Abstract
We have conducted the first meta-analyses for nonsyndromic cleft lip with or without cleft palate (NSCL/P) using data from the two largest genome-wide association studies published to date. We confirmed associations with all previously identified loci and identified six additional susceptibility regions (1p36, 2p21, 3p11.1, 8q21.3, 13q31.1 and 15q22). Analysis of phenotypic variability identified the first specific genetic risk factor for NSCLP (nonsyndromic cleft lip plus palate) (rs8001641; PNSCLP = 6.51 × 10−11; homozygote relative risk = 2.41, 95% confidence interval (CI) 1.84–3.16).
Nonsyndromic cleft lip with or without cleft palate is one of the most common birth defects in humans. Formal genetic and epidemiological studies have indicated a multifactorial etiology, with both genetic and environmental factors contributing to disease risk.
Four genome-wide association studies (GWAS) of NSCL/P have been published to date1. These identified five new susceptibility loci, thus adding to the previously identified susceptibility gene IRF6 (ref. 2). To further elucidate the genetic architecture of NSCL/P, we combined data from the two largest GWAS3,4. Data from the Baltimore study3 were retrieved from the database of Genotypes and Phenotypes (dbGaP) after approval was obtained for data access. After quality control (Supplementary Methods), 666 complete European trios (including European Americans) and 795 complete Asian trios from that study were combined with 399 cases and 1,318 controls of Central European origin (Bonn-II study)4. This combined sample represents approximately 95% of all previously reported individuals with NSCL/P. A likelihood ratio test (LRT; Supplementary Methods) was performed on 497,084 SNPs.
First, we combined all European case-control data with the subset of European-American trios. This meta-analysisEuro yielded 47 SNPs from six chromosomal regions that were associated with NSCL/P with genome-wide significance (Fig. 1a and Table 1). Three of these regions have been identified in previous reports (8q24, 10q25 and 17q22). Notably, 36 SNPs mapped to the key locus at 8q24 (lowest P value for rs987525, P = 3.94 × 10−34). Of the three new regions that were associated with genome-wide significance, two (2p21: rs7590268, P = 4.05 × 10−8; 13q31: rs8001641, P = 6.20 × 10−10) were implicated in the Bonn-II study, with subsequent replication of these associations in the independent EuroCran sample4. The third locus (15q22: rs1873147, P = 2.81 × 10−8) was also implicated in the initial sample of the Bonn-II study, although an attempt at independent replication was unsuccessful4. The present independent evidence obtained from the Baltimore European trio sample (PTDT_Euro = 2.37 × 10−3; Table 1) again suggests that this locus contributes to NSCL/P.
Figure 1.
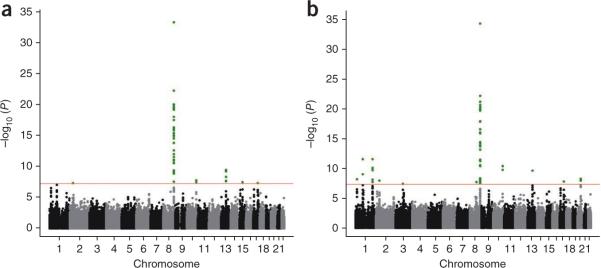
Manhattan plots for meta-analyses. (a,b) Genome-wide meta-analyses for NSCL/P in Europeans (a) and combined European and Asian samples (b). Negative log10 P values are shown for all SNPs that passed quality control. Genome-wide significance (P < 5 × 10−8) is indicated by the red line. SNPs surpassing this threshold are shown in green.
Table 1.
