Abstract
To subvert host defenses, some microbial pathogens produce proteins that interact with conserved motifs in variable regions of B-cell antigen receptor shared by large sets of lymphocytes, which define the properties of a superantigen. As the clonal composition of the lymphocyte pool is a major determinant of immune responsiveness, this study was undertaken to examine the in vivo effect on the host immune system of exposure to a B-cell superantigen, protein L (PpL), a product of the common commensal bacterial species, Finegoldia magna, which is one of the most common pathogenic species amongst Gram-positive anaerobic cocci. Libraries of variable kappa (Vκ) light chain transcripts were generated from the spleens of control and PpL-exposed mice, and the expressed Vκ rearrangements were characterized by high-throughput sequencing. A total of 120,855 sequencing reads could be assigned to a germline Vκ gene, with all 20 known Vκ subgroups represented. In control mice, we found a recurrent and consistent hierarchy of Vκ gene usage, as well as patterns of preferential Vκ-Jκ pairing. PpL exposure induced significant targeted global shifts in repertoire with reduction of Vκ that contain the superantigen binding motif in all exposed mice, with significant targeted reductions in the expression of clonotypes encoded by 14 specific Vκ genes with the predicted PpL binding motif. These rigorous surveys document the capacity of a microbial protein to modulate the composition of the expressed lymphocyte repertoire, which also has broad potential implications for host-microbiome and host-pathogen relationships.
Keywords: High-throughput sequencing, BCR repertoire, Protein L, Immunoglobulin kappa light chain, 454 sequencing
Introduction
Co-evolution of microbial species with the host immune system has given rise to diverse mechanisms by which commensals and pathogens can evade and subvert, or at times reinforce immunological defenses. The adaptive immune system provides special host advantages, with the capacity to select and expand antigen-specific lymphocyte clones and generate memory for rapid recall responses. However, there are bacteria and viruses that in turn have developed high avidity proteins with specificity for conserved and highly represented framework-associated motifs within the variable regions of antigen receptors, which are distinct from conventional antigen binding pockets. While these proteins may not be critical for the microbe's metabolic pathways for growth and survival, in several cases they have been shown to act as virulence factors. The best characterized of the naturally occurring superantigens that target B cells are Protein A (SpA) produced by Staphylococcus aureus, and Protein L (PpL) by Finegoldia magna, also termed Peptostreptococcus magnus (1) (reviewed in (2)).
F. magna is a common component of commensal human flora that colonizes epithelial surfaces of the skin, mucosal surfaces, and the gastrointestinal tract. It is also one of the most common pathogenic species amongst Gram-positive anaerobic coccoids, and cause of serious clinical infections of bone and joints, as well as wound infections and abscesses (3, 4). The bacterial virulence of F. magna isolates correlates with expression of PpL (5), a 76 to 106 kDa protein composed of four to five homologous Fab-binding domains that recognize a conserved framework-associated site (Table I) in the variable regions of many immunoglobulin light chain (Vκ) gene products (6-9).
Table I. Variations in amino acid residue expression in murine VL at positions implicated in PpL binding.
| Residuea: | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 18 | 20 | 22 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| predicted PpL binding motif from human κ-light chains | T | Q | S | P | S,A,D | T,S | L | S,A | A,L,V | R | T | T,S,N | R,G,K | |
| VL subgroupb | PpL motif | |||||||||||||
| Vκ1 (10) | - | T | Q |
|
P |
|
T,S | L |
|
V,A |
|
|
S |
|
| Vκ2 (4) | - | T | Q |
|
|
|
S |
|
|
V |
|
|
S | R |
| Vκ3 (9) | + | T | Q |
|
P | A | S | L | A | V | R | T |
|
|
| Vκ4 (32) | - |
|
Q | S | P |
|
|
|
|
A |
|
T | S,T |
|
| Vκ5 (4) | + | T | Q | S | P | A |
|
L | S | V |
|
|
S | R |
| Vκ6 (6) | - | T | Q |
|
|
|
|
|
|
|
R |
|
T,S | K |
| Vκ7 (1) | - | T | Q | S | P |
|
|
L | A | V |
|
T | S |
|
| Vκ8 (11) | + |
|
Q | S |
|
P,S | S | L |
|
|
|
T |
|
K |
| Vκ9 (4) | + |
|
Q | S | P | S | S |
|
|
A | R | S | T,S | R |
| Vκ10 (3) | - | T | Q |
|
|
S | S | L | S | A | R | T | S |
|
| Vκ11 (1) | - |
|
Q | S | P | S | S | L | S | A |
|
T | T |
|
| Vκ12 (7) | + | T | Q | S | P | A | S |
|
S,A | A,V |
|
T | T |
|
| Vκ13 (2) | - | T | Q | S |
|
S |
|
|
S |
|
R | T | T | R |
| Vκ14 (3) | + | T | Q | S | P | S | S |
|
|
V,A |
|
|
T,S |
|
| Vκ15 (1) | - |
|
Q | S | P | S | S | L | S | A |
|
T | T |
|
| Vκ16 (1) | - | T | Q | S | P | S | Y | L | A | A |
|
T | T,N | R |
| Vκ17 (2) | - | T | Q | S | P | A | S | L | S |
|
|
T |
|
|
| Vκ18 (1) | - | T | Q | A | P | A | S | L | S |
|
|
T | S | R |
| Vκ19 (1) | + | T | Q | S | P | S | S | L | S | A |
|
T | T | K |
| Vκ20 (1) | - | I | Q | S | P |
|
|
L | S | A | R | T | S | K |
|
| ||||||||||||||
| Vλ1 (1) | - | T | Q |
|
|
S |
|
L |
|
|
|
T | T | R |
| Vλ2 (1) | - | T | Q |
|
|
S |
|
L |
|
|
|
|
T | R |
| Vλ3 (1) | - | T | Q |
|
P | S | S |
|
S |
|
|
|
T |
|
| Vλ4;5;7 (3) | - | T | Q |
|
P | S | S |
|
S |
|
|
|
S | K |
| Vλ6;8 (2) | - | T | Q |
|
P | S | S |
|
S |
|
|
|
|
K |
The PpL binding motif has been previously described (7, 9). Based on deduced amino acid residues implicated in the PpL-Fab co-crystal, sequences belonging to Vκ subgroups highlighted in bold possess the conserved PpL primary binding motif as observed in the human Fab antibody PpL co-complex structure. Positions 6 and 7 in italics are not directly involved in the binding interface but contribute to important structural differences between kappa and lambda chains. The Fab-PpL binding interaction is dominated by the contribution of residues at position 8-12. In the above prediction of PpL binders, proline at position 8 was deemed essential while the leucine to methionine substitution at position 11 was deemed a conservative change. Asterisks (*) indicate missing residues. Residues underlined in grey are non-conserved residues compared to the consensus human Vκ sequences of known PpL binders.
The number of germ-line sequences in each subgroup, as reported in the IMGT database, is indicated in parentheses. Non-functional ORFs, pseudogenes and vestigial genes are not included.
In vivo challenge studies have revealed that PpL can cause activation-induced apoptotic cell death resulting in the loss of more than 40% of splenic B cells (10). The greatest targeted depletion occurs amongst splenic marginal zone B cells and B-1 cells, which provide innate-like antigen-specific B-cell defenses against infectious pathogens (10, 11). To better understand the immunobiologic implications of interactions with a B-cell targeting superantigen, we performed high-throughput sequencing of immunoglobulin light chain gene rearrangements to investigate how a limited in vivo exposure of PpL can affect the expressed B-cell repertoire.
Material and Methods
Mice and immunogens
C57BL/6 mice were obtained from The Jackson Laboratory (Bay Harbor, ME, USA) and bred under specific pathogen-free conditions under the supervision of the University of California San Diego (UCSD) Animal Subjects Program. All animal protocols were approved by the UCSD Institutional Animal Care and Use Committee. Adapting a well studied regimen developed for the evaluation of responses to putative B-cell superantigens (10-15), on day 0, one group of four 8-10 week old C57BL/6 mice received 0.5 mg endotoxin-free recombinant PpL (Biovision, Mountain View, CA, USA) in 500 μl PBS by intraperitoneal injection, which was repeated on day 4. Four sex- and age-matched control mice received injections of saline alone. On day 7 after the initial dose, mice were sacrificed and the spleen from each mouse was harvested and divided, with 50% used for immediate ex vivo flow cytometry analysis and 50% for RNA extraction.
Flow cytometry analysis
Spleens were dissociated into single cell suspensions and red blood cells lysed using ACK Lysing Buffer (Lonza, Walkersville, MD, USA). Adapting previously reported methods (10), subsets of splenocytes were identified with fluorochrome-labeled antibodies specific for B220, CD3, lambda and kappa light chains, using isotype control antibodies, as appropriate. PpL-binding cells were detected with biotinylated recombinant PpL (Biovision) and fluorochrome-labeled streptavidin (BD Biosciences, San Diego, CA, USA). Staining was performed in the presence of Fc-block (BD Biosciences). Data were acquired using a FACSCalibur (BD Biosciences) and analyzed with FloJo software (Treestar, Ashland, OR, USA).
