Abstract
Retroviruses are highly successful intracellular parasites, and as such they are found in nearly all branches of life. Some are relatively benign, but many are highly pathogenic and can cause either acute or chronic diseases. Therefore, there is tremendous selective pressure on the host to prevent retroviral replication, and for this reason cells have evolved a variety of restriction factors that act to inhibit or block the viruses. This review is a survey of the best-characterized restriction factors capable of inhibiting retroviral replication and aims to highlight the diversity of strategies used for this task.
Keywords: Trim5α, Trim28, Zap, Tetherin, APOBEC, intrinsic immunity
INTRODUCTION
Retroviruses are extremely successful pathogens affecting virtually all branches of life. These viruses are champions of persistence, maintained as proviral DNAs integrated into the genome of somatic cells and even entering into the germ line. Infection can result in cell death, or in oncogenic transformation by insertional mutagenesis. Thus, there is tremendous selective pressure to block or prevent retrovirus replication. In recent years, it has become apparent that mammalian cells have evolved a number of powerful mechanisms to limit or restrict virus replication, constituting novel aspects of intrinsic immunity. These mechanisms act at many diverse steps in the life cycle. The potential importance of these restriction factors is highlighted by the fact that many retroviruses, in turn, have evolved mechanisms to inactivate or overcome the blocks to infection. The picture now emerging is one of an ongoing battle between virus and host.
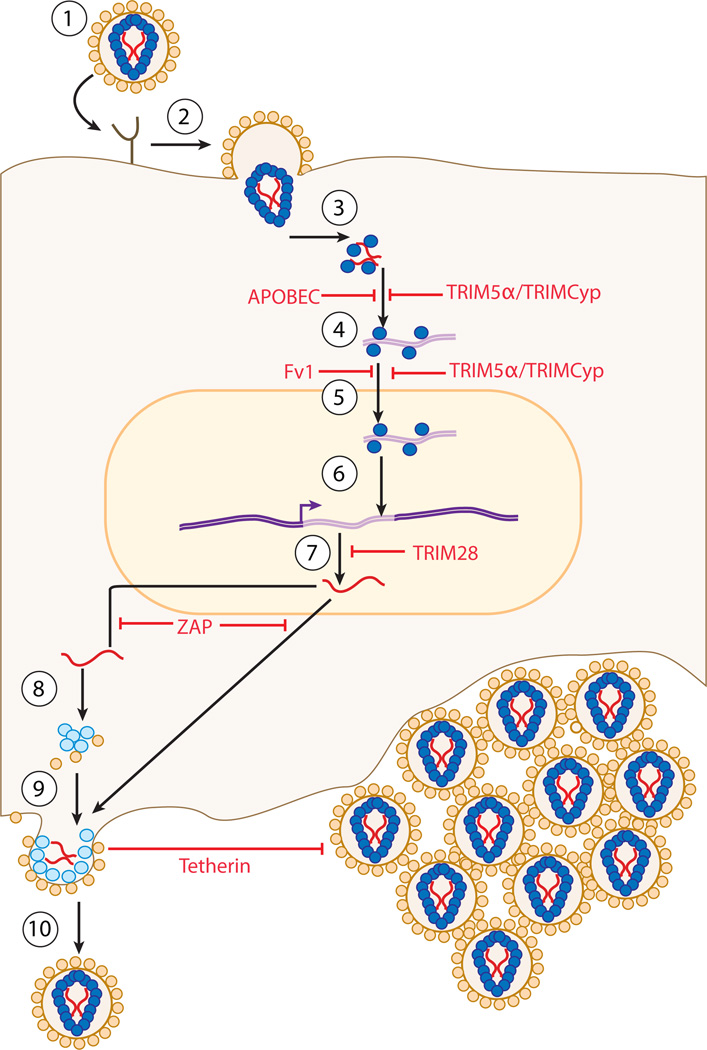
In this review, we summarize what is known about several key restriction factors, those providing the best understood and perhaps most powerful blocks to infection. Proceeding from early to late in the course of viral infection, we discuss the following: the APOBECs (apolipoprotein B mRNA-editing catalytic polypeptides), cytidine deaminases attacking the viral DNA as it is synthesized; Fv1, TRIM5α, and TRIMCyp, all attacking the viral capsid (CA) soon after entry into the cell; TRIM28, blocking viral transcription; ZAP (zinc-finger antiviral protein), directing the degradation of the viral RNAs; and tetherin, trapping the virions on the surface of the producer cell. For an overview of the retroviral life cycle and the restriction factors discussed herein see Figure 1.
Figure 1.
