Abstract
Pathogenic bacteria using a type III secretion system (T3SS)1,2 to manipulate host cells cause many different infections including Shigella dysentery, typhoid fever, enterohaemorrhagic colitis and bubonic plague. An essential part of the T3SS is a hollow needle-like protein filament through which effector proteins are injected into eukaryotic host cells3–6. Currently, the three-dimensional structure of the needle is unknown because it is not amenable to X-ray crystallography and solution NMR, as a result of its inherent non-crystallinity and insolubility. Cryo-electron microscopy combined with crystal or solution NMR subunit structures has recently provided a powerful hybrid approach for studying supramolecular assemblies7–12, resulting in low-resolution and medium-resolution models13–17. However, such approaches cannot deliver atomic details, especially of the crucial subunit–subunit interfaces, because of the limited cryo-electron microscopic resolution obtained in these studies. Here we report an alternative approach combining recombinant wild-type needle production, solid-state NMR, electron microscopy and Rosetta modelling to reveal the supramolecular interfaces and ultimately the complete atomic structure of the Salmonella typhimurium T3SS needle. We show that the 80-residue subunits form a right-handed helical assembly with roughly 11 subunits per two turns, similar to that of the flagellar filament of S. typhimurium. In contrast to established models of the needle in which the amino terminus of the protein subunit was assumed to be α-helical and positioned inside the needle, our model reveals an extended amino-terminal domain that is positioned on the surface of the needle, while the highly conserved carboxy terminus points towards the lumen.
Wild-type S. typhimurium T3SS needles were obtained by in vitro polymerization of recombinantly produced full-length PrgI protomers. The diameter of the resulting needle-like filaments (Supplementary Fig. 1b) agrees with that of T3SS needles from natural sources3. The addition of wild-type PrgI protomers also elongated needles of isolated S. typhimurium T3SSs up to several micrometres in length (Supplementary Fig. 2), showing the functionality of the recombinant protein (as previously reported for PrgI* (ref. 18)). Recently we demonstrated that excellent-quality solid-stateNMR(ssNMR) spectra of PrgI needles can be obtained by 13C spin dilution19, which allowed us to achieve the complete ssNMR resonance assignment. As determined from conformation-dependent chemical shifts, PrgI adopts a rigid conformation (except for Met 1 and Ala 2) comprising four distinct structural elements (Fig. 1a): an N-terminal extended domain (Thr 3–Tyr 8), an α-helix (α1) (Leu 9–Ala 35), a loop (Ala 36–Pro 41) and a C-terminal α-helix (α2) (Ala 42–Arg 80). Very narrow 13C linewidths (~0.15–0.3 p.p.m.) and the presence of a unique set of NMR signals exclude the possibility of conformational heterogeneity in the wild-type needle state, whereas in X-ray structures of soluble, C-terminally truncated protein17 or the double-mutant PrgI* (V65A/V67A)18 the first ~18 N-terminal residues are disordered (Supplementary Fig. 1c). In particular, ssNMR reveals a highly ordered N-terminal segment (Thr 3–Tyr 8) with unusual chemical shifts that do not correspond to regular α-helical or β-sheet conformation. Akink (Val 20–Asn 22) interrupts the helix α1 that is not observed in the monomeric X-ray crystal structures18. In PrgI* needles, β-strand conformation was observed at the C terminus18 (Supplementary Fig. 1c), in contrast with the wild-type needle preparation. However, the observed structural differences could represent different functional states of the filament, as suggested previously6.
Figure 1. Structural data from ssNMR.
a, Secondary structure analysis. Positive and negative secondary chemical shifts are indicative of α-helical and β-sheet structure, respectively. b–f, Collection of ssNMR distance restraints. b, 13C–13C PDSD spectra of [1-13C]Glc-labelled T3SS needles. c, 13C–13C spectra of [2-13C]Glc-labelled (in magenta) and [(1/2)-13C]Glc-labelled (in blue) T3SS needles. Intersubunit restraints can be obtained from the [(1/2)-13C]Glc-labelled spectrum, for example, Gly 19Cα–Ala 35Cβ and Gly 19Cα–Ala 36Cβ. d, 13C–13C spectra of [1-13C]Glc-labelled (in green), [2-13C]Glc-labelled (in magenta) needles. e, 13C–13C spectra of [(1/2)-13C]Glc-labelled (in blue) T3SS needles. Intersubunit restraints are encoded in the [(1/2)-13C]Glc-labelled spectrum: Pro 41Cδ–Tyr 8Cβ and Ser 39Cα–Tyr 8Cβ. f, 15N–13C PAIN-CP spectrum of [2-13C]Glc-labelled T3SS needles.