Preparation of VL amplicon libraries
Immediately after harvest, spleens were stored in RNALater (Qiagen, Hilden, Germany) and RNA extracted using Qiagen RNA extraction kit following manufacturer's instructions. RNA concentration was determined with an ND-1000 (Nanodrop, Thermo Fisher Scientific, Wilmington, DE, USA). Isolated RNA (1 μg) was used for first-strand cDNA synthesis and amplification by Rapid Amplification of cDNA Ends (RACE) from the 5′end (5/3′RACE 2nd generation kit, Roche) using a specific reverse primer annealing in the constant part of the kappa light chain gene (Cκ). Purified first strand cDNA was polyA-tailed, then amplified by PCR using a nested Cκ-annealing reverse primer and an Oligo(dT) anchor forward primer that added a 3′ anchor sequence. For each library, we prepared a 50μl PCR reaction, consisting of 0.25 mM of forward and reverse primer mixes, 1 ul of deoxynucleotide mix (10 mM), 5 μl 10× High Fidelity reaction buffer (Roche, Indianapolis, IN, USA), 5 μl of purified cDNA, 0.5 μl of Fast start High Fidelity polymerase (Roche) and 36.5 μl of double-distilled H2O. The thermocycle program was: 94°C for 2 min; 30 cycles of 94°C for 30 s, 56 °C for 1 min, 72°C for 1 min; then 72°C for 10 min; and 4°C storage. PCR products were gel purified (QiaEx; Qiagen), then used as template (2-6 μl) for a second PCR step, using two PCR reactions per library, and following the same conditions as above, with the addition of library-specific individual Multiplex Identifiers (MID) tags and sequencing primer keys. Final amplicon libraries were then separately gel purified. No differences were found in the concentration or purity of PCR products obtained for each of the libraries. Sequences for oligonucleotides used in library generation are compiled in Supplemental Table I.
High-throughput sequencing
The sequences of transcripts in the eight Vκ gene cDNA libraries from four PpL-treated mice and four control-treated mice were determined by pyrosequencing with a 454 GS-FLX instrument (Roche) following the manufacturer's protocol.
Bioinformatics analysis
For rapid processing of the sequence data, we developed a custom bioinformatics pipeline (Supplemental Figure 1) based on two published approaches originally developed in the context of HIV-1 sequence analysis. Briefly, all reads were examined for the presence of one of the eight library-specific MIDs, allowing up to one nucleotide mismatch. Reads were then filtered based on the presence of at least 100 consecutive nucleotides with q-scores of ≥20 (corresponding to ≤1% per base error rates) as previously described (16). Vκ-Jκ rearrangements were assigned to individual reads using a modification of the phylogenetic algorithm (SCUEAL) for classifying recombinant HIV-1 strains (17). We refer to this modification as Ig-SCUEAL. A collection of 100 germline Vκ genes, excluding pseudogenes and genes with large deletions or stop codons, and 5 Jκ genes, was taken from the international immunogenetics (IMGT) data base (http://www.imgt.org/textes/vquest/refseqh.html, September 9 2011) (18), aligned in amino-acid space using MUSCLE (19), and mapped to corresponding nucleotide sequences to yield a codon-based alignment.
We next reconstructed maximum likelihood phylogenetic trees for the Vκ and Jκ germline sequences using the GTR+G+I model in Garli v2.0 (http://code.google.com/p/garli/) (20). Vκ and Jκ alignments (and the 318 bp sequence of the 5′ end of the constant region) and corresponding trees were concatenated to form the reference alignment for rearrangement mapping using Ig-SCUEAL. The most recent common ancestors (MRCA) of Vκ and Jκ genes were reconstructed using codon-based maximum likelihood models (17) and used for homology screening. This step served the dual purpose of removing reads that were insufficiently similar to germline genes and rapidly correcting for 454 instrument specific errors in regions with homopolymers. We developed a computationally parsimonious approach using a modification of the Smith-Waterman algorithm that generates an alignment of two codon sequences and allows for penalized frameshifts. The advantage of this algorithm over nucleotide-based read mapping is that it permits for the alignment of divergent sequences that contain stretches of lower nucleotide homology, yet encode for amino acid sequences that are similar. Furthermore, when a read contains a frameshift due to a homopolymer length miscall (a commonly observed technical shortcoming of 454 instruments), the subsequent protein alignment can also be unreliable as only a part of the read may be in-frame. A codon-based alignment can explicitly correct for such frameshifts, while being cognizant of the amino-acid homology. To remove reads with poor homology even after correcting for possible frameshifts, we applied the requirement that each read have a per-codon alignment score that exceeded by a factor of five that of a random sequence with the same average amino-acid composition, as previously validated (16). Sequences were also determined for the reverse complement and we retained the direction with the highest homology to germline genes/alleles.
For each read that passed the homology screen, Ig-SCUEAL identified the most closely related Vκ and Jκ germline genes using a genetic algorithm, which provided phylogenetic assignments, with likelihood-based confidence estimates and alternative assignments. Unlike a purely homology based approach (e.g. IgBLAST (http://www.ncbi.nlm.nih.gov/igblast/) or IMGT V-Quest (21), this approach takes phylogenetic relatedness of germline genes into account and uses an evolutionary model to correct for nucleotide substitution biases, unequal base frequencies and site-to-site substitution rate variation.
Related transcript reads were assigned to a single clonotype that covered the Vκ-Jκ junction and mapped to a particular rearrangement with >70% confidence, if two conditions were met:
Each had the exact same in-frame junctional sequence at the nucleotide level.
Each included no more than 0.05 substitutions/base distant from any other read assigned to the same clonotype, i.e. the maximal pairwise nucleotide distance did not exceed 0.05 (approximate 95% homology). Distances were estimated using pairwise codon-based alignments and the maximum likelihood fitting of a substitution model that allows for unequal base frequencies and transition/transversion rates (22).
All analyses were implemented in the HyPhy package (23) and executed in parallel on an MPI cluster. The current pipeline throughput is ∼100,000 sequences/day on 200 CPUs, with the majority of time consumed by Ig-SCUEAL – a processor intensive phylogenetic approach.
Statistical analysis
For each study, the specific statistical test is indicated, with significance based on p < 0.05.
Results
PpL depleted a subset of Vκ bearing B cells in vivo
To investigate the impact on the B-cell repertoire, we modified a previously reported protocol (10-15), in which groups of four adult mice received injections of the VL-targeting superantigen, PpL, or control treatments with saline. While PpL injections did not result in significant differences in spleen weights, the total number of mononuclear splenocytes was significantly lower in the PpL-exposed mice compared to control mice (mean 56×106 compared to 73×106 cells, p = 0.038). There was also a significant decrease in the ratio of B cells (B220-positive cells) versus T cells (CD3-positive cells) in the PpL-exposed mice (p= 0.008; Figure 1A).
Figure 1. In vivo PpL exposure induces selective loss of a subset of κ-expressing B cells in the spleen.
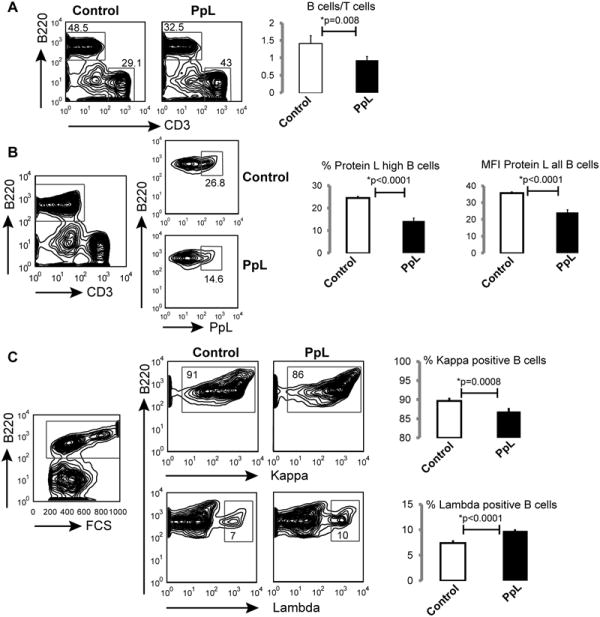
A. Flow cytometry analyses of splenocytes from PpL-treated or saline-treated mice demonstrate induced changes in the ratio of B cells (B220 positive)/T cells (CD3 positive). B. PpL binding to B220 positive B cells is reduced by PpL treatment. C. % kappa L chain positive versus lambda L chain positive B cells in the spleens of saline-treated and PpL-treated mice. n=4 in each group, p-values are from two-sided t-test. Statistical results and representative images are presented from mice used to generate Vκ libraries, which are representative of multiple previous studies (10).
