Abstract
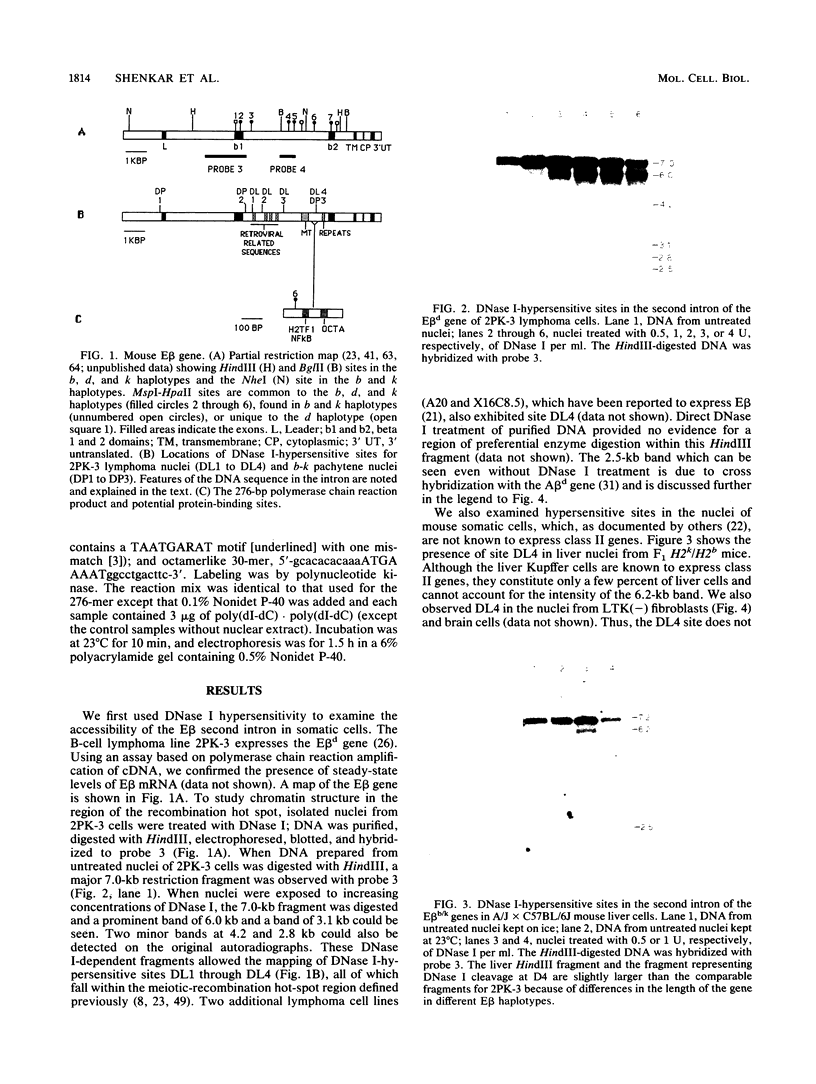
The second intron of the E beta gene in the mouse major histocompatibility complex is the site of a meiotic recombination hot spot. We detected two DNase I-hypersensitive sites in this intron in meiotic cells isolated from mouse testes. One site appears to be constitutive and is found in other tissues regardless of whether or not they express the E beta gene. Near this hypersensitive site are potential binding motifs for H2TF1/KBF1, NF kappa B, and octamer transcription factors. Gel retardation studies with mouse lymphoma cell nuclear extracts confirmed that each of these motifs is capable of binding protein. The binding of transcription factors may contribute to the enhancement of recombination potential by altering chromatin structure and increasing the accessibility of the DNA to the recombination machinery.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin A. S., Jr, Sharp P. A. Binding of a nuclear factor to a regulatory sequence in the promoter of the mouse H-2Kb class I major histocompatibility gene. Mol Cell Biol. 1987 Jan;7(1):305–313. doi: 10.1128/mcb.7.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A. S., Jr, Sharp P. A. Two transcription factors, NF-kappa B and H2TF1, interact with a single regulatory sequence in the class I major histocompatibility complex promoter. Proc Natl Acad Sci U S A. 1988 Feb;85(3):723–727. doi: 10.1073/pnas.85.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumruker T., Sturm R., Herr W. OBP100 binds remarkably degenerate octamer motifs through specific interactions with flanking sequences. Genes Dev. 1988 Nov;2(11):1400–1413. doi: 10.1101/gad.2.11.1400. [DOI] [PubMed] [Google Scholar]
- Begovich A. B., Jones P. P. Free Ia E alpha chain expression in the E+ alpha : E- beta recombinant strain A.TFR5. Immunogenetics. 1985;22(6):523–532. doi: 10.1007/BF00430300. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Alt F. W. Molecular characterization of the lymphoid V(D)J recombination activity. J Biol Chem. 1989 Jun 25;264(18):10327–10330. [PubMed] [Google Scholar]
- Blackwell T. K., Moore M. W., Yancopoulos G. D., Suh H., Lutzker S., Selsing E., Alt F. W. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986 Dec 11;324(6097):585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- Boehm T., Mengle-Gaw L., Kees U. R., Spurr N., Lavenir I., Forster A., Rabbitts T. H. Alternating purine-pyrimidine tracts may promote chromosomal translocations seen in a variety of human lymphoid tumours. EMBO J. 1989 Sep;8(9):2621–2631. doi: 10.1002/j.1460-2075.1989.tb08402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock P., Miller J., Botchan M. Effects of poly[d(pGpT).d(pApC)] and poly[d(pCpG).d(pCpG)] repeats on homologous recombination in somatic cells. Mol Cell Biol. 1986 Nov;6(11):3948–3953. doi: 10.1128/mcb.6.11.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Chandley A. C., Hotta Y., Stern H. Biochemical analysis of meiosis in the male mouse. I. Separation of DNA labelling of specific spermatogenic stages. Chromosoma. 1977 Jul 8;62(3):243–253. doi: 10.1007/BF00286046. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W., Kröger B., Goller M., Horak I. A recombination hotspot in the LTR of a mouse retrotransposon identified in an in vitro system. Cell. 1989 Jun 16;57(6):937–946. doi: 10.1016/0092-8674(89)90332-2. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S., Vasavada H. A., Weissman S. M. Multiple enhancer-like sequences in the HLA-B7 gene. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5247–5251. doi: 10.1073/pnas.86.14.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Israël A., Kimura A., Kieran M., Yano O., Kanellopoulos J., Le Bail O., Kourilsky P. A common positive trans-acting factor binds to enhancer sequences in the promoters of mouse H-2 and beta 2-microglobulin genes. Proc Natl Acad Sci U S A. 1987 May;84(9):2653–2657. doi: 10.1073/pnas.84.9.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kim K. J., Kanellopoulos-Langevin C., Merwin R. M., Sachs D. H., Asofsky R. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J Immunol. 1979 Feb;122(2):549–554. [PubMed] [Google Scholar]