The sparse [1-13C]Glc or [2-13C]Glc labelling significantly improves the quality of 13C–13C ssNMR spectra19, and numerous cross-peaks can readily be assigned to long-range distance restraints (see Supplementary Figs 3– 7 and Methods). The [1-13C]Glc-labelled20 sample (Fig. 1b) provides many methyl-based long-range restraints. Similarly, the [2-13C]Glc-labelled21 sample (Fig. 1c) shows excellent spectral resolution in the aromatic region, permitting the collection of unambiguous restraints. In total, 521 restraints were assigned (including 247 long-range ones; see Supplementary Table 1 and Methods), including long-range 15N–13C restraints (detected in a PAIN-CP (proton-assisted insensitive nuclei cross-polarization) spectrum22; Fig. 1f). Roughly twice as many unambiguous long-range restraints were found per residue than in the current benchmark in ssNMR structural studies, the HET-s prion domain23.
To distinguish intrasubunit from intersubunit distance restraints, we compared spectra of [1-13C]Glc-labelled, [2-13C]Glc-labelled and mixed [(1/2)-13C]Glc-labelled PrgI needle samples24 (Fig. 1d, e) and identified 162 intersubunit and 359 intrasubunit interactions that together define the overall organization of the T3SS needle assembly (Fig. 2).
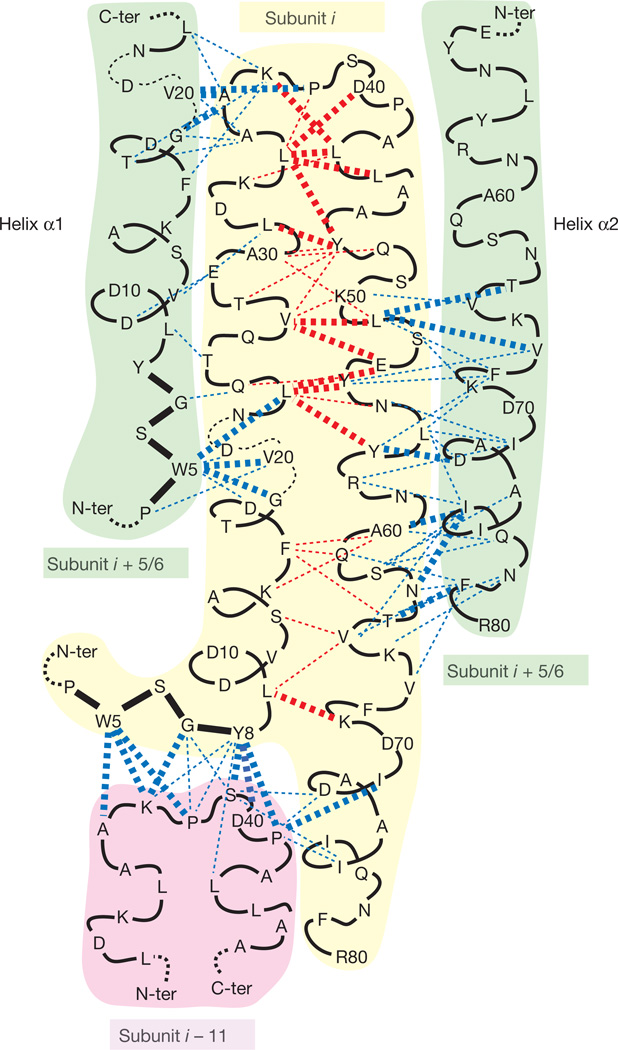
Figure 2. Architecture of the T3SS needle assembly as determined by ssNMR.
The dashed lines represent the intrasubunit (in red) and intersubunit (in blue) ssNMR distance restraints. Bold lines indicate the detection of more than three distance restraints. Lateral interactions between subunits i and i + 5 or i + 6 are not distinguishable at this stage of the analysis, but only after first rounds of Rosetta calculations (see Methods). N-ter, N terminus; C-ter, C terminus.