In naïve mice, there are both weak and strong PpL binders amongst κ-bearing B cells, while λ-bearing B cells do not bind PpL (10). After exposure to PpL, both the representation of PpLhigh binding B cells, as well as the mean fluorescent intensity of overall PpL-binding by B cells, were significantly lower (Figure 1B; p< 0.0001). In addition, PpL exposure resulted in a significant decrease in the overall representation of κ-bearing B cells, and an increase in the proportion of λ-bearing B cells (Figure 1C). Taken together, these findings demonstrate that in vivo exposure to PpL results in the significant depletion of B cells that bind PpL, which is likely mediated by a BCR-mediated intrinsic pathway of apoptotic death (10). However, the direct impact on the composition of the polyclonal B-cell repertoire has not been previously investigated.
High-throughput sequencing shows a shift in the expressed Vκ repertoire after PpL exposure
We characterized the expressed Vκ light chain repertoire in control (i.e. saline) treated animals compared to those exposed to PpL. In mice (M. musculus), the Vκ reportoire includes 177 Vκ germline genes, assigned by sequence similarity to 20 subgroups ((18, 24) http://www.imgt.org). While 170,821 reads passed initial quality filtering (see Methods), a total of 120,885 reads were found to be homologous to one of the Vκ germline genes, of which 84,771 spanned a productive junction and contained sufficient phylogenetic signal to be reliably assigned to a unique rearrangement/clonotype.
From each of the libraries, 3,979 to 23,425 of the sequences derived from the control mice were successfully assigned as Vκ-Jκ rearrangements, and 649 to 15,146 sequences from libraries derived from the PpL-treated mice (Supplemental Table II). While there were intragroup variations, the libraries from the mice in the control group shared similar expression patterns at a Vκ subgroup level, which was distinctly different from those generated from the PpL-treated mice. Specifically, in the control mice, the majority (59.5% to 68.4 %, and mean of 62.5%) of all transcripts were encoded by only five dominant Vκ subgroups; Vκ1, Vκ4, Vκ8, Vκ6 and Vκ12.
From comparisons of the expression levels of specific families within the libraries, we found that after PpL treatment there were significant reductions of the families Vκ3, Vκ8, Vκ9, Vκ14 and Vκ15 (Supplemental Table II). In fact, we found PpL treatment was associated with significant increases only for the Vκ13 family, which was otherwise uncommonly expressed, representing 0.2% ± 0.2% in control-treated libraries (Supplemental Table II). Taken together, these findings documented non-random PpL-induced changes in the expressed Vκ-repertoire.
The conserved PpL binding motif identifies Vk gene usage susceptible to negative selection
The structural basis for PpL binding has been best explored in the human system, where PpL is bound by Ig of some, but not other, Vκ subgroups (9). The crystallographic solution of the structure of a PpL co-complex with a Fab antibody demonstrated two binding interfaces, in which the primary interface had a binding interaction that was estimated to be the stronger by more than an order of magnitude (7). This primary interface is largely determined by ten conserved residues located in framework 1 (FR1) of the Vκ region, with residues at positions 8 to 12 appearing to be the most important (Table I). The evolutionary conservation of this surface likely explains reports of PpL-binding interactions with Ig from other mammalian species (25), and in many cases Vκ genes from mice may have common evolutionary ancestors with human analogs. We therefore assessed the homology of the deduced amino acid sequences of the murine Vκ subgroups with the amino acid sequence residues implicated in the human consensus PpL binding motif (7, 9) (Supplemental Table II). Our analysis predicted conservation of the PpL primary binding motif in products of the murine Vκ subgroups 3, 5, 8, 9, 12, 14 and 19, with impaired or absent binding predicted for the other Vκ subgroups or with Vλ light chains.
In light of the predicted PpL interactions, we next reconsidered Vκ expression within libraries, and found that the representation of the sum of all seven Vκ subgroups containing the conserved PpL-binding motif in the control libraries was significantly reduced by PpL exposure (mean 44.5% versus 18.5%; p=0.02)(Figure 2, Supplemental Table II. When each of these subgroups was examined separately, PpL exposure resulted in significant decreases in four of these seven individual subgroups associated with the conserved PpL binding motif (Vκ3; 8; 9; 14) (Supplemental Table II), with similar trends in two other Vκ subgroups that did not attain significance. Conversely, the representation of the sum of the Vκ subgroups without predicted PpL binding was significantly increased in the mice that received this bacterial superantigen compared to the control mice (p=0.02) (Supplemental Table II).
Figure 2. Protein L treatment results in significant reductions in the representation of Vκ subgroups that include the PpL binding motif.
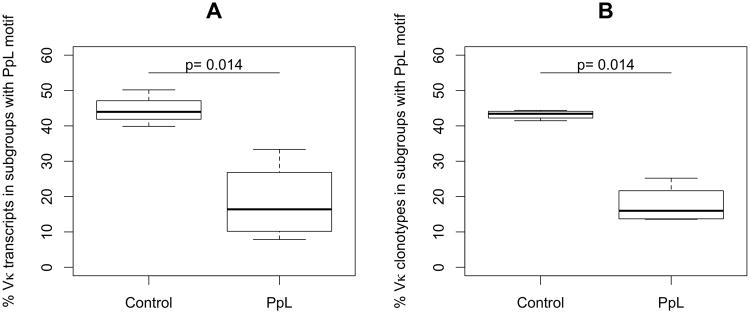
Results are shown from 454 sequencing of Vκ transcripts in mice that received either saline or the superantigen, PpL. The representation of Vκ subgroups is shown, and organized based on whether the Vκ subgroup is predicted to have the conserved PpL binding motif, with comparisons of representation between the two treatment groups (n=4/group), using a one-sided Wilcoxon test. A. Results from analysis of all transcripts. B. Results for data sets are reduction for comparisons of unique Vκ clonotypes that are each counted only once.
In the above-described surveys, each transcript was individually counted, without regard to whether there were multiple reads of the same or a clonally-related transcript in the same library. In the generation of the libraries we did not employ cell sorting, which could have restricted our samplings to include only B cells of distinct subsets, maturation level or activation state, but which could also have introduced other biases. Our surveys of relative Vκ expression could still be influenced by differences in transcript abundance amongst lymphocytes, and especially by the inclusion of end-differentiated plasma cells that may have 1000-fold more Ig transcripts per cell than a resting B cell. To attempt to correct for these influences, using rules outlined in the methods and methods section (supplemental Figure 1), we repeated the analysis after assigning reads to unique Vκ clonotypes (Figure 2, Table II, and Supplemental Figures 1-2). In the libraries from the control mice, we found that the number of unique clonotypes ranged from 1158 to 4101, while in the PpL-treated mice, they ranged from 258 to 2684. Importantly, an analysis of the distribution of Vκ subgroup assignments in control versus PpL-exposed mice at the clonotype level identified similar differences. Hence, the representation of Vκ subgroups with the conserved PpL-binding motif amongst clonotypes was still significantly reduced by PpL exposure, compared to the control group (Figure 2; Table II, mean 17.7% compared to 43.1%, p=0.003).
Table II. Vκ subgroup assignments for unique Vκ clonotypes.