Summary of the retroviral life cycle and points of action of restriction factors. Cartoon showing the simplified life cycle of a retrovirus. Stages of life cycle are numbered and are as follows: 1. Envelope-mediated binding of retrovirus to target cell receptor. 2. Fusion of retroviral envelope and target cell membrane, leading to core viral particle entering cell. 3. Uncoating of core viral particle. 4. Reverse transcription of viral ssRNA genome into dsDNA, and formation of preintegration complex (PIC). 5. Nuclear entry of PIC. 6. Integration of viral genome into target cell genome. 7. Production of new viral RNA genomes and mRNA molecules by transcription. 8. Nuclear export of viral RNA species followed by either translation into viral proteins, or packaging into forming viral particles at a cellular membrane. 9. New translation of viral proteins and RNAs congregating at membrane and induction of membrane curvature as viral particles form. 10. Budding of fully formed viral particles from the membrane, and protease-mediated cleavage of viral proteins, leading to release of the fully mature virion. Dark blue circles denote protease cleaved viral proteins. Light blue circles denote viral proteins that have not been cleaved by protease. Small empty black circles denote viral envelope proteins. Wavy red lines denote viral RNAs; green wavy lines denote viral DNAs. Straight red lines denote point of interference in retroviral life cycle by restriction factors.
APOBECs: CYTIDINE DEAMINASES FOR VIRAL DNAs
The existence of the APOBEC restriction system was uncovered through the study of the viral infectivity factor (Vif) of HIV-1. It was long known that the Vif protein, encoded by one of the so-called accessory genes of the virus, was dispensable for viral replication in certain “permissive” cell lines such as CEM-SS and SupT1, but essential for replication in “nonpermissive” cells such as primaryCD4+ T-cells, monocyte-derived macrophages, and certain T cell leukemia lines such as CEM (44, 48, 134, 151, 154). The effect of Vif deletion appeared to be dependent on the cell line in which the virus was produced. Vif-minus virions produced by nonpermissive cells were poorly infectious, whereas Vif-minus virions produced in permissive cells were able to infect both permissive and nonpermissive cell types (48, 134, 151). These observations coupled with heterokaryon fusion experiments between permissive and nonpermissive cells suggested the existence of a dominantly acting antiviral factor that Vif was able to neutralize (96, 147). The identity of this factor was uncovered in 2002 through a cDNA subtraction screen for transcripts specifically expressed in nonpermissive cells (144). Of the large number of such genes identified, only one, originally known as CEM15, could convert a permissive cell into a nonpermissive one (144). Furthermore, CEM15 was found to be highly expressed in nonpermissive cells and weakly expressed or absent in permissive cells. This gene was subsequently identified as a member of the small APOBEC gene family and was named APOBEC3G.
APOBEC proteins constitute a family of polynucleotide cytidine deaminases, named after apolipoprotein B mRNA-editing catalytic polypeptide 1 (APOBEC1), the first protein of this class identified (163). All proteins in this family are characterized as having a His-X-Glu-X23–28-Pro-Cys-X2–4-Cys catalytic motif (71) that, based on studies of yeast and bacterial homologs, coordinates a Zn2+ ion (73, 79). Catalysis by the enzyme results in the hydrolytic deamination at the C4 position of the cytosine base of either DNA or RNA, thereby converting cytidine (C) to uridine (U). APOBEC1 was cloned because of its ability to specifically deaminate cytidine 6666 of the APOB mRNA, generating a stop codon and leading to the production of a truncated APO B protein. Another prominent member of the APOBEC family, AID, is expressed in activated B cells and is essential for several processes involved in antibody gene diversification, including somatic hypermutation (102).
The cytidine deaminase activity of the restriction factor neutralized by Vif strongly suggested that the restriction it conveyed could be due to specific deamination of either viral RNA or DNA. This prediction was quickly borne out by several groups who showed that APOBEC3G restriction correlated with the presence of G-to-A mutations in the sense strand of the retroviral DNA formed after infection of nonpermissive cells (60, 97, 193). This suggested that APOBEC3G was deaminating cytosines in the minus or antisense DNA strand during the first steps of reverse transcription. Such deaminated DNA could be subject to removal of these uracil bases by the uracil N-glycosidase (UNG) and subsequent cleavage by DNA repair enzymes such as apurinic/apyrimidinic endonuclease-1 (APE1) (183). Should such a modified provirus escape degradation and become integrated, it would most likely be inactive due to the G-to-A mutagenesis (189). Such inactivation would be the result of multiple mutation events, including missense changes, mutation of start codons, the introduction of new splice sites, and premature termination codons (189). Selective pressure to evade deamination by APOBEC proteins may explain why HIV-1 genes have atypical codon frequencies (9).
Consistent with the model that APOBEC3G-mediated antiviral activity is generated in the cell in which the viral particle is produced, APOBEC3G was found to be incorporated into the HIV-1 particle (60, 97, 144). This incorporation was attributed to a specific interaction between APOBEC3G and the nucleocapsid (NC) portion of the virus group-specific antigen (gag) protein (3, 28, 38, 137, 191). It has also been proposed that the NC-APOPEC3G interaction is dependent on RNA (137, 161, 191), and this assertion is plausible as both NC and APOBEC3G bind RNA (10, 86). The details of how APOBEC3G becomes encapsidated remain controversial, and more biophysical analysis is required for resolution of this question. It has been suggested that the reason mouse APOBEC3 is unable to restrict murine leukemia virus (MLV) is the absence of an APOBEC3-NC interaction, which results in no APOBEC3 being incorporated into MLV virions (37). However, some studies have shown that APOBEC3 is incorporated into MLV particles, and it has been suggested that instead MLV protease is able to inactivate APOBEC3 (1, 100).