Intrasubunit restraints (Fig. 2, red lines) reveal a helical hairpin motif in PrgI. The kink segment (Val 20–Asn 22) disrupts the α1–α 2 interactions, with no intrasubunit long-range restraints detected for residues Asp 17–Asn 22 and their counterpart Arg 58–Asn 59. The N-terminal part of α1 is again interacting with the C-terminal part of α2. Multiple restraints (Fig. 2, bold lines) were detected between Leu 9 and Lys 69, the most N-terminal and C-terminal residues that take part in the helical hairpin motif. No intrasubunit long-range restraints were detected for the N-terminal (Thr 3–Tyr 8) and C-terminal (Asp 70–Arg 80) segments, but the high intensity of their NMR signals19 clearly demonstrates their rigidity and well-defined structure, indicating that these residues are stabilized through intersubunit contacts as described below.
The intersubunit distance restraints (Fig. 2, blue lines) reveal a lateral interface, mostly formed by helix–helix packing, and an axial interface. The lateral interface connects neighbouring PrgI subunits by means of α1–α1 and α2–α2 helix–helix packing in an arrangement in which the subunits are staggered along the filament axis with an axial translation of ~24Å(indicated for example by restraints involving Ala 60 of one molecule and Ile 76 of an adjacent one). The last five C-terminal residues that are known to be essential for the needle formation and infectivity, in particular Ile 76 and Phe 79, are involved in multiple intersubunit restraints. The central part of α1 helices (Asp 17–Leu 23, including the kink) interacts with adjacent subunits through the extended N terminus, in particular the Trp 5 aromatic ring, and also through the end of helix α1 (Ala 35–Lys 37) of the adjacent subunit. The ssNMR restraints also reveal strong intersubunit interactions between the extended N-terminal domain (Thr 3–Tyr 8) and the loop region (Ala 36–Pro 41), defining the axial packing between subunits on top of each other (Fig. 2).
According to previous scanning transmission electron microscopy (STEM) measurements25, the axial rise per subunit in PrgI needles is ~4.2Å. The intermolecular distance restraints show that laterally adjacent subunits are axially translated from one turn to the next by ~24Å, which divided by ~4.2Å yields ~5.7 subunits per turn. These values are consistent with a helical arrangement in which roughly 11 subunits are arranged in two turns. To generate three-dimensional models of the filament, rather than docking preformed monomeric structures which would not capture possible conformational changes on assembly, we extended the Rosetta symmetric fold-and-dock protocol26 to helical symmetry. Starting from a helically arrayed set of extended polypeptide chains, we optimized the total energy of the system including the ssNMR restraints and conformation-dependent chemical shifts by simultaneously sampling both the internal degrees of freedom of the monomers and the five rigid body degrees of freedom (the radius of the needle was restrained using the previously published cryo-electron microscopy map25). Models were generated for 9-start, 11-start, 13-start and 15-start helices (see Methods); the 11-start models had helical parameters more consistent with the STEM data25 and had more favourable ssNMR restraint energies (Supplementary Table 3).
In an 11-start helical arrangement, interactions between subunits i and i ± 11 constitute the axial interface, and subunit i is laterally surrounded by subunits i ± 5 and i ± 6. We used first-round models generated with only the unambiguous axial and intramolecular restraints to resolve the (5/6) ambiguities in the lateral restraints in two successive rounds of structure calculations (Supplementary Fig. 8). Whereas the inner α2 helices are parallel in the final models, the α1 helices present a cross-fenced pattern that emerges from the inclusion of two clusters of restraints, one involving Trp 5–Val 20 and the other Val 20–Ala 36 lateral interactions (Fig. 3; alternative assignments of these restraints resulted in significantly higher energy structures). The cross structure is further corroborated by the chemical shift data, which indicate a kink at Val 20 (Fig. 1a) as well as the lack of long-range intramolecular restraints for the same residue (Fig. 2). The combination of the ssNMR restraints with all-atom Rosetta refinement may yield near-atomic resolution; for example, the turn connecting the two helices has a backbone root mean squared deviation (r.m.s.d.) to the crystal structure of the monomer of less than 0.5Å (Supplementary Fig. 8g).
Figure 3. Complete atomic model of the T3SS needle.
a, b, Ribbon representation showing different subunits: side perspective (a); top view (b). The N-terminal domains (N-ter) are highlighted by red dashed circles. c–e, Selections of the subunit–subunit interfaces. c, The axial interface between subunits i and i − 11. d, The lateral interface between subunits i and i + 6. e, The lateral interface between subunits i and i + 5. Blue dashed lines represent ssNMR restraints.