| % Vκ clonotypes assigned to subgroupa: | Control Treated Mice | PpL Treated Mice | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| With conserved PpL motif | 1 | 2 | 3 | 4 | Mean (±SD) | 5 | 6 | 7 | 8 | Mean (±SD) | p-value (two tailed t-test)b |
| Vκ3 | 9.7 | 8.9 | 11.4 | 11.7 | 10.4±1.3 | 0.8 | 3.8 | 5.5 | 1.3 | 2.9±2.2 | 0.004 |
| Vκ5 | 4.6 | 3.2 | 5.1 | 2.8 | 3.9±1.1 | 0.8 | 4.2 | 1.2 | 1.9 | 2.0±1.5 | 0.10 |
| Vκ8 | 10.3 | 14.3 | 12.7 | 12.7 | 12.5±1.6 | 4.3 | 4.7 | 7.5 | 5.9 | 5.6±1.4 | 0.002 |
| Vκ9 | 3.8 | 4.4 | 3.2 | 3.6 | 3.8±0.5 | 0.4 | 1.5 | 2.0 | 1.0 | 1.2±0.7 | 0.002 |
| Vκ12 | 9.3 | 8.0 | 4.3 | 7.3 | 7.2±2.1 | 3.5 | 2.3 | 7.0 | 1.8 | 3.7±2.3 | 0.07 |
| Vκ14 | 4.8 | 3.3 | 3.7 | 3.7 | 3.9±0.6 | 2.7 | 1.2 | 1.4 | 1.8 | 1.8±0.7 | 0.006 |
| Vκ19 | 1.8 | 0.9 | 1.0 | 1.9 | 1.4±0.5 | 1.2 | 0.4 | 0.6 | 0.2 | 0.6±0.4 | 0.06 |
| Total % conserved PpL | 44.3 | 43 | 41.4 | 43.7 | 43.1±1.3 | 13.7 | 18.1 | 25.2 | 13.9 | 17.7±5.4 | 0.003 |
|
| |||||||||||
| Without conserved PpL motif | |||||||||||
| Vκ1 | 13.4 | 15.2 | 9.9 | 11.1 | 12.4±2.4 | 39.5 | 18.8 | 19.6 | 22.7 | 25.2±9.7 | 0.08 |
| Vκ2 | 2.8 | 3.5 | 5.3 | 3.4 | 3.8±1.1 | 9.7 | 10.7 | 4.8 | 10.9 | 9.0±2.9 | 0.04 |
| Vκ4 | 19.9 | 17.6 | 17.1 | 15.5 | 17.5±1.8 | 13.2 | 24.5 | 23.3 | 23.8 | 21.2±5.4 | 0.28 |
| Vκ6 | 9.7 | 12.6 | 18.8 | 17.1 | 14.6±4.2 | 14.3 | 20.9 | 20.8 | 20.0 | 19.0±3.2 | 0.15 |
| Vκ7 | 0.3 | 0.2 | 0.9 | 0.4 | 0.5±0.3 | 0.4 | 0.1 | 0.0 | 0.2 | 0.2±0.2 | 0.20 |
| Vκ10 | 2.5 | 2.5 | 2.3 | 2.8 | 2.5±0.2 | 6.2 | 3.0 | 2.9 | 4.5 | 4.2±1.6 | 0.13 |
| Vκ11 | 0.3 | 0.1 | 0.1 | 0.0 | 0.1±0.1 | 0.8 | 0.2 | 0.1 | 0.2 | 0.3±0.3 | 0.33 |
| Vκ13 | 0.7 | 0.4 | 0.6 | 0.3 | 0.5±0.2 | 0.8 | 1.0 | 0.8 | 1.3 | 1.0±0.2 | 0.03 |
| Vκ15 | 1.3 | 2.0 | 1.4 | 2.4 | 1.8±0.5 | 0.4 | 0.3 | 1.2 | 0.2 | 0.5±0.5 | 0.02 |
| Vκ16 | 0.4 | 0.6 | 0.5 | 1.5 | 0.8±0.5 | 0.0 | 0.4 | 0.3 | 0.6 | 0.3±0.3 | 0.21 |
| Vκ17 | 3.8 | 1.9 | 1.0 | 0.8 | 1.9±1.4 | 1.2 | 0.8 | 0.6 | 0.6 | 0.8±0.3 | 0.22 |
| Vκ18 | 0.2 | 0.1 | 0.0 | 0.3 | 0.2±0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0±0.0 | - |
| Vκ20 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0±0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.1±0.1 | - |
| Total % without a PpL motif | 55.3 | 56.7 | 57.9 | 55.6 | 56.4±1.2 | 86.5 | 80.7 | 74.5 | 85.1 | 81.7±5.4 | 0.003 |
|
| |||||||||||
| Total Vκ unique clonotypes: | 2152 | 2809 | 4101 | 1158 | 258 | 2684 | 1449 | 1147 | |||
Results from analysis of 454 sequencing of 4 libraries from four control treated mice (Library 1-4) and 4 libraries from four PpL treated mice (Library 5-8). Results are presented as % unique clonotypes into which all transcripts were grouped. Two transcripts were binned as the same clonotype if three conditions were met (see text for details): both were assigned to the same Vκ-Jκ rearrangement, both shared exactly the same nucleotide CDR3-V-J junction, and the transcripts were no more than 5% distant at the nucleotide level outside the junction region.
The two groups were compared by two-tailed Welch's t-test assuming unequal variances.
Compared to their human counterparts, the number of murine Vκ genes is much greater, and these are more structurally diverse. Furthermore, some murine germline encoded amino acid residue variations in FR1 are not represented in human sequences (26). We therefore performed analyses of each of the individual Vκ genes in subgroups with PpL-associated binding motifs (Table III). We found that compared to control-treated libraries, there were consistent and significant reductions of rearrangements of 14 of these 39 individual genes in the PpL-exposed mice, while many, but not all of the other related genes also showed decreases that did not reach significance in our studies. However, sequence analyses did not find evidence of increased representation of FR1 replacement mutations after PpL exposure that might contribute to inefficient negative selection of otherwise susceptible Vκ genes (data not shown).
Table III. Analysis of changes after PpL exposure in individual murine genes in subgroups predicted to bind PpL.
| Residuea: | % Vκ clonotypes assigned to VL gene | CDR3 length, aa | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control treated mice | PpL Treated Mice | Control | PpL | |||||||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||||||
| VL subgroupb | VL gene | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 18 | 20 | 22 | 24 | 1 | 2 | 3 | 4 | Mean | 5 | 6 | 7 | 8 | Mean | p-valuec | Mean (±SD) | Mean (±SD) | p-valuec |
| Vκ3 | Vκ3-1 | T | Q | S | P | A | S | L | A | V | R | T | S | R | 0.97 | 0.25 | 0.44 | 0.52 | 0.55 | 0.00 | 0.07 | 0.07 | 0.00 | 0.04 | ↓0.05 | 9.12±0.11 | 9.67±0.33 | ↑0.02 |
| Vκ3-2 | T | Q | S | P | A | S | L | A | V | R | T | S | R | 1.21 | 0.89 | 1.15 | 1.47 | 1.18 | 0.00 | 0.11 | 0.69 | 0.33 | 0.28 | ↓0.006 | 9.18±0.17 | 9.06±0.06 | 0.28 | |
| Vκ3-3 | T | Q | S | P | A | S | L | A | V | R | T | F | R | 0.09 | 0.11 | 0.00 | 0.00 | 0.05 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ND | 9±0 | ND | ND | |
| Vκ3-4d | T | Q | S | P | A | S | L | A | V | R | T | S | K | 3.06 | 1.88 | 3.00 | 2.85 | 2.70 | 0.78 | 0.60 | 2.40 | 0.42 | 1.05 | ↓0.04 | 9.21±0.26 | 9.05±0.16 | ↓0.04 | |
| Vκ3-5 d | T | Q | S | P | A | S | L | A | V | R | T | S | R | 0.60 | 0.75 | 1.46 | 1.47 | 1.07 | 0.00 | 1.38 | 1.17 | 0.00 | 0.64 | 0.37 | 9.24±0.22 | 9.04±0.04 | ↓0.005 | |
| Vκ3-7 | T | Q | S | P | A | S | L | A | V | R | T | S | R | 0.60 | 0.53 | 0.73 | 1.90 | 0.94 | 0.00 | 0.11 | 0.27 | 0.08 | 0.12 | 0.09 | 9.3±0.42 | 9.38±0.27 | 0.82 | |
| Vκ3-9 d | T | Q | S | P | A | S | L | A | V | R | T | S | Q | 0.14 | 0.14 | 0.20 | 0.00 | 0.12 | 0.00 | 0.07 | 0.00 | 0.00 | 0.02 | 0.09 | 9.2±0.31 | 9±0 | 0.64 | |
| Vκ3-10 | T | Q | S | P | A | S | L | A | V | R | T | S | R | 1.21 | 1.60 | 1.58 | 1.30 | 1.42 | 0.00 | 0.19 | 0.07 | 0.17 | 0.11 | ↓0.0003 | 9.24±0.23 | 9.5±0.57 | 0.31 | |
| Vκ3-12 | T | Q | S | P | A | S | L | A | V | R | T | S | R | 0.46 | 0.60 | 0.37 | 0.78 | 0.55 | 0.00 | 0.26 | 0.14 | 0.08 | 0.12 | ↓0.02 | 9.31±0.22 | 9.1±0.1 | 0.18 | |