- Kobori J. A., Strauss E., Minard K., Hood L. Molecular analysis of the hotspot of recombination in the murine major histocompatibility complex. Science. 1986 Oct 10;234(4773):173–179. doi: 10.1126/science.3018929. [DOI] [PubMed] [Google Scholar]
- Lafuse W. P., Berg N., Savarirayan S., David C. S. Mapping of a second recombination hot spot within the I-E region of the mouse H-2 gene complex. J Exp Med. 1986 Jun 1;163(6):1518–1528. doi: 10.1084/jem.163.6.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuse W. P., David C. S. Recombination hot spots within the I region of the mouse H-2 complex map to the E beta and E alpha genes. Immunogenetics. 1986;24(6):352–360. doi: 10.1007/BF00377952. [DOI] [PubMed] [Google Scholar]
- Lanier L. L., Warner N. L., Ledbetter J. A., Herzenberg L. A. Quantitative immunofluorescent analysis of surface phenotypes of murine B cell lymphomas and plasmacytomas with monoclonal antibodies. J Immunol. 1981 Oct;127(4):1691–1697. [PubMed] [Google Scholar]
- Larhammar D., Hammerling U., Denaro M., Lund T., Flavell R. A., Rask L., Peterson P. A. Structure of the murine immune response I-A beta locus: sequence of the I-A beta gene and an adjacent beta-chain second domain exon. Cell. 1983 Aug;34(1):179–188. doi: 10.1016/0092-8674(83)90148-4. [DOI] [PubMed] [Google Scholar]
- Lenardo M. J., Baltimore D. NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989 Jul 28;58(2):227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- Liou H. C., Polla B. S., Aragnol D., Leserman L. D., Griffith I. J., Glimcher L. H. A tissue-specific DNase I-hypersensitive site in a class II A alpha gene is under trans-regulatory control. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2738–2742. doi: 10.1073/pnas.85.8.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Araki K., Kitamura D., Wang J., Watanabe T. Nuclear factors binding to the human immunoglobulin heavy-chain gene enhancer. Nucleic Acids Res. 1987 Apr 10;15(7):2851–2869. doi: 10.1093/nar/15.7.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M., Hunkapiller T., Hood L. Nucleotide sequence of a light chain gene of the mouse I-A subregion: A beta d. Science. 1983 Aug 19;221(4612):750–754. doi: 10.1126/science.6410508. [DOI] [PubMed] [Google Scholar]
- Mauxion F., Sen R. Alteration of a single nucleotide allows efficient binding of H2TF1/KBF1 to the immunoglobulin kappa enhancer B motif. Mol Cell Biol. 1989 Aug;9(8):3548–3552. doi: 10.1128/mcb.9.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin S. M., Lyamichev V. I., Kumarev V. P., Kobzev V. F., Nosikov V. V., Vologodskii A. V. The energetics of the B-Z transition in DNA. J Biomol Struct Dyn. 1987 Aug;5(1):79–88. doi: 10.1080/07391102.1987.10506376. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Nickol J., Behe M., Felsenfeld G. Effect of the B--Z transition in poly(dG-m5dC) . poly(dG-m5dC) on nucleosome formation. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1771–1775. doi: 10.1073/pnas.79.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A., Treco D., Schultes N. P., Szostak J. W. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989 Mar 2;338(6210):35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parslow T. G., Blair D. L., Murphy W. J., Granner D. K. Structure of the 5' ends of immunoglobulin genes: a novel conserved sequence. Proc Natl Acad Sci U S A. 1984 May;81(9):2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin B. M., Hardy K. J., Larsen A. S. Chromatin structure of the HLA-DR alpha gene in different functional states of major histocompatibility complex class II gene expression. Mol Cell Biol. 1987 May;7(5):1967–1972. doi: 10.1128/mcb.7.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B. K., Cullen S. E. Molecular mapping of murine I region recombinants: crossing over in the E beta gene. J Immunol. 1986 Feb 1;136(3):1112–1116. [PubMed] [Google Scholar]