The resulting T3SS needle model (Fig. 3 and Supplementary Fig. 9) consists of a ~80-Åouter diameter tube with a ~25-Åaxial lumen (Fig. 3b). The N-terminal domain was proposed to be α-helical in previous studies of the homologous Shigella flexneri needle17,27 and to form the inner shell of the filament. Our atomic model of the T3SS needle clearly diverges from this prediction: the N-terminal domain is not α-helical and is located on the exterior surface of the needle, not on the inner side.
To corroborate this finding, we performed immunoelectron microscopy on both S. typhimurium cells and needles polymerized in vitro (Supplementary Figs 10 and 11). S. typhimurium prgI-knockout cells complemented with prgI fused to an upstream Strep-tag II were labelled with monoclonal anti-Strep antibodies (Supplementary Figs 11 and 13). The Strep-PrgI is fully functional in HeLa cell invasion assays of complemented S. typhimurium prgI-knockout cells (Supplementary Fig. 14). Similarly, in vitro needles of an N-terminal Histag fusion with wild-type PrgI bind to an anti-His-tag monoclonal antibody that decorates the exterior surface of such needles (Supplementary Figs 10 and 12b). These results confirm that the PrgI N terminus is surface-exposed, an arrangement similar to that of the flagellar filament14, which shares a genetic origin with the T3SS needle.
The rigid extended conformation of the N-terminal residues reported here clearly differs from previous models. The two aromatic residues Trp 5 and Tyr 8 form the basis of the axial N-terminal-domain–loop interface (Fig. 3c). Indeed, in vivo invasion assays show that the single point mutant Y8A leads to decreased invasiveness of S. typhimurium (Supplementary Fig. 14), and the double mutation W5A/Y8A completely abrogates host cell invasion (Supplementary Fig. 14). Trp 5 also seems to have a crucial function in the lateral i to i + 6 interface (Fig. 3d; and W5A/V20A double mutant in Supplementary Fig. 14). Furthermore, the atomic model reveals that the lateral i to i ± 5,6 interface strongly involves α2–α2 helix–helix contacts (Fig. 3e) and that the last C-terminal residues Ile 75, Ile 76 and Phe 79 are essential. This is confirmed by the non-invasiveness of the I76A/F79A PrgI double mutant (Supplementary Fig. 14). In addition, we compared S. typhimurium wild-type and prig-knockout cells either complemented with wild-type or mutated prgI by using effector secretion assays and transmission electron microscopy (TEM) of a negatively stained specimen. The results obtained (Supplementary Fig. 16) agree well with the invasion assays shown in Supplementary Fig. 14. SDS–PAGE and western blot analysis showed that the double mutations (W5A/Y8A and I76A/F79A), but not the single mutations (W5A and Y8A), abolish effector secretion (Supplementary Fig. 16a, b). However, the W5A/V20A double mutant can secrete effector proteins, which is also in agreement with the results previously obtained by bacterial invasion assays. TEM analysis of these strains confirmed needles only in wild-type and prgI-knockout cells complemented with wild-type prgI (Supplementary Fig. 16c, d) but not in the mutant strains, which might be an indication of decreased needle stability.
Residues conserved between S. typhimurium, Shigella flexneri, Yersinia pestis and Escherichia coli T3SS needle proteins are mostly facing towards the lumen (Supplementary Figs 15 and 17). Weakly or non-conserved residues are exposed to the needle surface and are thus accessible from outside the pathogen, which could reflect a bacterial strategy to evade a host-cell immune response. The inner surface of the channel consists mainly of polar residues (see Supplementary Fig. 18 for the electrostatic surface potential of the T3SS channel). In a similar manner to the situation in the flagellar filament, the most C-terminal residue Arg 80 (in flagellin this is Arg 494 (ref. 14)) places a positive charge into the channel.
Our approach opens an avenue for the investigation of the subunit interfaces of other homo-oligomeric assemblies at atomic resolution in their native-like state28–30. For larger or more complex systems, such as the flagellar filament14, our combination of atomic-level ssNMR structural restraints and cryo-electron microscopy data could bridge the gap between X-ray or NMR structures of monodisperse subunits and cryo-electron microscopy data of the filamentous state.