|
| ||||||||||||||||||||||||||||
| Vκ5 | Vκ5-37 d | T | Q | S | P | A | T | L | S | V | T | S | S | R | 0.00 | 0.00 | 0.07 | 0.09 | 0.04 | 0.00 | 0.00 | 0.07 | 0.08 | 0.04 | 0.96 | 9±0 | 9±0 | ND |
| Vκ5-39 d | T | Q | S | P | A | T | L | S | V | R | S | S | R | 1.81 | 0.57 | 0.76 | 0.78 | 0.98 | 0.00 | 2.31 | 0.21 | 0.92 | 0.86 | 0.85 | 9.08±0.14 | 9.04±0.32 | 0.92 | |
| Vκ5-43 | T | Q | S | P | A | T | L | S | V | S | S | S | R | 0.51 | 0.67 | 0.59 | 0.26 | 0.51 | 0.78 | 0.11 | 0.00 | 0.00 | 0.22 | 0.24 | 9.11±0.27 | 9.4±0.3 | 0.12 | |
| Vκ5-45 d | T | Q | S | P | A | T | L | S | V | R | S | S | R | 0.14 | 0.00 | 0.29 | 0.09 | 0.13 | 0.00 | 0.11 | 0.14 | 0.00 | 0.06 | 0.40 | 9±0 | 9±0 | ND | |
| Vκ5-48d | T | Q | S | P | A | I | L | S | V | R | S | S | R | 1.67 | 1.49 | 1.93 | 1.04 | 1.53 | 0.00 | 0.37 | 0.69 | 0.83 | 0.47 | ↓0.01 | 9.03±0.31 | 8.93±0.44 | 0.43 | |
|
|
|
|||||||||||||||||||||||||||
| Vκ8 | Vκ8-16 d | T | Q | S | I | P | S | L | T | V | R | T | S | K | 0.28 | 0.07 | 0.12 | 0.00 | 0.12 | 0.00 | 0.19 | 0.07 | 0.17 | 0.11 | 0.87 | 8.77±0.36 | 9.25±0.21 | 0.05 |
| Vκ8-18 d | T | Q | S | P | S | S | L | A | V | K | T | R | K | 0.00 | 0.04 | 0.00 | 0.00 | 0.01 | 0.00 | 0.04 | 0.14 | 0.00 | 0.04 | 0.38 | 12±n/a | 12± 0 | n/a | |
| Vκ8-19 d | T | Q | S | P | S | S | L | T | V | K | T | S | K | 0.37 | 0.71 | 0.10 | 0.86 | 0.51 | 0.00 | 0.07 | 0.00 | 0.08 | 0.04 | 0.07 | 9.05±0.34 | 8.67±0.33 | 0.29 | |
| Vκ8-21d | S | Q | S | P | S | S | L | A | V | K | T | S | K | 1.11 | 1.24 | 2.15 | 1.04 | 1.38 | 0.39 | 0.19 | 0.14 | 0.25 | 0.24 | ↓0.02 | 8.89±0.39 | 8.73±0.22 | 0.23 | |
| Vκ8-24d | T | Q | S | P | S | S | L | A | M | K | T | S | K | 2.79 | 3.62 | 1.76 | 3.02 | 2.80 | 0.00 | 0.41 | 2.47 | 0.67 | 0.89 | ↓0.04 | 9.05±0.41 | 9.11±0.18 | 0.76 | |
| Vκ8-26 d | T | Q | S | P | S | S | L | A | V | K | T | S | K | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.08 | 0.02 | ND | ND | 12±0 | ND | |
| Vκ8-27d | T | Q | S | P | S | S | L | A | V | K | T | S | K | 1.62 | 2.59 | 2.88 | 3.37 | 2.62 | 0.39 | 1.01 | 1.24 | 1.42 | 1.01 | ↓0.02 | 8.64±0.44 | 8.47±0.32 | ↓0.04 | |
| Vκ8-28 d | T | Q | S | P | S | S | L | S | V | K | T | S | K | 0.37 | 0.89 | 0.88 | 0.78 | 0.73 | 0.78 | 0.22 | 0.21 | 0.33 | 0.38 | 0.11 | 9±0.19 | 9.14±0.13 | 0.26 | |
| Vκ8-30 d | S | Q | S | P | S | S | L | A | V | K | T | S | K | 1.30 | 2.45 | 1.85 | 1.47 | 1.77 | 0.39 | 0.93 | 1.65 | 0.67 | 0.91 | 0.07 | 8.9±0.38 | 8.86±0.27 | 0.33 | |
| Vκ8-34 d | T | Q | S | P | S | S | L | T | V | K | T | S | K | 0.14 | 0.11 | 0.54 | 0.17 | 0.24 | 0.00 | 0.04 | 0.07 | 0.00 | 0.03 | 0.13 | 9.04±0.044 | 9±0 | 0.88 | |
|
| ||||||||||||||||||||||||||||
| Vκ9 | Vκ9-120d | T | Q | S | P | S | S | L | S | A | R | S | T | R | 2.14 | 2.20 | 1.90 | 2.07 | 2.08 | 0.39 | 0.93 | 1.65 | 0.58 | 0.89 | ↓0.02 | 9.14±0.17 | 9.11±0.24 | 0.40 |
| Vκ9-123 | I | Q | S | P | S | S | M | F | A | R | S | S | R | 0.05 | 0.32 | 0.24 | 0.17 | 0.20 | 0.00 | 0.15 | 0.14 | 0.08 | 0.09 | 0.20 | 9±0 | 9±0 | ND | |
| Vκ9-124 | T | Q | S | P | S | S | L | S | A | R | S | T | R | 1.02 | 1.10 | 0.68 | 1.04 | 0.96 | 0.00 | 0.30 | 0.14 | 0.17 | 0.15 | ↓0.0008 | 9.13±0.12 | 9.25±0.39 | 0.67 | |
| Vκ9-129 | T | Q | S | P | S | S | L | S | A | R | S | T | R | 0.32 | 0.43 | 0.07 | 0.00 | 0.21 | 0.00 | 0.04 | 0.00 | 0.00 | 0.01 | 0.15 | 9.09±0.09 | 10±0 | ↑0.01 | |
|
| ||||||||||||||||||||||||||||
| Vκ12 | Vκ12-38 d | T | Q | S | P | A | S | L | A | A | V | T | T | R | 0.37 | 0.21 | 0.22 | 0.09 | 0.22 | 0.00 | 0.07 | 0.00 | 0.08 | 0.04 | 0.06 | 9±0.1 | 8.67±0.33 | 0.15 |
| Vκ12-41 d | T | Q | S | P | A | S | L | S | A | V | T | T | R | 2.60 | 1.21 | 0.66 | 1.12 | 1.40 | 0.78 | 0.04 | 0.07 | 0.08 | 0.24 | 0.06 | 9.15±0.25 | 9±0 | 0.45 | |
| Vκ12-44 | T | Q | S | P | A | S | L | S | A | V | T | T | R | 3.16 | 2.66 | 1.12 | 2.76 | 2.43 | 0.39 | 0.52 | 0.34 | 0.17 | 0.35 | ↓0.02 | 9.19±0.22 | 9.18±0.16 | 0.90 | |
| Vκ12-46 d | T | Q | S | P | A | S | L | S | V | V | T | T | R | 2.04 | 1.81 | 1.17 | 1.30 | 1.58 | 0.39 | 0.52 | 3.16 | 0.17 | 1.06 | 0.53 | 9.2±0.16 | 9.05±0.05 | ↓0.007 | |
| Vκ12-89 | T | Q | S | P | A | S | L | S | A | V | T | T | G | 0.74 | 1.10 | 0.68 | 1.21 | 0.93 | 0.78 | 0.37 | 0.34 | 0.25 | 0.44 | ↓0.04 | 8.98±0.10 | 8.82±0.15 | ↓0.03 | |
| Vκ12-98 d | T | Q | S | P | A | S | Q | S | A | V | T | T | L | 0.28 | 0.75 | 0.27 | 0.60 | 0.47 | 1.16 | 0.75 | 2.61 | 0.92 | 1.36 | 0.14 | 9±0 | 9.03±0.03 | 0.26 | |
| Vκ12-e | T | Q | S | P | A | S | L | S | V | V | T | T | R | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ND | ND | ND | ND | |
|
|
|
|||||||||||||||||||||||||||
| Vκ14 | Vκ14-100 | T | Q | S | P | S | S | M | S | L | V | T | T | H | 1.16 | 0.57 | 1.54 | 1.12 | 1.10 | 1.94 | 0.30 | 0.27 | 0.83 | 0.84 | 0.58 | 9.1±0.15 | 9.04±0.12 | 0.43 |
| Vκ14-111d | T | Q | S | P | S | S | M | Y | L | V | T | T | K | 3.57 | 2.70 | 2.12 | 2.50 | 2.73 | 0.78 | 0.75 | 1.10 | 1.00 | 0.91 | ↓0.01 | 9.07±0.19 | 9.06±0.15 | 0.84 | |
| Vκ14-130 d | T | Q | S | P | S | S | M | S | L | I | T | T | Q | 0.05 | 0.07 | 0.05 | 0.09 | 0.06 | 0.00 | 0.11 | 0.00 | 0.00 | 0.03 | 0.32 | 8.83±0.17 | 9±0 | 0.64 | |
|
| ||||||||||||||||||||||||||||
| Vκ19 | Vκ19-93 d | T | Q | S | P | S | S | L | S | A | V | T | T | K | 1.81 | 0.89 | 1.05 | 1.90 | 1.41 | 1.16 | 0.37 | 0.62 | 0.17 | 0.58 | 0.06 | 8.52±1.28 | 8.71±0.39 | 0.63 |
Amino acid residues implicated in the primary frame work 1 binding interface in the PpL-Fab co-crystal as described (7).
Individual functional Vκ genes in subgroups with conserved PpL binding motif. In all cases *01 alleles of indicated genes were used.
p-value from 2-tailed t-test with Welch correction assuming unequal variances..
Reference sequence from IMGT was originally reported in C57BL/6 (18).
The IMGT sequences for Vκ5-48, Vκ8-30 and Vκ14-111 has also been reported in the literature in C57BL/6 as annotated in IMGT (18).
Genes marked in bold and highlighted in gray were found to be significantly reduced in the PpL treated mice compared to controls.
Residues in gray font were not consistent with the predicted human PpL motif.