- Saito H., Maki R. A., Clayton L. K., Tonegawa S. Complete primary structures of the E beta chain and gene of the mouse major histocompatibility complex. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5520–5524. doi: 10.1073/pnas.80.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes N. P., Szostak J. W. A poly(dA.dT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler H. R., Balling R., Hatzopoulos A. K., Suzuki N., Gruss P. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 1989 Sep;8(9):2551–2557. doi: 10.1002/j.1460-2075.1989.tb08393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler H. R., Hatzopoulos A. K., Balling R., Suzuki N., Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 1989 Sep;8(9):2543–2550. doi: 10.1002/j.1460-2075.1989.tb08392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroishi T., Hanzawa N., Sagai T., Ishiura M., Gojobori T., Steinmetz M., Moriwaki K. Recombinational hotspot specific to female meiosis in the mouse major histocompatibility complex. Immunogenetics. 1990;31(2):79–88. doi: 10.1007/BF00661217. [DOI] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Minard K., Horvath S., McNicholas J., Srelinger J., Wake C., Long E., Mach B., Hood L. A molecular map of the immune response region from the major histocompatibility complex of the mouse. Nature. 1982 Nov 4;300(5887):35–42. doi: 10.1038/300035a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Recombination between poly[d(GT).d(CA)] sequences in simian virus 40-infected cultured cells. Mol Cell Biol. 1985 Jun;5(6):1247–1259. doi: 10.1128/mcb.5.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. E., Peterlin B. M. Transcriptional enhancers in the HLA-DQ subregion. Mol Cell Biol. 1987 Sep;7(9):3315–3319. doi: 10.1128/mcb.7.9.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Thuriaux P. Is recombination confined to structural genes on the eukaryotic genome? Nature. 1977 Aug 4;268(5619):460–462. doi: 10.1038/268460a0. [DOI] [PubMed] [Google Scholar]
- Treco D., Arnheim N. The evolutionarily conserved repetitive sequence d(TG.AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol Cell Biol. 1986 Nov;6(11):3934–3947. doi: 10.1128/mcb.6.11.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu Y., Fischer Lindahl K., Steinmetz M. The same MHC recombinational hot spots are active in crossing-over between wild/wild and wild/inbred mouse chromosomes. Immunogenetics. 1988;27(2):96–101. doi: 10.1007/BF00351082. [DOI] [PubMed] [Google Scholar]
- Uematsu Y., Kiefer H., Schulze R., Fischer-Lindahl K., Steinmetz M. Molecular characterization of a meiotic recombinational hotspot enhancing homologous equal crossing-over. EMBO J. 1986 Sep;5(9):2123–2129. doi: 10.1002/j.1460-2075.1986.tb04475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil R. L., Roeder G. S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987 Mar 27;48(6):1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Wahls W. P., Wallace L. J., Moore P. D. The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol Cell Biol. 1990 Feb;10(2):785–793. doi: 10.1128/mcb.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Larsen A. S., Peterlin B. M. A tissue-specific transcriptional enhancer is found in the body of the HLA-DR alpha gene. J Exp Med. 1987 Sep 1;166(3):625–636. doi: 10.1084/jem.166.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widera G., Flavell R. A. The nucleotide sequence of the murine I-E beta b immune response gene: evidence for gene conversion events in class II genes of the major histocompatibility complex. EMBO J. 1984 Jun;3(6):1221–1225. doi: 10.1002/j.1460-2075.1984.tb01956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]