METHODS
Expression, purification and polymerization of 15N-labelled and 13C-labelled wild-type PrgI protein
The synthetic full-length PrgI encoding DNA was purchased from GeneArt and cloned into a modified pET16b vector (Novagen). The resulting construct codes for a fusion protein with an N-terminal His7 tag followed by a tobacco etch virus (TEV) protease cleavage recognition sequence that connects it to the PrgI protein sequence. Thus, after protease cleavage the released PrgI protein contains the non-native N-terminal residues glycine and histidine. The construct was transformed into E. coli strain BL21(DE3). Expression of labelled PrgI protein was performed in minimal medium with 15NH4Cl as nitrogen source and d-[U-13C6]glucose, d-[1-13C]glucose or d-[2-13C]glucose as carbon source. An overnight minimal medium culture was inoculated 1:25 (v/v) into 1 l of minimal medium. The culture was shaken at 37 °C until a D600nm of 0.7. Protein expression was induced by adding 0.7 mM isopropyl β-d-thiogalactoside; 5 h after induction the cells were harvested and stored at −80 °C.
The cell pellet of a 1-litre expression culture was dissolved in 50 ml of lysis buffer (100 mM sodium phosphate, 10mM Tris-HCl, pH 8.0, 8M urea, 0.5 mM phenylmethylsulphonyl fluoride (PMSF)) and stirred for 1 h at room temperature. Subsequently, the suspension was centrifuged for 30 min at 33,000g and the supernatant was incubated on a rocking shaker for 1 h with 3 ml of Ni2+-nitrilotriacetate (Qiagen) resin that had been pre-equilibrated with the lysis buffer. The resin suspension was filled into a 10-ml disposable plastic column (Pierce). The column was washed with 60 ml of lysis buffer and the protein was eluted with 8 × 2 ml of elution buffer 1 (100 mM sodium phosphate, 10 mM Tris-HCl, pH5.9, 8M urea, 0.5 mM PMSF) and 8 × 2 ml of elution buffer 2 (100 mM sodium phosphate, 10 mM Tris-HCl, pH4.5, 8M urea, 0.5 mM PMSF). The elution fractions were analysed on a 17.5% SDS–PAGE gel. Fractions containing the fusion protein were combined and concentrated with a Vivascience 5-kDa molecular mass cut-off concentrator (Sartorius Stedim Biotech) to a final volume of 4 ml. Subsequently, 2-ml aliquots of the sample were loaded onto a 8 mm × 250 mm semipreparative HPLC column (RP18 Eurospher 100; Knauer) that had been pre-equilibrated with 0.1% aqueous trifluoroacetic acid (TFA). The column was washed with ten column volumes of 0.1% TFA, and the protein was eluted at a flow rate of 3 ml min−1 with a 150-ml linear gradient of 0–100% acetonitrile/0.1% TFA. The elution peaks were analysed by electrospray ionization mass spectrometry (ESI-MS). Elution fractions containing the fusion protein without any further detectable impurities were combined and freeze-dried.
The freeze-dried protein was dissolved in ice-cold water by vortex-mixing and then incubated for 1 h on ice. Subsequently, using cold stock solutions, the following buffer was adjusted in this aqueous protein solution: 50 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 1 mM dithiothreitol, 0.5 mM PMSF. To this buffered protein solution TEV protease was added at a final ratio of 3 mg of TEV protease per 100 mg of fusion protein. The digestion was performed for 20 h at 4 °C. The released PrgI protein was further purified by reverse-phase HPLC as described above for the fusion protein. Elution fractions containing pure PrgI protein according to ESI-MS analysis were combined and freeze-dried.
The freeze-dried protein was dissolved at 0.085 mM concentration (ε280 = 10,810 M−1 cm−1) in ice-cold water by vortex-mixing and dialysed at 4 °C against 20 mM MES, pH 5.5, 0.02% sodium azide. Finally, the sample was concentrated with a Vivascience 5-kDa molecular mass cut-off concentrator to a final concentration of 0.1 mM.
The polymerization of the PrgI protein took place by incubation at room temperature for 1 week without shaking the sample. Thereafter the needles were centrifuged for 30 min at 52,000g. The protein pellet was washed extensively with 20 mM MES pH 5.5 before transfer to the ssNMR rotor.
Invasion assays
HeLa cell invasion assays were performed as described in ref. 18. In brief, cells were infected with S. typhimurium SL1344 and PrgI mutants at multiplicity of infection of 10:1 after inducing their expression with 0.2 µg ml−1 anhydrotetracycline for 1 h. After 20 min of infection, the cells were washed with PBS, and fresh medium supplemented with 100 µg ml−1 gentamicin was added. Cells were incubated for a further 2 h to allow intracellular survival and replication of bacteria. They were then lysed with 0.1% Triton X-100 and the lysates were plated on tryptic soy agar plates to count the number of viable bacteria after 16 h.