Boxed residues are likely to considerably change the motif properties and be intolerant to PpL binding.
ND: Non-determined.
Arrow down (↓) indicates that the mean was significantly lower by 2-tailed t-test with Welch correction in the PpL treated mice compared to control mice.
Arrow up (↑) indicates that the mean was significantly higher by 2-tailed t-test with Welch correction in the PpL treated mice compared to control mice.
Despite overall homology to human Vκ families associated with PpL-binding activity, due to germline sequence variations not all murine genes within the analyzed subgroups were predicted to bind PpL, and sequence analyses found no reductions in the representation of these genes in PpL-exposed animals. Our studies therefore identified several germline-associated amino acid variations that correlate with altered properties of the motif. For example, in the presence of threonine or serine at position 18, and arginine at position 22 there was no significant PpL-associated depletion, which suggested that these variations greatly reduced or abolished the capacity for PpL binding. In contrast, other common murine variations appeared to be permissive of PpL-associated depletion, such as methionine at residue 11, and lysine or valine at position 18, which we did not appear to adversely affect the PpL binding interface (Table III). Based on evidence of the murine Vκ genes that were significantly reduced by the PpL exposure, we formulated a primary sequence consensus motif for non-immune binding of PpL for the murine immune system (Figure 4).
Figure 4. Consensus sequence of the murine Vκ primary PpL binding motif.
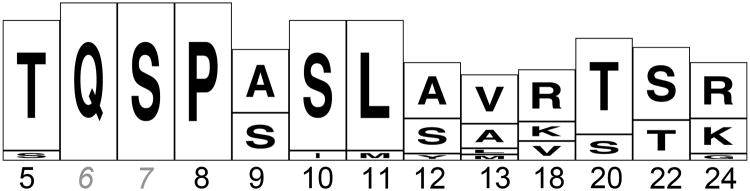
The consensus sequence is based on the relative representation of amino acid variations at specific positions in the PpL binding motif (7) in the 14 germline Vκ gene that were significantly reduced in libraries from PpL-treated mice compared to control mice (see Table III). These data are presented as a WebLogo image (54). The image represents the alignment of each residue by a stack of letters, where the height of each letter is proportional to the observed frequency of the corresponding amino acid, and the overall height of each stack is proportional to the sequence conservation, at that position.
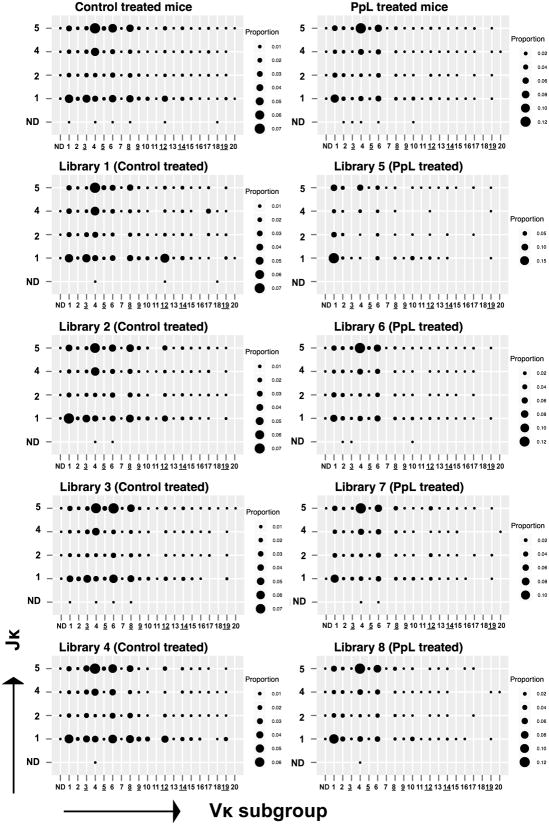
We also characterized the diversity within our libraries with regard to specific Jκ usage for different Vκ subgroups (Figure 3). These analyses demonstrated a remarkable conservation of preferential Vκ-Jκ association between the individual mice within the treatment groups. As previously well documented, immunologically intact mice display a general bias in Jκ usage, with preferentially uses in favor of Vκ-proximal Jκ elements, leading to a frequency hierarchy of Jκ1>Jκ2> Jκ4>Jκ5, as the murine Jκ3 is a pseudogene (27). In our studies, clonotypes from the Vκ1, Vκ3 and Vκ4 subgroups showed a consistently higher level of Jκ1 rearrangements. Notably, for some of the other Vκ subgroups we found very different patterns, and the Vκ4 subgroup displayed the highest frequency of Jκ5-pairing in all eight libraries, which is consistent with previous reports (28-31). These patterns may reflect differences in Jκ preferences in primary rearrangements, or result from secondary rearrangements due to receptor editing (reviewed in (32)).
Figure 3. Igκ V-J repertoire in C57BL/6 mice with and without exposure to the superantigen PpL.
Results are depicted for the frequency of clonotypes with a specific V-J pairing in four control mice (libraries 1-4) and four PpL exposed mice (libraries 5-8) either when evaluated for treatment groups or individually. Underlined Vκ subgroups were predicted to have PpL interactions with products of one or more associated members. ND, represents the small number of clonotypes that could not be assigned to a specific Vκ subgroup or Jκ gene. The results are based on analyses of a total of 84,771 unique clonotypes from the eight libraries.
We also looked for variations in the distribution of CDR3 length in the libraries. Overall, in comparisons of clonotypic sets in libraries from saline-treated and PpL-treated mice, we did not find significant differences in the distribution of CDR3 length (data not shown). In addition, we examined the Vκ genes in subgroups associated with the PpL binding motif, but did not find consistent differences in CDR3 length in rearrangements identified by specific Vκ gene usage (Table III). Taken together these findings suggest that there was no relationship between the overall size of the CDR3, the somatically generated subdomain with the greatest structural diversity, and the non-immune binding interactions with PpL.
Discussion
In the current studies we examined the in vivo outcome of exposure to a bacterial virulence factor, PpL, which has the functional properties of an Ig light chain targeted superantigen. In control mice, amongst Igκ-bearing B cells there was a continuous distribution of binding capacities to PpL, with some B cells displaying strong binding and others with none detectable. After PpL exposure, cellular analyses documented a decrease in the B-cell/T-cell ratio and in overall kappa-bearing B cells, which was consistent with a specific targeted depletion of B cells with the strongest PpL-binding capacity. As the effect of such a microbial toxin on the expressed BCR repertoire at a molecular level was unknown, we compared the representation of Igκ transcripts in libraries made from the spleens of control mice and PpL-exposed mice. In control mice raised under specific pathogen-free conditions, we found an unexpectedly high degree of conservation in their repertoire composition, as there was a recurrent and conserved hierarchy of Vκ gene usage with predominance of the Vκ1, Vκ4, Vκ8, Vκ6 and Vκ12 subgroups. In addition, fine analysis of the Igκ rearrangements also demonstrated conserved patterns of preferential pairing of Vκ-Jκ. Therefore, using an approach technically similar to a recently reported characterization of libraries made from cells from pooled donors (33), our investigations provide novel insights into the conservation of overall expression patterns within the overall kappa light chain repertoire, for V-J joining patterns in individual mice within the different groups, and the outcome of superantigen exposure.
The principal finding in our studies was striking evidence of induced global shifts in the repertoire of each of the four PpL-exposed mice, with significant reductions of the Vκ subgroups that include the superantigen-binding motif in the Vκ framework region that has been characterized in crystallographic studies of a Fab-PpL co-complex (7). These findings were remarkably consistent between members of the same group. Moreover, each of the seven families predicted to encode for PpL binding showed reductions, and it was statistically significant in four of these families (Vκ3, Vκ8, Vκ9 and Vκ14), and with similar trends in the remaining three (Vκ5, Vκ12 and Vκ19). There was also a significant reduction in the Vκ15 family, which remains unexplained. Interestingly, the cellular analysis confirmed evidence from Vκ gene analyses that some PpL binding to a subset of B cells can still be detected in the PpL exposed mice. This may suggest that some B cells, with weaker binding interactions with PpL, were not completely depleted. However, it is possible that this could be related to differences in in vivo PpL availability, the B cell surface density of BCR and other H chain isotypes, or even receptor dilution that may be prevalent in marginal zone B cells. In the future, more detailed studies using single cell sorting and BCR cloning of these remaining cells with lower PpL binding activity will be needed to further investigate this topic.
We interpret our findings as consistent with negative supraclonal selection of PpL-binding B cells, which results from strong B-cell superantigen-mediated BCR-mediated first signal in the absence of a balanced and sufficient second signal (12, 13). In other settings, if there is impaired BCR-mediated death signaling or in the presence of second signals (12, 13), which could occur during active infection, we predict that PpL-binding B-cell clones could be greatly expanded to further dominate the immune repertoire. Notably, these superantigen-induced effects were completely different from the outcome commonly associated with immunization with a conventional antigen as there was no evidence of expansions of specific clonotypes in the PpL-exposed animals (see Supplemental Figure 2). Our surveys therefore also demonstrate that even limited exposure to this bacterial virulence factor can globally shift the immune repertoire in a manner that is highly diagnostic of the influence of a superantigen-BCR interaction. We have earlier reported evidence that similar superantigen-induced changes can alter immune responsiveness to new microbial threats as well as for recall immune responses (15).
