waters JK, Hughes BL, 2nd, Purcell LC, Gerhardt KO, Mawhinney TP, Emerich DW. 1998. Alanine, not ammonia, is excreted from N2-fixing soybean nodule bacteroids. Proceedings of the National Academy of Sciences, USA 95, 12038–12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Prell J, James EK, Poole P. 2007. Nutrient sharing between symbionts. Plant Physiology 144, 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Lohaus G, Heldt W. 1992. Phloem transport of amino acids in relation to their cytosolic levels in barley leaves. Plant Physiology 99, 996–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.