To prevent APOBEC3G-mediated restriction, the Vif protein must be able to neutralize it during viral particle production. The Vif protein likely achieves this primarily by inducing proteasomal-mediated degradation of the APOBEC3G protein (31, 91, 101, 106, 145, 153). It induces degradation by bridging an interaction between APOBEC3G and a ubiquitin E3 ligase complex consisting of Elongin B, Elongin C, Cullin 5, and Ring Box-1 (190). This interaction leads to the polyubiquination of the APOBEC3G protein and its subsequent degradation by the proteosome (190). There is also some evidence that APOBEC3G is excluded fromHIV-1 virions directly through an interaction with Vif (75, 100). The interaction between APOBEC3G and Vif is species specific, and a single amino acid difference between African green monkey (AGM) and humanAPOBEC3G at residue 128 is sufficient to render the AGM APOBEC3G resistant to HIV-1 Vif-mediated degradation (17, 98, 139, 180). Conversely, the Vif protein from the AGM simian immunodeficiency virus (SIVAGM) is able to degrade AGM APOBEC3G but not human (17, 139). The species specificity of Vif for its APOBEC targets is in part responsible for the host range of the virus that carries it (63). This interaction therefore represents a therapeutic target, and the identification of pharmacological agents able to disrupt this interaction could lead to novel anti-HIV-1 therapeutics.
APOBEC3G is not the only cytidine deaminase to have specific antiretroviral function; multiple studies have shown that APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3F, and APOBEC3D/E are all, to varying degrees, able to restrict HIV-1 in the absence of Vif (16, 24, 33, 88, 132). It is therefore important to ascertain which of these proteins is most likely to be influencing HIV-1 replication in vivo. There is as yet no definitive answer to this question, but several lines of evidence point to APOBEC3G and APOBEC3F as being the predominant restriction factors during HIV-1 infection. First, Vif can only substantially inhibit APOBEC3G and APOBEC3F (17, 36, 84, 100, 145, 153, 174), which implies that these are the enzymes likely to be most potent in their ability to restrict HIV-1. Second, APOBEC3G and APOBEC3F are expressed in a number of tissues appropriate for HIV replication, including CD4+ T-cells and macrophages (88, 174), whereas many of the other APOBEC family members are not (16, 30, 37). Finally, different members of the APOBEC family have different substrate specificities. APOBEC3B, -3C, and -3F all have similar preferences for 5′-TC dinucleotides on the antisense strand, which manifest as 5′-GA to AA mutations in the sense strand of HIV-1, and patient-derived HIV-1 DNA sequences have been shown to carry these mutations frequently (23, 45, 171). The same data also show many 5′-GG to AG mutations, which are characteristic of APOBEC3G and which result from its preference for modifying CC dinucleotides on the antisense strand. These results make it probable that during a productive infection in humans, HIV-1 is encountering at least two of these enzymes: APOBEC3G and either APOBEC3B, -3C, or -3F.
Recent evidence suggests that the deaminase activity of the APOBEC enzymes is sometimes dispensable for their restrictive activity (116), which raises the question as to whether these proteins have an additional mode of retroviral restriction. Subsequent studies have shown that many of the APOBEC proteins mutated in their cytidine deaminase domains are still able to reduce the accumulation of reverse transcription products during the early stages of viral replication (15, 67). More recent studies have shown that APOBEC3G is able to specifically inhibit all stages of reverse transcription in vitro (70). The precise relevance of these in vitro effects to the in vivo situation is uncertain as several groups have demonstrated that the restriction caused by APOBEC family members in the absence of deaminase activity is negligible when the mutant proteins are expressed in cells at levels similar to those found physiologically (109, 142).
The APOBEC enzymes are likely responsible for innate or “intrinsic” immunity toward retroviruses but also toward other cellular pathogens, as well as acting as a guard against endogenous retroelements. APOBEC3G is able to block hepatitis B virus (HBV) replication (168), although no significant nucleotide changes were detected in the viral cDNA, and the cytidine deaminase activity of APOBEC3G was completely dispensable for this restriction. It appears rather that APOBEC3G is able to restrict HBV by interfering with the packaging of pregenomic RNA into subviral particles (168). Additional studies have shown that APOBEC3F and 3B can also restrict HBV, and some of these studies have shown the presence of hypermutation in the HBV cDNA, presumably due to cytidine deaminase activity. Therefore, in a situation similar to HIV-1, the exact mechanism of the APOBECs restriction of HBV remains under debate (20, 120, 121, 133, 160). The APOBEC enzymes are also able to inhibit a variety of retrotransposons such as MusD and IAP (intracisternal A-Particle) and non-LTR (long terminal repeat) retrotransposons such as LINE-1, and Alu elements (18, 30, 40, 41, 69, 111). Though many of the APOBECs appear to have some ability to restrict these retroelements, APOBEC3A and APOBEC3B appear to be the most potent and therefore most likely to be the most physiologically relevant to this activity (18, 19).