Meta-analysis of NSCL/P
| Locus information |
European population |
Combined European and Asian populations |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dbSNP ID (top SNP) | Allelea | Chr. | Position (Mb) | GWS SNPs at locus | P case-control | P TDT_Euro | P meta_Euro | RR het (95% CI) | RR hom (95% CI) | P TDT_all | P meta_all | RR het (95% CI) | RR hom (95% CI) |
| SNPs at previously confirmed loci | |||||||||||||
| rs560426 | G/A | 1p22.1 | 94.32–94.35 | 2 | 5.25 × 10−3 | 4.11 × 10−5 | 1.02 × 10−6 | 1.344 (1.126–1.604) | 1.722 (1.383–2.144) | 6.70 × 10−11 | 3.14 × 10−12 | 1.420 (1.243–1.623) | 1.862 (1.556–2.228) |
| rs861020 | A/G | 1q32.2 | 208.00–208.12 | 5 | 8.20 × 10−5 | 3.92 × 10−3 | 1.78 × 10−6 | 1.436 (1.224–1.685) | 1.707 (1.240–2.348) | 7.76 × 10−9 | 3.24 × 10−12 | 1.443 (1.273–1.635) | 2.039 (1.598–2.602) |
| rs987525 | A/C | 8q24 | 129.77–130.30 | 36 | 3.09 × 10−21 | 2.59 × 10−15 | 3.94 × 10−34 | 2.074 (1.755–2.450) | 4.681 (3.581–6.120) | 1.06 × 10−16 | 5.12 × 10−35 | 1.919 (1.660–2.218) | 4.384 (3.393–5.666) |
| rs7078160b | A/G | 10q25 | 118.81–118.83 | 2 | 9.50 × 10−6 | 4.24 × 10−4 | 2.81 × 10−8 | 1.459 (1.238–1.719) | 2.214 (1.555–3.153) | 3.08 × 10−7 | 3.96 × 10−11 | 1.383 (1.213–1.576) | 1.941 (1.579–2.385) |
| rs227731 | C/A | 17q22 | 52.12 | 1 | 1.58 × 10−5 | 5.58 × 10−4 | 4.26 × 10−8 | 1.274 (1.069–1.519) | 1.838 (1.480–2.284) | 1.02 × 10−4 | 1.78 × 10−8 | 1.228 (1.078–1.400) | 1.669 (1.400– 1.989) |
| rs13041247 | C/T | 20q12 | 38.70–38.71 | 3 | 2.71 × 10−1 | 3.22 × 10−4 | 7.41 × 10−4 | 0.921 (0.789–1.076) | 0.605 (0.468–0.782) | 6.40 × 10−10 | 6.17 × 10−9 | 0.837 (0.742–0.944) | 0.545 (0.447–0.663) |
| SNPs at newly identified loci | |||||||||||||
| rs742071 | T/G | 1p36 | 18.85 | 1 | 4.88 × 10−3 | 7.85 × 10−6 | 2.63 × 10−7 | 1.248 (1.049–1.486) | 1.800 (1.444–2.244) | 1.43 × 10−7 | 7.02 × 10−9 | 1.316 (1.126–1.537) | 1.878 (1.519–2.323) |
| rs7590268 | G/T | 2p21 | 43.39 | 1 | 1.11 × 10−5 | 5.81 × 10−4 | 4.05 × 10−8 | 1.419 (1.212–1.662) | 2.040 (1.510–2.756) | 1.59 × 10−4 | 1.25 × 10−8 | 1.415 (1.225–1.636) | 1.978 (1.474–2.656) |
| rs7632427 | C/T | 3p11.1 | 89.61 | 1 | 1.47 × 10−2 | 8.99 × 10−4 | 4.20 × 10−5 | 0.787 (0.673–0.920) | 0.627 (0.494–0.797) | 5.21 × 10−7 | 3.90 × 10−8 | 0.731 (0.644–0.830) | 0.609 (0.490–0.757) |
| rs12543318 | C/A | 8q21.3 | 88.93 | 1 | 6.83 × 10−3 | 3.05 × 10−5 | 1.02 × 10−6 | 1.255 (1.068–1.475) | 1.832 (1.446–2.321) | 7.72 × 10−7 | 1.90 × 10−8 | 1.272 (1.106–1.463) | 1.676 (1.400–2.007) |
| rs8001641 | A/G | 13q31.1 | 79.57–79.60 | 6 | 2.83 × 10−7 | 3.36 × 10−4 | 6.20 × 10−10 | 1.461 (1.199–1.781) | 2.033 (1.617–2.556) | 6.16 × 10−5 | 2.62 × 10−10 | 1.307 (1.130–1.511) | 1.863 (1.537–2.258) |
| rs1873147 | C/T | 15q22.2 | 61.09 | 1 | 7.04 × 10−7 | 2.37 × 10−3 | 2.81 × 10−8 | 1.467 (1.251–1.719) | 1.890 (1.444–2.474) | 9.98 × 10−3 | 7.92 × 10−7 | 1.431 (1.230–1.666) | 1.652 (1.340–2.037) |
Relative risks (RRs) are given with the major allele set as baseline. Pcase-control is the P value from the Bonn-II GWAS. PTDT is the statistic from the Baltimore study, recalculated for the present meta-analyses. P values are in bold if genome-wide significance was reached. Chr., chromosome; het, heterozygous; hom, homozygous; TDT, transmission disequilibrium test; GWS, genome-wide significant.