In the murine and human immune systems, immune competence is in part dependent on the formation of a clonally diverse preimmune repertoire. Murine BCR are formed and molded by somatic mechanisms that can access a large number of germline Vκ genes, which vary considerably in the canonical structures of their CDR (34). Presumably there has been selective pressure for the retention of these diverse inherited basic building blocks for the formation of antigen-receptors that can bind very different types of antigens. In fact, a deficiency in the inheritance of even a single specific Vκ germline gene can result in impaired immune defenses and serious and recurrent infections from a common bacterial pathogen (35).
Our findings support emerging evidence that a B-cell superantigen can effectively target a large genetically and structurally defined set of Vκ-expressing adaptive immune cells for depletion. We have previously reported that in vivo exposure to a superantigen can induce preferential targeted depletion of innate-like marginal zone B cells and B-1 cells, which can significantly impair immune responses to certain microbial antigens (14, 15). A B-cell superantigen can cause activation-induced apoptosis in vivo(10, 13-15) by a process that requires the induction of the pro-apoptotic Bim member of the Bcl-2 family (12). Hence, these microbial products appear to have co-evolved with the host's immune system to enable the hijacking of BCR-associated signaling pathways, in a manner similar to that shown in transgenic mouse models of regulated negative selection of auto-reactive B cells by introduction of neo-antigens (28, 36) or a synthetic Igκ-reactive macro-self antigen (37). Notably, in the current studies, adapting a regimen widely used to evaluate responses to putative B-cell superantigens (10-15), mice received two 500 μg doses of PpL. However with another superantigen, SpA, systemic doses as low as 2 μg were shown to significantly decrease the in vivo frequency of superantigen-binding B cells (38). While the level of in vivo production of this bacterial product during colonization or active infection is currently unknown, based on the dramatic effects herein demonstrated for systemic doses of protein L, we postulate that even proportionately smaller doses locally produced during invasive infection could have substantial local effects in draining lymph nodes wherein defensive immune responses are initiated.
The current investigation therefore contributes to emerging data that a B-cell superantigen bacterial product can induce a targeted supraclonal hole in the repertoire (15). Such superantigens can represent key virulence factors necessary for the persistence of the pathogen in host tissue (39), and this effect may facilitate the microorganism's escape from host immune recognition. In accordance, in experimental infection models with S. aureus, one of the most common causes of invasive and life-threatening infections, immunization of mice with a modified non-toxogenic form of the SpA superantigen was shown to result in induction of neutralizing antibodies and protection against highly virulent staphylococcal strains (40). Hence, microbial expression of a B-cell superantigen may provide advantages to the pathogen by blocking the capacity of the immune system to limit a bacterial infection and to form recall responses that protect from reinfection, while neutralization of this superantigen influence can restore immune defenses (40, 41). Yet, while both S. aureus and F. magna are usually commensal species and part of the micobiome in healthy individuals, nonetheless each can become invasive pathogens, presumably after local breaches in defenses.
The microbiome refers to the vast collection of symbiotic microorganisms in the human body and their collective interacting genomes. It performs numerous key biochemical functions for the host, and disorders of the microbiome are associated with many human disease processes (42). In a recent report, Matzinger and coworkers showed that immune homeostasis depends on a three-way interaction between the B-cell compartment of the immune system, gut-associated microbiota, and innate responses from the intestinal epithelium. In the absence of B cells, or of their IgA products, which is the predominant antibody isotype of secreted antibodies in the gut, the intestinal epithelium launches its own protective mechanisms after exposure to the microbiota. This results in upregulation of interferon-inducible immune response pathways and altered metabolic functions (43). Moreover, changes in the Ig repertoire expressed in gut-associated lymphoid tissue (GALT) can alter the balance amongst species within the microbiome, and thereby may affect global immune homeostasis. The intestinal microbiome is dominated by anaerobic microbes and the true prevalence of different species in health and during infections is now being reconsidered in light of recently developed culture independent methods of identification (44). Knight and coworkers have shown that intestinal bacterial species may commonly express proteins with superantigen properties, and when introduced into an otherwise sterile gut such microbial isolates can induce B-cell proliferation in gut-associated lymphoid tissue (GALT) and contribute to the development and expansion of a new “preimmune” repertoire (45). Therefore, while in some settings the B-cell targeted effects of a superantigen can represent a pathogenic factor that interferes with host defenses during infection, these or related factors may also provide a pathway by which intestinal flora may selectively mold the physiologic immune repertoire (46). The outcome of immune exposure to a superantigen is likely to be highly dependent on the context of intestinal colonization and the balance amongst commensals within the overall microbiome. However, as part of an invasive infection, secretion of a superantigen, along with potentially stimulatory co-expressed factors, may instead impair immune defenses.
In our investigations, we have applied recently developed technologic approaches that incorporate high-throughput sequencing and in-house developed bioinformatics tools to investigate the representation of expressed light chain rearrangements. As the immune system contains more than 1011 B and T lymphocytes that derive from a large number of parental clones, with an estimated 3-9 million B-cell clones with different Ig heavy chains in the circulation at any given moment (47), traditional Sanger DNA sequencing technology can provide only limited insights into the true clonal diversity within the repertoire. By contrast, next-generation sequencing platforms have greatly expanded our capacity to more accurately assess the complex diversity of gene sequences that encode for lymphocyte antigen receptors (47-53). Our studies have provided a more complete insight into the effects on B-cell clonal diversity of exposure to a single microbial factor.
These findings therefore shed new light on the ability of pathogens to globally disrupt or bias the clonal distribution within the B-cell compartment that affects host defenses. Furthermore, our methodologic approach, which applied one of the most rigorous methods now available, provided a practical means to survey for potential effects on lymphocyte clonal diversity within host defenses. In the future, high-throughput sequencing of immune repertoires should be extensively used to advance our understanding of the microbiome-host relationship and to guide the development of new and more effective vaccines, including those against pathogens that produce superantigens.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Doug Richman, Dr. Nick Webster, and UCSD Center for AIDS Research for the opportunity to use the Roche 454 instrument. For sample preparation and sequence determinations we thank the UCSD GeneChip Microarray Core. For assistance in sequence analysis, we thank the UCSD CFAR BIT Core. We thank Professor Marie-Paule Lefranc for helpful advice.
This work was supported by grants R01 AI090118, AI068063, AI47745, AI57167, GM093939, AI40305, P01 AI074621 and P30 AI036214 from the National Institutes of Health, and grants from the Alliance for Lupus Research.
Abbreviations
- PpL
Protein L
- SpA
Protein A
- F. magna
Finegoldia magna
- Cκ
Immunoglobulin kappa constant region
- Vκ
Kappa light chain variable region
- Vλ
Lambda light chain variable region
- MID
Multiplex identifiers
Footnotes
The authors have no conflicting financial interests.