EARLY POSTENTRY BLOCKS TO INFECTION: FV1, TRIM5α, AND TRIMCYP
Two related retroviral restriction factors that block the retroviral life cycle subsequent to cell entry but before nuclear entry are Fv1 and Trim5α proteins. The Fv1 gene was first identified as a mouse locus that determined susceptibility to the Friend murine leukemia virus (89, 127, 128), and was subsequently shown to also convey resistance to other murine leukemia viruses (MLVs). Lilly and coworkers went on to describe two major naturally occurring Fv1 alleles among inbred mouse strains: the Fv1b allele from Balb/c mice, which allowed replication of a subset of MLVs named B-tropic and blocked replication of MLVs named N-tropic, and the Fv1n allele from NIH Swiss mice, which allowed replication of N-tropic MLVs and blocked B-tropic MLVs. The alleles were shown to be dominant, and thus Fv1n/b heterozygous mice restrict both N- and B-tropic MLVs. A third set of MLVs termed NB-tropic are able to infect animals with both Fv1b or Fv1n alleles; the well-characterized Moloney-MLV (Mo-MLV) falls into this class. The presence of a restrictive Fv1 allele during MLV infection leads to replication being blocked largely after reverse transcription, but before nuclear translocation of the preintegration complex (PIC) (74, 184). PICs isolated from an infected restrictive cell remain competent for integration when assayed in vitro, suggesting that the Fv1-mediated restriction prevents their translocation in the nucleus (129). The determinant of viral sensitivity to Fv1 was shown to lie in the Gag protein (34, 50, 68), specifically at residue 110 of the capsid (CA) polypeptide (21, 22, 80). This fact remains the best evidence for an active role of the CA protein during the early events of the retroviral life cycle.
The Fv1 gene was identified in 1996 by a positional cloning strategy (12) and was found to encode a Gag-like protein with sequence similarity to the ERV-L family of endogenous retroviruses (12). The expression of the appropriate Fv1 gene is sufficient to induce restriction of a sensitive MLV strain (14), and this restriction is thought to function via a direct interaction between the Fv1 protein and the CA protein of the incoming MLV virions. This interaction has never been directly observed, and perhaps the best evidence for its existence are experiments showing that Fv1-mediated restriction is saturable by infection with very high multiplicities of virus (8, 39). Importantly, this saturation can also be achieved by viral particles that are defective for replication, but only if they are of a tropism restricted by the Fv1 they are being used to saturate (8). Such saturation of Fv1-mediated restriction by an excess of restricted viral particles is termed “abrogation” and suggests that Fv1 is being specifically bound by incoming CA and titrated in these experiments.
These studies of Fv1-mediated restriction became of considerably greater interest to the general retroviral community when it was discovered that human cell lines exhibited a resistance to N-tropic MLV similar to that observed in cells expressing Fv1b (11, 164). Amazingly, the determinant for susceptibility to this restriction was also amino acid 110 of CA, though the factor in human cells appeared to block infection at a slightly earlier stage than Fv1, i.e., before reverse transcription. This restriction, named Ref1, was found to be saturable in a manner similar to Fv1 (165). Further interest in this restriction was generated by the discovery that many cell lines originally deemed to be nonpermissive for HIV-1 replication (65, 146) were in fact restricting HIV-1 in a saturable manner (66, 164). As for Ref1 and Fv1, susceptibility to the HIV-1 restriction was determined by sequences within the HIV-1 CA protein (32, 61, 122). The protein mediating both of these restrictions was determined to be TRIM5α (62, 76, 125, 157, 186).
The TRIM5α gene was identified by a cDNA library screen to identify the factor mediating HIV-1 resistance in rhesus macaque lung fibroblasts and had no homology to the Fv1 gene (157). Rather, the TRIM5α gene product is a member of the tripartite motif (TRIM) family of proteins characterized as having three domains usually at the N terminus of the protein: a RING domain, either one or two B-boxes, and a coiled-coil domain (118). The RING domain is a cysteine-rich zinc binding domain, which is commonly found in E3 ubiquitin ligases, and indeed there is evidence to suggest that TRIM5α is a ubiquitin ligase (35, 181). The B-box domains are thought to act as an interaction domain and thus determine RING box ubiquitin ligase substrate specificity (103, 104). The coiled-coil domain has been shown to be involved in homo- and heteromultimerization of the TRIM proteins (123, 130). TRIM5α forms trimers in solution and its coiled-coil domain is required for this self-assembly (72, 107, 124, 192). The C terminus of TRIM5α contains a B30.2 domain; this domain binds to CA molecules of incoming retroviruses, and therefore its sequence determines which retroviruses a specific TRIM5α will restrict (62, 113, 124, 125, 143, 158, 159, 187).
Subsequent to the cloning of rhesus macaque TRIM5α, TRIM5α genes have been cloned from a variety of species and tested for their ability to restrict different retroviruses (62, 124, 135, 138, 148, 157, 186, 188). It was found that species variation of TRIM5α sequences (specifically in the B30.2 domain) led to differences in their ability to restrict HIV-1 and other retroviruses. For instance, human TRIM5α potently restricts N-tropic MLV but not B-tropic MLV, HIV-1, or SIVmac, whereas rhesus macaque TRIM5α potently restricts HIV-1 N-tropic MLV but not SIVmac (62, 124, 135, 148, 157, 186). In this way, TRIM5α is a major determinant of retroviral species tropism (63).