The minor allele is given first. The risk allele is shown in bold.
rs7078160 was chosen as the top SNP at this locus on the basis of the initial findings in ref. 4. A second marker, rs4752028, is in high LD with rs7078160 and may also be described as a top marker.
In a second step, we added the Asian trios (meta-analysisall) to determine which of the loci identified in the European population also confer risk in Asian populations and to identify susceptibility regions that are common to European and Asian ancestry groups but which may have escaped detection in meta-analysisEuro due to limited power. Fifty SNPs showed associations that reached genome-wide significance (Fig. 1b). Five of the six loci identified in meta-analysisEuro had even smaller association P values in meta-analysisall, suggesting that they contribute to NSCL/P in both European and Asian populations (Table 1). One locus (at 15q22) showed association that did not reach statistical significance, suggesting that it may only be implicated in cases from a defined ancestry group. Notably, inclusion of the Asian sample yielded six additional regions that associated with genome-wide significance (Table 1), including three previously reported regions (1p22.1: rs560426, P = 3.14 × 10−12; 1q32.2 (IRF6): rs861020, P = 3.24 × 10−12; 20q12: rs13041247, P = 6.17 × 10−9). In addition, three new loci that associated with NSCL/P with genome-wide significance were detected. The locus at 1p36 (rs742071, P = 7.02 × 10−9) was implicated at a suggestive level of significance in the Baltimore study. The other two new loci (3p11.1: rs7632427, P = 3.90 × 10−8; 8q21.3: rs12543318, P = 1.90 × 10−8) have not been reported previously. For each of the newly identified loci associated with genome-wide significance, data for the Asian trios only are presented in Supplementary Table 1.
To exclude genotyping errors as a confounding factor in the identification of new loci, we subsequently imputed the six genomic regions using IMPUTE2 (see URLs) and performed locus-specific meta-analyses based on the best-guess genotypes (Supplementary Methods). We observed highly significant association P values for both imputed and genotyped SNPs in high linkage disequilibrium (LD) in meta-analysisEuro and meta-analysisall, respectively (Supplementary Table 2). This suggests that the single markers shown to associate in the GWAS are true positives. Association results within the respective genomic contexts are shown in Supplementary Figure 1.
Several lines of evidence support the hypothesis that these newly identified loci contribute to orofacial clefting. The associated single SNP at 1p36 is located in an intron in the PAX7 gene (encoding paired box 7). PAX7 has already been functionally implicated in craniofacial development5. A further study investigated seven PAX7 variants in NSCL/P case-parent trios from various populations6, and two of these variants showed a strong parent-of-origin effect. Analysis of rs742071 in the trio data from the Baltimore study provided no support for a parent-of-origin effect (data not shown).
The rs7632427 SNP at 3p11.1 is located approximately 3 kb downstream of the EPHA3 gene (encoding ephrin receptor A3). Members of the ephrin receptor family are involved in the regulation of cell shape and cell-cell contacts7. A strong methylation site was detected approximately 200 bp downstream of rs7632427 (ref. 8). This represents a starting point for investigations into the role of methylation patterns in craniofacial development.
For three additional regions, possible regulatory functions can be hypothesized. The single associated marker at 2p21 is located within the THADA gene (encoding thyroid adenoma associated). Reported THADA functions9 are not implicated in craniofacial development. However, variants within THADA have been associated with a range of diseases. This may be related to the large size of THADA (370 kb) or to the effects of regulatory elements. The latter hypothesis is supported by the presence of transcription factor–binding sites described in the Encyclopedia of DNA Elements (ENCODE; see URLs). We also propose a regulatory effect for the 13q31.1 markers mapping to an intergenic region. Adjacent genes include SPRY2 (encoding sprouty homolog 2), which is an excellent candidate for orofacial clefting according to animal models10,11. The SNP associated with genome-wide significance at 15q22.2, which was not associated in Asians but was strongly associated in Europeans, is located approximately 20 kb upstream of the TPM1 gene (encoding tropomysin 1). The associated variant maps to a regulatory region containing both strong enhancer and promoter signatures and multiple transcription factor–binding sites, as predicted by ENCODE. For the associated marker at 8q21.3, which maps to an intergenic region, no functional annotation related to orofacial clefting is currently available. All SNPs associated with P of <1 × 10−4 in meta-analysisEuro or in meta-analysisall are listed in Supplementary Table 3.