Secretion assays
Single colonies of S. typhimurium SL1344 and PrgI mutants were inoculated in Luria–Bertani medium with 0.04 µg ml−1 anhydrotetracycline and incubated with shaking at 37 °C for 16 h. The cultures were centrifuged at 4,354g for 10 min to remove the bacteria. The supernatants were filtered through 0.22-mm filters and precipitated with trichloroacetic acid. Pellets were washed once with acetone, dried, resuspended in SDS loading buffer and analysed by SDS–PAGE.
Preparation of cells for electron microscopy analysis
Cultures of S. typhimurium were grown as described above, and 1 ml aliquots were fixed with 2% para-formaldehyde for 1 h. The fixed cells were washed once with PBS and resuspended in 200 µl of PBS for electron microscopy analysis.
ssNMR spectroscopy and detection of distance restraints
ssNMR experiments were conducted on 14.1-T and 20.0-T (1H resonance frequencies of 600 and 850 MHz, respectively) wide-bore spectrometers (Bruker Biospin) equipped with 4mm triple-resonance (1H, 13C and 15N) MAS probes. Samples were packed in 4-mm MAS rotors, using protein quantities of ~10 mg. Spectra were recorded at MAS spinning rates of 10.75 or 11 kHz and were calibrated with 4,4-dimethyl-4-silapentane-1-sulphonic acid (DSS) as an internal reference. The temperature-dependent position of the water proton resonance was used to measure the temperature inside the MAS rotor31. All experiments were conducted at a sample temperature of 278 K. High-power 1H–13C decoupling (SPINAL-64)32 with a radiofrequency amplitude of 83 kHz was applied during evolution and detection periods. An initial ramped cross-polarization was used to transfer magnetization from 1H to 13C with a contact time of 1.5 ms. 13C–13C polarization transfer was achieved by means of PDSD33 with mixing times as indicated in the text and in the supplementary figures.
A 15N–13C PAIN-CP spectrum was recorded as described in ref. 19. Total experimental times were 4–9 days depending on the number of scans (maximum acquisition times t1 of 12 ms for [1-13C]Glc-labelled and [2-13C]Glc-labelled samples, and 7 ms for the mixed-labelled sample were used). Distance restraints with a unique assignment (spectrally unambiguous) were collected in a first round of analysis (corresponding to ~70% of the total number of restraints) and used to clarify the assignment of additional ambiguous restraints (with four or fewer assignment possibilities). Intermolecular distance restraints were first assigned in the [(1/2)-13C]Glc-labelled sample by direct comparison with [1-13C]Glc-labelled and [2-13C]Glc-labelled samples, as described in ref. 24. On the basis of this first set of restraints, additional intermolecular distances were later identified in spectra of the [1-13C]Glc-labelled and [2-13C]Glc-labelled samples. NMR spectra were analysed with CcpNmr34.
Modelling
A modification of the fold-and-dock protocol26, now part of the standard Rosetta 3.3 distribution, was used for all model calculations. All symmetric subunits within the needle helix are treated explicitly in a 29-subunit system. The protocol consists of backbone fragment insertion trials that are replicated among symmetric subunits and symmetrical rigid body trials of adjacent subunits using a coarse-grain representation of the system, followed by side-chain packing and all-atom refinement of the side chain, backbone and rigid body degrees of freedom, using a physically relevant energy function35.
The chemical shift assignments of the backbone 15N and 13C atoms from ssNMR experiments were used to bias the selection of overlapping three-residue and nine-residue fragments of backbone conformations. Using these fragments and the intrasubunit and intersubunit restraints, we performed a series of model calculations, starting from a helix of extended chains. A pseudo-energy term was used to restrain the calculations according to the agreement with the experimental distances, while optimizing the Rosetta energy function. ssNMR cross-peaks were converted to internuclear C–C and C–N distance restraints with an upper limit of 5Å and a penalty that grew exponentially with distance.
Intersubunit distance restraints were first classified into two categories: the ‘axial’ restraints corresponding to i to i ± 11 (equivalently i to i ± n for an n-start arrangement in the more general case) intersubunit interactions and the ‘lateral’ restraints corresponding to i to i ± 5/6 intersubunit interactions. The resolution of (5/6) ambiguities was made through the manual identification of self-consistent clusters of contacts between subunits i and i + 5 or between subunits i and i + 6.