The mechanism of TRIM5α-mediated restriction remains to be fully elucidated. Under normal circumstances, it blocks retroviral replication early in the life cycle, after viral entry but before reverse transcription (65, 146). Inhibition of the proteosome during infection allows reverse transcription to take place; however, the PICs are still not able to enter the nucleus and thus infectivity is not rescued by such treatment (5, 179). As TRIM5α is an E3 ubiquitin ligase it is tempting to speculate that the effect is due to a TRIM5α–specific targeting of CA or another viral protein for degradation. However, it has been shown that disruption of E3 ligase activity by point mutation does not completely abolish the ability ofTRIM5αto induce restriction (35) and therefore TRIM5α certainly also induces restriction in a proteosome-independent fashion. TRIM5α has been observed using a capsid sedimentation assay to promote the rapid uncoating of incoming HIV-1 capsids that may be detrimental to PIC formation (158). Also, TRIM5α induces the degradation of CA in a proteosome-independent manner that may also contribute to its ability to restrict HIV-1 (29). Perhaps all of these mechanisms contribute to TRIM5α-mediated restriction.
Trimeric TRIM5α interacts with capsids via its B30.2 domain (143, 158), and a wealth of evidence intimates an interaction between capsid and the Fv1 protein (21, 22, 80). Both proteins have no sequence homology and yet seem to bind to the same very small area of the CA protein centered around amino acid 110. The fact that this interaction and a subsequent early block to retroviral restriction have evolved independently at least twice in mammals suggests that CA has a conserved and vital role in early events of retroviral replication that host cells can disrupt to induce restriction. Lending further credence to this hypothesis was the discovery of the TRIM5α-cyclophilin A (CypA) fusion protein in owl monkey cells named TRIMCyp (136). CypA is a peptidyl prolyl isomerase and had been previously identified as a protein that specifically interacts with HIV-1 CA (95). This interaction is important for HIV-1 replication and can be disrupted by either the G89Vmutation inHIV-1CAor by competition with the drug cyclosporine A (CSA), which is a competitive inhibitor of CypA. The TRIMCyp fusion in owl monkey cells appears to have formed by the retrotransposition of a CypA pseudogene into the 3′ section of the TRIM5 gene and creates an in-frame fusion ofTRIM5α and CypA in which CypA replaces the B30.2 domain. In this way, the TRIMCyp molecule uses the CypA protein to bind specifically to CA and thereby restrict HIV-1 replication (136). In agreement with this theory, it has been demonstrated that CSA treatment of owl monkey cells disrupts HIV-1 restriction and allows replication to proceed normally (166). Very recently there has been yet another twist in this story with the discovery that the TRIM5α CypA fusion protein has itself evolved independently on two separate occasions. The second occurrence of the TRIMCyp fusion protein is found in several Old World monkeys, including both pigtailed and rhesus macaques. This second allele arose by convergent evolution as the CypA insertion occurred in a slightly different location in the TRIM5 gene (25, 87, 117, 173, 176).
The inhibition of the early stages of the retroviral replication cycle using a protein that is able to recognize CA has clearly occurred independently several times over the course of evolution. The restriction induced by these proteins is extremely potent and therefore enhancement of this restriction remains a tantalizing prospect for antiretroviral therapies.
DESTRUCTION OF VIRAL RNAs BY A ZINC-FINGER ANTIVIRAL PROTEIN
The zinc-finger antiviral protein (ZAP) was isolated using a screen for dominant-acting antiretroviral genes (49). RAT2 cells (which are highly permissive for MLV replication) were stably transduced with a rat cDNA library, and cDNAs that induced resistance to MLV infection were then selected after repeated infection with an MLV vector expressing the herpes thymidine kinase (TK) gene (49). Subsequent treatment of these cells with trifluorothymidine killed all cells that had become TK+ and thereby selected for cells which had remained TK-minus through resistance to infection. One of the resulting clones from this screen expressed the N-terminal portion of a novel protein ZAP, encompassing a cluster of four CCCH-type zinc fingers. CCCH zinc fingers are uncommon and are found only in a small group of RNA-binding proteins known as the tristetraprolin (TTP) tandem zinc finger (TZF) family (83). Members of the TTP protein family have been shown to bind specifically to AU-rich elements (ARE) in the 3′ untranslated regions of several cytokine mRNAs and to lead to their degradation (26, 27, 82, 83). This finding immediately suggested that ZAP would most likely interfere with an RNA component of the retroviral life cycle. In agreement with this notion, ZAP-expressing cells were found to block the MLV life cycle by preventing postintegration accumulation of viral RNA in the cytoplasm (49). The block was posttranscriptional, with no effect on the amounts of viral RNA in the nucleus but causing a dramatic decrease in steady-state levels in the cytoplasm (49). Further studies demonstrated that ZAP specifically interacted with the 3′LTR of MLV and that mutations that abolished this interaction (in either the second or fourth CCCH zinc finger) concomitantly abolished the ability of ZAP to mediate viral restriction (56). The 3′LTR of MLV contains no obvious AREs, and ZAP is also not able to specifically mediate the destruction of ARE-containing mRNA transcripts (56), suggesting that the CCCH motifs in ZAP recognize a different viral-specific motif. The question of what RNA feature ZAP specifically was recognizing was made more pertinent by the revelation that ZAP is also able to induce the restriction of alphaviruses such as Sindbis virus (SIN) (13, 56, 194) and filoviruses such as Ebola virus (EBOV) (112). Studies into ZAP-mediated restriction of these viruses suggest that ZAP is targeting viral RNA for degradation. In the case of SIN, the region of the viral genome with which ZAP is able to interact was mapped and showed no significant homology to the 3′LTR of MLV(56). The studies of ZAP-mediated restriction of other viruses has also led to the discovery that ZAP is an interferon-stimulated gene whose upregulation is partially required for interferon-induced cellular restriction of SIN (194).