References
- 1.Murdoch DA. Gram-positive anaerobic cocci. Clin Microbiol Rev. 1998;11:81–120. doi: 10.1128/cmr.11.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverman GJ, Goodyear CS. Confounding B-cell defences: lessons from a staphylococcal superantigen. Nat Rev Immunol. 2006;6:465–475. doi: 10.1038/nri1853. [DOI] [PubMed] [Google Scholar]
- 3.Bourgault AM, Rosenblatt JE, Fitzgerald RH. Peptococcus magnus: a significant human pathogen. Ann Intern Med. 1980;93:244–248. doi: 10.7326/0003-4819-93-2-244. [DOI] [PubMed] [Google Scholar]
- 4.Levy PY, Fenollar F, Stein A, Borrione F, Raoult D. Finegoldia magna: a forgotten pathogen in prosthetic joint infection rediscovered by molecular biology. Clin Infect Dis. 2009;49:1244–1247. doi: 10.1086/605672. [DOI] [PubMed] [Google Scholar]
- 5.Ricci S, Medaglini D, Marcotte H, Olsen A, Pozzi G, Bjorck L. Immunoglobulin-binding domains of peptostreptococcal protein L enhance vaginal colonization of mice by Streptococcus gordonii. Microb Pathog. 2001;30:229–235. doi: 10.1006/mpat.2000.0427. [DOI] [PubMed] [Google Scholar]
- 6.Bjorck L. Protein L. A novel bacterial cell wall protein with affinity for Ig L chains. J Immunol. 1988;140:1194–1197. [PubMed] [Google Scholar]
- 7.Graille M, Stura EA, Housden NG, Beckingham JA, Bottomley SP, Beale D, Taussig MJ, Sutton BJ, Gore MG, Charbonnier JB. Complex between Peptostreptococcus magnus protein L and a human antibody reveals structural convergence in the interaction modes of Fab binding proteins. Structure. 2001;9:679–687. doi: 10.1016/s0969-2126(01)00630-x. [DOI] [PubMed] [Google Scholar]
- 8.Kastern W, Sjobring U, Bjorck L. Structure of peptostreptococcal protein L and identification of a repeated immunoglobulin light chain-binding domain. J Biol Chem. 1992;267:12820–12825. [PubMed] [Google Scholar]
- 9.Nilson BH, Solomon A, Bjorck L, Akerstrom B. Protein L from Peptostreptococcus magnus binds to the kappa light chain variable domain. J Biol Chem. 1992;267:2234–2239. [PubMed] [Google Scholar]
- 10.Goodyear CS, Narita M, Silverman GJ. In vivo VL-targeted activation-induced apoptotic supraclonal deletion by a microbial B cell toxin. J Immunol. 2004;172:2870–2877. doi: 10.4049/jimmunol.172.5.2870. [DOI] [PubMed] [Google Scholar]
- 11.Viau M, Longo NS, Lipsky PE, Bjorck L, Zouali M. Specific in vivo deletion of B-cell subpopulations expressing human immunoglobulins by the B-cell superantigen protein L. Infect Immun. 2004;72:3515–3523. doi: 10.1128/IAI.72.6.3515-3523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodyear CS, Corr M, Sugiyama F, Boyle DL, Silverman GJ. Cutting Edge: Bim is required for superantigen-mediated B cell death. J Immunol. 2007;178:2636–2640. doi: 10.4049/jimmunol.178.5.2636. [DOI] [PubMed] [Google Scholar]
- 13.Goodyear CS, Silverman GJ. Death by a B cell superantigen: In vivo VH-targeted apoptotic supraclonal B cell deletion by a Staphylococcal Toxin. J Exp Med. 2003;197:1125–1139. doi: 10.1084/jem.20020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodyear CS, Silverman GJ. Staphylococcal toxin induced preferential and prolonged in vivo deletion of innate-like B lymphocytes. Proc Natl Acad Sci U S A. 2004;101:11392–11397. doi: 10.1073/pnas.0404382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman GJ, Cary SP, Dwyer DC, Luo L, Wagenknecht R, Curtiss VE. A B cell superantigen-induced persistent “Hole” in the B-1 repertoire. J Exp Med. 2000;192:87–98. doi: 10.1084/jem.192.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pacold M, Smith D, Little S, Cheng PM, Jordan P, Ignacio C, Richman D, Pond SK. Comparison of methods to detect HIV dual infection. AIDS research and human retroviruses. 2010;26:1291–1298. doi: 10.1089/aid.2010.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosakovsky Pond SL, Posada D, Stawiski E, Chappey C, Poon AF, Hughes G, Fearnhill E, Gravenor MB, Leigh Brown AJ, Frost SD. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol. 2009;5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwickl DJ. PhD thesis. University of Texas at Austin; 2006. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. [Google Scholar]
- 21.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic acids research. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial-DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 23.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Jean C, Folch G, Lefranc MP. Nomenclature and overview of the mouse (Mus musculus and Mus sp.) immunoglobulin kappa (IGK) genes. Exp Clin Immunogenet. 2001;18:255–279. doi: 10.1159/000049204. [DOI] [PubMed] [Google Scholar]
- 25.De Chateau M, Nilson BH, Erntell M, Myhre E, Magnusson CG, Akerstrom B, Bjorck L. On the interaction between protein L and immunoglobulins of various mammalian species. Scand J Immunol. 1993;37:399–405. doi: 10.1111/j.1365-3083.1993.tb03310.x. [DOI] [PubMed] [Google Scholar]
- 26.Thiebe R, Schable KF, Bensch A, Brensing-Kuppers J, Heim V, Kirschbaum T, Mitlohner H, Ohnrich M, Pourrajabi S, Roschenthaler F, Schwendinger J, Wichelhaus D, Zocher I, Zachau HG. The variable genes and gene families of the mouse immunoglobulin kappa locus. Eur J Immunol. 1999;29:2072–2081. doi: 10.1002/(SICI)1521-4141(199907)29:07<2072::AID-IMMU2072>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 27.Wood DL, Coleclough C. Different joining region J elements of the murine kappa immunoglobulin light chain locus are used at markedly different frequencies. Proc Natl Acad Sci U S A. 1984;81:4756–4760. doi: 10.1073/pnas.81.15.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adelstein S, Pritchard-Briscoe H, Anderson TA, Crosbie J, Gammon G, Loblay RH, Basten A, Goodnow CC. Induction of self-tolerance in T cells but not B cells of transgenic mice expressing little self antigen. Science. 1991;251:1223–1225. doi: 10.1126/science.1900950. [DOI] [PubMed] [Google Scholar]
- 29.Kalled SL, Brodeur PH. Preferential rearrangement of V kappa 4 gene segments in pre-B cell lines. J Exp Med. 1990;172:559–566. doi: 10.1084/jem.172.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medina CA, Teale JM. Restricted kappa chain expression in early ontogeny: biased utilization of V kappa exons and preferential V kappa-J kappa recombinations. J Exp Med. 1993;177:1317–1330. doi: 10.1084/jem.177.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milstein C, Even J, Jarvis JM, Gonzales-Fernandez A, Gherardi E. Non-random features of the repertoire expressed by the members of one V kappa gene family and of the V-J recombination. Eur J Immunol. 1992;22:1958. doi: 10.1002/eji.1830220745. [DOI] [PubMed] [Google Scholar]
- 32.Nemazee D. Receptor editing in lymphocyte development and central tolerance. Nat Rev Immunol. 2006;6:728–740. doi: 10.1038/nri1939. [DOI] [PubMed] [Google Scholar]
- 33.Ota M, Duong BH, Torkamani A, Doyle CM, Gavin AL, Ota T, Nemazee D. Regulation of the B cell receptor repertoire and self-reactivity by BAFF. J Immunol. 2010;185:4128–4136. doi: 10.4049/jimmunol.1002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 35.Feeney AJ, Atkinson MJ, Cowan MJ, Escuro G, Lugo G. A defective Vkappa A2 allele in Navajos which may play a role in increased susceptibility to haemophilus influenzae type b disease. J Clin Invest. 1996;97:2277–2282. doi: 10.1172/JCI118669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA, Pritchard-Briscoe H, Wotherspoon JS, Loblay RH, Raphael K, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 37.Ait-Azzouzene D, Gavin AL, Skog P, Duong B, Nemazee D. Effect of cell:cell competition and BAFF expression on peripheral B cell tolerance and B-1 cell survival in transgenic mice expressing a low level of Igkappa-reactive macroself antigen. Eur J Immunol. 2006;36:985–996. doi: 10.1002/eji.200535581. [DOI] [PubMed] [Google Scholar]
- 38.Goodyear CS, Sugiyama F, Silverman GJ. Temporal and dose-dependent relationships between in vivo B cell receptor-targeted proliferation and deletion-induced by a microbial B cell toxin. J Immunol. 2006;176:2262–2271. doi: 10.4049/jimmunol.176.4.2262. [DOI] [PubMed] [Google Scholar]
- 39.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HK, Cheng AG, Kim HY, Missiakas DM, Schneewind O. Nontoxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J Exp Med. 2010;207:1863–1870. doi: 10.1084/jem.20092514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HK, Kim HY, Schneewind O, Missiakas D. Identifying protective antigens of Staphylococcus aureus, a pathogen that suppresses host immune responses. FASEB J. 2011;25:3605–3612. doi: 10.1096/fj.11-187963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honda K, Littman DR. The Microbiome in Infectious Disease and Inflammation. Annu Rev Immunol. 2011 doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O, Orandle M, Mayer L, Macpherson AJ, McCoy KD, Fraser-Liggett C, Matzinger P. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med. 2011;17:1585–1593. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.La Scola B, Fournier PE, Raoult D. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe. 2011;17:106–112. doi: 10.1016/j.anaerobe.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Severson KM, Mallozzi M, Driks A, Knight KL. B cell development in GALT: role of bacterial superantigen-like molecules. J Immunol. 2010;184:6782–6789. doi: 10.4049/jimmunol.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhee KJ, Jasper PJ, Sethupathi P, Shanmugam M, Lanning D, Knight KL. Positive selection of the peripheral B cell repertoire in gut-associated lymphoid tissues. J Exp Med. 2005;201:55–62. doi: 10.1084/jem.20041849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnaout R, Lee W, Cahill P, Honan T, Sparrow T, Weiand M, Nusbaum C, Rajewsky K, Koralov SB. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One. 2011;6:e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyd SD, Gaeta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Collins AM. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010;184:6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang N, Weinstein JA, Penland L, White RA, 3rd, Fisher DS, Quake SR. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci U S A. 2011;108:5348–5353. doi: 10.1073/pnas.1014277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, Ellington AD, Georgiou G. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010;28:965–969. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- 51.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116:1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.