In detail, this was done in three stages. In stage I, using the axial and intrasubunit restraints alone, we identify two clusters of helical arrangements (Supplementary Fig. 8b). The first cluster corresponds to a left-handed helix of 5.6 subunits per turn that showed lateral interactions between subunits i to i + 5 and i to i + 6, and the second cluster is a right-handed helix with 11 subunits per turn that shows lateral interactions between subunits i to i − 1 and i to i + 1. This model is in disagreement with the estimates of subunits per turn of ~5.6 from STEM and ssNMR data (see the text). We therefore focus on the first cluster, using which we can easily assign two sets of lateral ssNMR restraints that report on the i to i + 5 and i to i + 6 interfaces between the inner (C-terminal) helices (Supplementary Fig. 8a). Using the axial, intrasubunit and the stage I assigned lateral restraints we perform a second set of calculations, one for each helical handedness as the same lateral interactions can be satisfied by a right-handed helix and by a left-handed helix (stage IIR and stage IIL). This is done by keeping the axial and intersubunit interactions fixed but interchanging the restraints assigned for the i to i + 5 and i to i + 6 interfaces (Supplementary Fig. 8d). Using this second round of models we can further assign two clusters of restraints that correspond to lateral interactions connecting the outer helices (Supplementary Fig. 8c). This corresponds to two clusters of interactions involving W5 from subunit i + 11 and A36 from subunit i with the V20 kink region of subunit i + 6 in the left-handed model, shown with red and yellow dashes, respectively, in Supplementary Fig. 8c. A final stage of calculations is then performed using all assigned lateral restraints for each helix (right-handed and left-handed) and the resulting ensembles are reported as the final models. The calculations converge within 2.1Å backbone r.m.s.d. for the right-handed and 2.3Å r.m.s.d. for the left-handed model (Supplementary Fig. 8e, f). Both models satisfy the experimental restraints with a minimal number of violations, although the right-handed model shows more favourable restraint and interface energies. Furthermore the two ensembles show very similar helical parameters (number of subunits per turn, radius and axial rise) and also full-atom interface energies (Supplementary Table 2). We note that the central part of the needle protein (Leu 34–Leu 43) is highly conserved between our atomic model and the crystal structure of PrgI* (ref. 18) (Supplementary Fig. 8g).
The radius of the needle was restrained during each calculation by using a pseudo-energy function that measures the correlation of the whole model to the previously published cryo-electron microscopy map25. In all calculations, no assumptions were made about the helical parameters of the needle (that is, the degree of angle rotation and unit translation along the helical axis). Instead, these parameters emerge in the final structural ensemble according to the interpretation of the intermolecular interactions. Calculations are repeated for different interpretations of the intersubunit distance restraints according to 9-start, 13-start and 15-start helical arrangements in addition to the 11-start presented here. These complementary calculations result in converged structural ensembles with similar axial and lateral interfaces but involving interactions between different symmetry-related monomers and helical parameters (Supplementary Table 3). Although no unambiguous distinction can be made between these geometries in terms of agreement with the ssNMR distance restraints alone, the symmetry considerations presented in the main text suggest that the 11-start arrangement agrees better with the complementary STEM measurements. This was the basis for selecting the 11-start needle as the final model.
Supplementary Material
Acknowledgements
We thank T. C. Marlovits and E. H. Egelman for providing the S. typhimurium T3SS needle cryo-electron microscopy density map; F. DiMaio and J.-P. Demers for discussions; and G. Wolf, B. Angerstein and G. Heim for technical help. This work was supported by the Max Planck Society (to C. Griesinger), the Deutsche Forschungsgemeinschaft (Emmy Noether Fellowship to A. Lange), the Fondation Bettencourt Schueller (to A. Loquet), the National Institutes of Health (1R01GM092802-01 to D.B.), EMBO (postdoctoral fellowship to A. Loquet), and the European Union Seventh Framework Program under Grant Agreement 261863 (Bio-NMR).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions A. Loquet performed ssNMR experiments. A. Loquet and A. Lange analysed ssNMR data. N.S. and D.B. performed structure calculations. K.G. and S.B. expressed, purified and polymerized in vitro T3SS needles. R.G. and M.K. performed the in vivo studies. C. Griesinger analysed NMR data. D.R. and C. Goosmann performed electron microscopy studies. A. Loquet and A. Lange wrote the paper; all authors discussed the results and commented on the manuscript.