A possible mechanism by which ZAP leads to the degradation of cytoplasmic RNAs is suggested by a recent study showing that ZAP is able to specifically interact with components of the exosome (57). The exosome is an evolutionarily highly conserved 3′–5′ exoribonuclease complex found both in the nucleus and cytoplasm of eukaryotes (4, 42, 108). Guo and coworkers showed that ZAP coimmunoprecipitates with exosome components Rrp40p and Rrp46p and crucially that RNAi-mediated knockdown of these proteins attenuates ZAP-mediated restriction of MLV (57). This finding strongly suggests that exosome activity is required for ZAP activity. Therefore, the current model for ZAP-mediated restriction is that it bridges an interaction between viral RNA species and the exosome and thereby targets them for degradation. A very recent study has also shown that a longer splice variant of ZAP (termed ZAP(L)) exists in both humans and rats that is more potent in its ability to restrict MLV. ZAP(L) contains a poly(ADP-ribose) polymerase (PARP) domain (77), and comparison of the domain from many primates suggests that this region has evolved under positive selection throughout primate evolution (77). Such positive selection is often attributed to host-pathogen interactions, suggesting that some viral proteins may bind to the PARP domain. No such positive selection was observed in the CCCH domain that binds to viral RNA (77). These data regarding ZAP(L) suggest that ZAP may have additional, as yet uncharacterized, antiviral activities.
PRIMER BINDING SITE (PBS)-MEDIATED RESTRICTION
Teich and colleagues observed in 1977 that murine leukemia viruses (MLVs) were unable to replicate in embryonic carcinoma (EC) cells (162). These pioneering studies reported that when EC cells were infected with MLV, integration of the proviruses proceeded normally; however, no viral mRNA transcripts could be detected (162). This study also revealed that EC cells, if forced to differentiate into a nonpluripotent state, were rendered permissive for MLV replication (162). Subsequently, it was demonstrated that such differentiation only rendered EC cells permissive to new infection by MLV but did not result in reactivation of viruses integrated prior to differentiation (119). To reactivate these silenced viruses, it was necessary to both differentiate the infected EC cells and also to treat them with the DNA demethylating agent 5′azacytidine (5–azaC) (119). These results strongly suggested that the mechanism by which MLV was silenced occurred in two stages: The first stage involved EC-specific factors, and the second involved DNA methylation that occurred subsequent to the initial silencing and persisted even in differentiated cells (119).
The nature of the EC-specific transcriptional silencing was delineated by several groups in a flurry of activity in the late 1980s. Both reduced transcription factor binding to the viral enhancers in the LTR (64, 90, 92, 152, 167) and the presence of repressive transacting factors were involved (2, 6, 43, 46, 52, 92, 167). The genomic site of action of one of these trans-acting factors was determined by a screen to identify MLV escape mutations to EC cell-mediated restriction. This screen identified a single base pair mutation (named B2) that greatly reduced this restriction (6). The B2 mutation is located within the primer binding site (PBS) of MLV, and further study identified 17 bp of the 18 bp of the PBS as essential for EC cell-specific restriction of MLV (6, 43, 92). The PBS of the MLV genome is complementary to 18 nucleotides at the 3′ end of the host proline tRNA and is critical for virus replication. The tRNApro is annealed to the PBS (PBSPro) in the RNA genome at the time of virus assembly, and upon infection is used as the primer for minus-strand DNA synthesis during reverse transcription (59). That the PBS is both a target for EC cell-specific restriction and is required for priming of reverse transcription suggests that this restriction could be occurring at the level of DNA or RNA. A DNA-binding trans-acting factor was postulated to exist based on a series of experiments characterizing the nature of PBS-mediated restriction in EC cells. It was observed that the PBSPro was able to silence LTR-driven transcription from reporter constructs in EC cells, independently of orientation and position, even when placed outside of the transcriptional unit (94, 126). It was also demonstrated that the MLV PBS functioned to silence transcription from promoters other than the MLV LTR and that the restriction activity was saturable by transfection of increasing amounts of DNA containing the PBSPro sequence (93). The presence of this transcriptional silencer was detected in EC cell nuclear extracts using a probe spanning the PBSPro sequence of MLV by exonuclease III protection assays (94). This exonuclease III protection footprint was reduced upon either differentiation of the EC cells or the use of a probe containing the B2 point mutation, suggesting the presence of a DNA-binding factor that correlates with this repression. Similar experiments performed using MLV PBSPro in electrophoretic mobility shift assays (EMSA) showed that DNA binding activity was enriched in nuclear extracts from EC cells when compared with differentiated cell lines (126).