Orofacial clefts show considerable phenotypic variability1. Epidemiological and embryological data support the hypothesis that NSCL/P can be subdivided into NSCLO, where only the lip is affected, and NSCLP, where both the lip and palate are affected12. Despite a substantial etiological overlap between these two anatomical malformations, epidemiological studies suggest that distinct genetic factors may be involved in their respective development13.
To address these etiological differences for the first time on a systematic, genome-wide level, we performed separate analyses of individuals with NSCLO and NSCLP (Supplementary Fig. 2 and Supplementary Methods). In total, 38 SNPs had associations that reached genome-wide significance. All of these 38 SNPs were among the SNPs that associated with genome-wide significance in at least one of the two meta-analyses on the combined NSCL/P group (Supplementary Table 4). For the 13q31.1 locus, association P values in the NSCLP group were lower than those obtained in both meta-analysisEuro and meta-analysisall, suggesting that this locus has a particularly strong effect on the development of a cleft palate following the development of a cleft lip. This finding was exclusive to NSCLP (Supplementary Table 5). To account for a possible bias secondary to the different sample sizes of the two subgroups, we tested the hypothesis that estimated relative risks for each of these 38 SNPs were identical in the NSCLO and NSCLP subgroups using heterogeneity LRT (Supplementary Methods). In the European analysis, a statistically significant difference in genotypic relative risk was observed between the two groups for two SNPs in the 13q31.1 region (Supplementary Tables 5 and 6). For the top marker rs8001641 (PNSCLP = 6.51 × 10−11; PNSCLO = 0.163), the homozygous relative risk was 2.41 (95% CI 1.84–3.16) in the NSCLP group compared to 1.33 (0.89–1.99) in the NSCLO group. Similar data were obtained in the combined European and Asian analysis (Supplementary Table 6). Two SNPs in the 13q31 region were associated in the replication step of the Bonn-II GWAS. Reanalysis of these data by NSCLO and NSCLP subgroups revealed even stronger association results when only NSCLP cases were considered (Supplementary Table 7). These data suggest that the 13q31 locus is the first identified genetic risk factor to contribute exclusively to the NSCLP subphenotype of clefting. This finding is different from previous reports on subgroup-specific contributing factors, such as IRF6 (ref. 2). Although these reports showed stronger association in one subgroup, association of at least nominal significance was also found in the second subgroup. Thus, our data now provide support for recent epidemiological observations at the molecular level.
The associated region at 13q31 maps downstream of SPRY2 (Supplementary Fig. 3). Although Spry2-knockout mice frequently have cleft palate14, the incidence of this anomaly is even higher in mice carrying a larger deletion of a region including Spry2 (ref. 11). This suggests that regulatory elements around SPRY2 contribute to palatal defects. Furthermore, a resequencing study of individuals with NSCL/P suggested the presence of rare and possibly detrimental variants in SPRY2 (ref. 15).
In conclusion, the present meta-analyses conclusively implicate six novel loci, associated with genome-wide significance, in the etiology of NSCL/P, five of which seem to be involved in both European and Asian populations. Future studies are warranted to identify specific causal variants for these associations. Furthermore, the present study is the first to our knowledge to identify a genetic locus that contributes exclusively to cleft palate in the presence of a cleft lip.
Supplementary Material
ACKNOWLEDGMENTS
We thank all affected individuals and their families for their participation in this study, as well as the German support group for people with cleft lip and/or palate (Deutsche Selbsthilfevereinigung für Lippen-Gaumen-Fehlbildungen e.V.). The study was supported by the Deutsche Forschungsgemeinschaft (FOR 423 and individual grants MA 2546/3-1, KR 1912/7-1, NO 246/6-1 and WI 1555/5-1) and the Austrian Cleft Palate Craniofacial Association (ACPCA). T.A. and E.N. are supported by grants from the Ministry of Higher Education, Syrian Arab Republic. The data sets used for the analyses described in this manuscript were obtained from dbGaP under accession phs000094.v1.p1. Additional acknowledgments are provided in the Supplementary Note.