Author Information The Salmonella typhimurium T3SS PrgI needle structure is deposited in the Protein Data Bank under the accession code 2LPZ. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Galan JE, Wolf-Watz H. Protein delivery intoeukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 2.Cornelis GR. The type III secretion injectisome. Nature Rev. Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 3.Kubori T, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 4.Kimbrough TG, Miller SI. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl Acad. Sci. USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamano K, et al. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO. J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blocker AJ, et al. What’s the point of the type III secretion system needle? Proc.Natl Acad. Sci. USA. 2008;105:6507–6513. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nogales E, Grigorieff N. Molecular machines: putting the pieces together. J. Cell Biol. 2001;152:F1–F10. doi: 10.1083/jcb.152.1.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volkmann N, Hanein D. Docking of atomic models into reconstructions from electron microscopy. Methods Enzymol. 2003;374:204–225. doi: 10.1016/S0076-6879(03)74010-5. [DOI] [PubMed] [Google Scholar]
- 9.Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining X-ray crystallography and electron microscopy. Structure. 2005;13:355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spreter T, et al. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nature Struct. Mol. Biol. 2009;16:468–476. doi: 10.1038/nsmb.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker ML, Zhang J, Ludtke SJ, Chiu W. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nature Protocols. 2010;5:1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schraidt O, Marlovits TC. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science. 2011;331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- 13.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 14.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 15.Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig L, et al. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Deane JE, et al. Molecular model of a type III secretion system needle: implications for host-cell sensing. Proc. Natl Acad. Sci. USA. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poyraz O, et al. Protein refolding is required for assembly of the type three secretion needle. Nature Struct. Mol. Biol. 2010;17:788–792. doi: 10.1038/nsmb.1822. [DOI] [PubMed] [Google Scholar]
- 19.Loquet A, Lv G, Giller K, Becker S, Lange A. 13C spin dilution for simplified and complete solid-state NMR resonance assignment of insoluble biological assemblies. J. Am. Chem. Soc. 2011;133:4722–4725. doi: 10.1021/ja200066s. [DOI] [PubMed] [Google Scholar]
- 20.Hong M. Determination of multiple w-torsion angles in proteins by selective and extensive13C labeling and two-dimensional solid-state NMR. J. Magn. Reson. 1999;139:389–401. doi: 10.1006/jmre.1999.1805. [DOI] [PubMed] [Google Scholar]
- 21.Lundstrom P, et al. Fractional13C enrichment of isolated carbons using [1-13C]- or [2-13C]-glucose facilitates the accurate measurement of dynamics at backbone Ca and side-chain methyl positions in proteins. J. Biomol. NMR. 2007;38:199–212. doi: 10.1007/s10858-007-9158-6. [DOI] [PubMed] [Google Scholar]
- 22.Lewandowski JR, De Paepe G, Griffin RG. Proton assisted insensitive nuclei cross polarization. J. Am. Chem. Soc. 2007;129:728–729. doi: 10.1021/ja0650394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasmer C, et al. Amyloid fibrils of the HET-s(218–289) prion forma beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 24.Loquet A, Giller K, Becker S, Lange A. Supramolecular interactions probed by13C–13C solid-state NMR spectroscopy. J. Am. Chem. Soc. 2010;132:15164–15166. doi: 10.1021/ja107460j. [DOI] [PubMed] [Google Scholar]
- 25.Galkin VE, Schmied WH, Schraidt O, Marlovits TC, Egelman EH. The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J. Mol. Biol. 2010;396:1392–1397. doi: 10.1016/j.jmb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das R, et al. Simultaneous prediction of protein folding and docking at high resolution. Proc. Natl Acad. Sci. USA. 2009;106:18978–18983. doi: 10.1073/pnas.0904407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenjale R, et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 28.Goldbourt A, Gross BJ, Day LA, McDermott AE. Filamentous phage studied by magic-angle spinning NMR: resonance assignment and secondary structure of the coat protein in Pf1. J. Am. Chem. Soc. 2007;129:2338–2344. doi: 10.1021/ja066928u. [DOI] [PubMed] [Google Scholar]
- 29.Han Y, et al. Solid-state NMR studies of HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 2010;132:1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jehle S, et al. Solid-state NMR and SAXS studies provide a structural basis for the activation of αB-crystallin oligomers. Nature Struct. Mol. Biol. 2010;17:1037–1042. doi: 10.1038/nsmb.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bockmann A, et al. Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR. 2009;45:319–327. doi: 10.1007/s10858-009-9374-3. [DOI] [PubMed] [Google Scholar]
- 32.Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 33.Szeverenyi NM, Sullivan MJ, Maciel GE. Observation of spin exchange by two-dimensional Fourier-transform C-13 cross polarization-magic-angle spinning. J. Magn. Reson. 1982;47:462–475. [Google Scholar]
- 34.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 35.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.