Further study of the PBS-mediated restriction of MLV showed that it was not limited to EC cells but also occurred in ES cells (55) as well in several hemapoietic cells lines (58). PBSPro-mediated restriction was also found to occur in human hematopoietic cells, either transformed or primary cells from cord blood (58). Another PBS sequence, corresponding to tRNALys1,2 (PBSLys1,2), utilized by retroviruses such as visna, spuma, and Mason-Pfizer monkey virus (105, 149, 150), also leads to restriction in EC cells (110, 182). A binding complex is detected by EMSA in restrictive cells, and in a manner similar to the B2 mutation in PBSPro, a single point mutation in the PBSLys1,2 sequence is able to relieve the restriction (6, 110, 182). These observations suggest that the PBSLys1,2 may recruit silencing machinery similar to the PBSPro. As a whole, these data suggest that PBS-mediated restriction occurs in many cell types from multiple species and targets multiple different retroviruses.
Recently, our laboratory identified TRIM28 as a factor required for PBSPro-mediated restriction of MLV in EC cells. TRIM28 was identified by biochemical purification of the PBSPro binding complex as observed by EMSA (178). TRIM28 (also known as Kap-1, or Tif1-beta) is a transcriptional corepressor that is recruited to its target genes by interactions with the Kr uppel associated box (KRAB) zinc finger DNA-binding proteins (47). This interaction is mediated by the KRAB box domain and leads to the complex acting as a sequence-specific transcriptional repressor (169). TRIM28 recruits several factors involved in transcriptional silencing and heterochromatin formation, including the histone H3 K9 methyltransferase ESET, the NuRD histone deacetylase complex, and the heterochromatin-associated protein HP1 (85, 140, 141). TRIM28 is required for PBSPro-mediated restriction in EC and is recruited to sites of proviral integration in a PBSPro-dependent manner (178). TRIM28 appears to specifically recruit the HP1 protein to the provirus, and this recruitment is absolutely required for the silencing of MLV (177, 178).
TRIM28 is expressed in many nonpluripotent cell types that do not exhibit PBSPro-mediated restriction, and thus TRIM28 expression appears to be necessary but not sufficient to induce restriction. This observation, combined with the fact that TRIM28 has no known DNA binding activity, suggests that a DNA-binding factor capable of binding the PBSPro sequence is differentially expressed between EC and differentiated cells. A KRAB zinc finger DNA-binding protein makes an attractive candidate for this DNA-binding activity, as these proteins interact with TRIM28 and through this interaction induce transcriptional silencing (169).
PBS-mediated silencing of retroviruses in pluripotent cells is likely to have evolved to protect the embryo from infection, but also from the reactivation of endogenous retroviruses and retrotransposons that could cause damaging mutations in the germ line and early progenitor cells. There is a rationale for the evolution of a repression machinery that targets the PBS elements: The PBS is essential for the priming of virus reverse transcription, and furthermore during reverse transcription the PBS sequence on one strand of the viral DNA is synthesized by reverse transcription of the cellular tRNA itself. Thus, the viral PBS sequence is a uniquely effective target site for repression by the host, since any escape point mutations in the PBS sequence would quickly revert to the original sequence during viral replication. The identification of TRIM28 as a component of this restriction machinery will likely aid greatly in the dissection of the molecular mechanism of this type of restriction.
INHIBITION OF RETROVIRAL PARTICLE RELEASE BY TETHERIN
Tetherin (also variously known as BST-2/HM1.24/CD317) was recently identified as a restriction factor that prevents retroviral particle release from the surface of producer cells (115, 170). Much as the APOBEC enzymes were discovered through the study of Vif (144), so tetherin was discovered through characterization of theHIV-1 accessory protein Vpu (115, 170). Vpu was first identified as a small transmembrane phosphoprotein expressed by HIV-1 that enhanced release of virions from infected cells (78, 155, 156). It was later shown that Vpu could also lead to the degradation of the HIV-1 receptor CD4 (175). This degradation is mediated through an interaction between Vpu and the F-box protein βTrCP, which targets CD4 for ubiquitination and subsequent proteasomal degradation (99). These two functions of Vpu are distinct, as Vpu can promote release of HIV-1 and other retroviral virions in cell lines that do not express CD4 (54, 185). The enhancement of virion release from HIV-1 infected cells by Vpu was found to be cell-type dependent (51, 54, 134), and heterokaryon fusion experiments demonstrated this dependence was due to a dominant acting restriction factor (172). This restriction factor is only effective against Vpu-minus HIV-1, and as such wild-type HIV-1 replication is unaffected by it (172). This restriction factor was also found to be upregulated in many permissive cell lines upon stimulation of interferon alpha (IFNα), which rendered these cells resistant to infection with Vpu-minus HIV-1 without significantly affecting wild-type HIV-1 replication (114). Production of Vpu-minus HIV-1 virions in cells expressing this restriction factor led to a striking phenotype, namely large numbers of fully formed viral particles bound to the extracellular leaflet of the cell’s outer membrane (114). These bound or “tethered” virions are fully infectious, and protease treatment of cells with such tethered virions causes a substantial increase in infectious particle release (114).