Footnotes
URLs. Database of Genotypes and Phenotypes (dbGaP), http://www.ncbi.nlm.nih.gov/gap; UCSC Genome Browser, http://genome.ucsc.edu/; SNAP, http://www.broadinstitute.org/mpg/snap/; the R Project for Statistical Computing, http://www.r-project.org/; IMPUTE2, http://mathgen.stats.ox.ac.uk/impute/impute_v2.html; Encyclopedia of DNA Elements (ENCODE; accessed from the UCSC Genome Browser), http://www.genome.gov/10005107.
Accession codes. References sequences for the relevant genes are available at GenBank, including for IRF6: NM_006147.3 (NM_001206696.1), PAX7: NM_001135254.1 (NM_002584.2, NM_013945.2), THADA: NM_022065.4 (NM_001083953.1), EPHA3: NM_005233.5 (NM_182644.2), WDR21C: NM_005842.2, SPRY2: NM_005842.2 and TPM1: NM_001018020.1 (NM_001018007.1, NM_ 001018006.1, NM_001018005.1, NM_001018004.1, NM_000366.5). Alternative transcripts are given in parentheses.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS E.M., F.-J.K., T.F.W., P.P. and M.M.N. initiated the study. E.M., S.N., K.U.L., P.H., M.K., M.R., P.A.M. and M.M.N. contributed to the study design. M.M.N., E.M., S.C., P.H. and K.U.L. coordinated the work. K.U.L., E.M., M.K. and M.M.N. prepared the manuscript, with feedback from the other authors. S.N., H.R., A.P., C. Lauster, B. Braumann, R.H.R., A.H., S.P., B. Blaumeiser, N.D., T.K., R.P.S.-T., F.-J.K., M.R. and P.A.M. clinically characterized the families with cleft lip and collected the blood samples. K.U.L., R.H., E.N., T.A., S.B., A.C.B., N.K., M.A.A. and J.B. prepared the DNA and performed the molecular genetic analyses in the Bonn-II study. J.C.M., M.L.M., I.R., A.F.S. and T.H.B. contributed data from the Baltimore study. M.K., S.H., C. Lange and M.M. conducted the statistical analyses. M.M.N., E.M., M.K., K.U.L., M.R. and P.P. analyzed and interpreted the data.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Mangold E, Ludwig KU, Nöthen MM. Trends Mol. Med. 2011;17:725–733. doi: 10.1016/j.molmed.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Rahimov F, et al. Nat. Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaty TH, et al. Nat. Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangold E, et al. Nat. Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 5.Mansouri A, Stoykova A, Torres M, Gruss P. Development. 1996;122:831–838. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- 6.Sull JW, et al. Eur. J. Hum. Genet. 2009;17:831–839. doi: 10.1038/ejhg.2008.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Himanen JP, Saha N, Nikolov DB. Curr. Opin. Cell Biol. 2007;19:534–542. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maunakea AK, et al. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drieschner N, et al. Gene. 2007;403:110–117. doi: 10.1016/j.gene.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 10.Goodnough LH, Brugmann SA, Hu D, Helms JA. Dev. Dyn. 2007;236:1918–1928. doi: 10.1002/dvdy.21195. [DOI] [PubMed] [Google Scholar]
- 11.Welsh IC, Hagge-Greenberg A, O'Brien TP. Mech. Dev. 2007;124:746–761. doi: 10.1016/j.mod.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jugessur A, Farlie PG, Kilpatrick N. Oral Dis. 2009;15:437–453. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- 13.Grosen D, et al. J. Med. Genet. 2010;47:162–168. doi: 10.1136/jmg.2009.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumura K, et al. Biochem. Biophys. Res. Commun. 2011;404:1076–1082. doi: 10.1016/j.bbrc.2010.12.116. [DOI] [PubMed] [Google Scholar]
- 15.Vieira AR, et al. PLoS Genet. 2005;1:e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