Tetherin was identified by comparative microarray analysis of cells that either did or did not require Vpu for efficient virion production, coupled with similar analysis of IFNα stimulated genes (115). Tetherin was one of fewer than ten genes whose expression correlated with a dependence on Vpu for efficient partial release and was shown to specially inhibit Vpu-minus HIV-1 production when expressed in the permissive 293T cell line (115). Tetherin is a transmembrane protein with a highly unusual topology including a N-terminal cytoplasmic tail, a single membrane-spanning helix, an extracellular coiled-coil domain, and a C-terminal glycosylphosphatidylinositol (GPI) membrane anchor (81, 131).With the possible exception of a minor variant of the prion protein, there exists no other mammalian protein with similar topology (131). It is not yet known how tetherin adheres virions to the extracellular membrane of cells. Its unusual topology and ability to dimerize (53) suggest several possibilities. One hypothesis would be that tetherin bridges an interaction between the cellular membrane and the viral envelope with one of its two lipid-bilayer anchoring domains in each membrane (115). Alternatively, the tethering could be mediated by dimerization of two tetherin molecules, one in the viral envelope and the other in the cell membrane (115). Further biophysical and biochemical analysis is required to determine which of these possibilities is correct.
The mechanism by which Vpu disrupts tetherin-mediated restriction is also not fully elucidated. Van Damme and colleagues have shown that expression of Vpu leads to a proteasome-independent downregulation of tetherin from the extracellular membrane of a target cell (170). It has also been noted that overexpressed Vpu leads to decreased steady state levels of tetherin (7). Furthermore, like Vif, Vpu may be species specific in its ability to inactivate tetherin, as HIV-1 Vpu is unable to relieve what is presumed to be tetherinmediated restriction of HIV-1 particle production in IFNα–treated African green monkey cells (114).
The nature of tetherin-mediated restriction suggests that tetherin might restrict a wide range of enveloped viruses. As yet, there is only sparse evidence to support this notion, though it has been shown that IFNα treatment of cells leads to a specific block in Ebola virus–like particle release from cells, and that this block can be abrogated by the expression of Vpu (114). Suggestive findings come from experiments showing that the Kaposi sarcoma–associated herpesvirus (KSHV) K5 protein is able to decrease the cellular steady-state levels of tetherin (7). This implies that tetherin may also be detrimental to KSHV replication. Further studies into this rapidly expanding field will likely shed light on these interesting questions.
SUMMARY AND CONCLUSIONS
Much of the early work on retroviruses focused on elucidating the life cycle and determining how the virally encoded machinery drove its progression. Recently, there has been a growing realization that no viral processes take place in the absence of interactions with cellular host factors. Over the course of evolutionary time, the host and retrovirus have been locked in a constant struggle for survival. This has resulted in retroviruses evolving the capability of exploiting a multitude of cellular proteins to promote their life cycle. Reciprocally it has led to the host cell developing many restriction factors to inhibit the retroviral life cycle. As discussed in this review, the study of hostretroviral interactions has recently uncovered a number of these restriction factors and has been extremely illuminating to the field of intrinsic immunity as well as to molecular biology. The future study of these interactions will continue to provide insight into these endogenous cellular processes. Finally, a greater understanding of the factors mediating intrinsic immunity may lead to the development of pharmacological agents that can boost their potency and thereby lead to treatments for viral disease.
SUMMARY POINTS.
A large number of cellular proteins have evolved to interfere with the replication cycle of retroviruses. These proteins are known collectively as restriction factors.
Restriction factors have been shown to interfere with many stages of the retroviral life cycle.
Retroviral restriction factors are often able to restrict other nonretroviral viruses.
Several restriction factors have been identified through the study of HIV-1 accessory gene function. Accessory genes Vif and Vpu are both able to specifically inhibit cellular restriction factors.
Several other restriction factors have been identified by studying differences between permissive and nonpermissive cell lines.
FUTURE ISSUES.
There are likely many other cellular restriction factors that remain unidentified; further characterization of the retroviral life cycle and cellular factors that inhibit it will, it is hoped, lead to their discovery.
As yet there has been no effective modification or stimulation of a cellular restriction factor to prevent a clinical disease. It is hoped that in the future such modification will be achieved either pharmacologically or genetically and that this will lead to effective clinical treatments for viral diseases.
ACKNOWLEDGMENTS
We are grateful to Helen Nickerson for critical reading of this manuscript and Gloria Arriagada for help with construction of the figure. This work was supported by PHS grant R37 CA 30488 from the National Cancer Institute. D.W. is an Associate, and S.P.G. is an Investigator of the Howard Hughes Medical Institute.
Glossary
- Restriction factor
a host cell protein or factor whose expression restricts or inhibits the viral life cycle
- APOBEC
apolipoprotein B mRNA-editing catalytic polypeptide
- Capsid (CA)
the major structural constituent of the retroviral core, synthesized as a proteolytic cleavage product of the Gag precursor protein
- ZAP
zinc-finger antiviral protein
- Accessory gene
one of several small open reading frames found in complex retroviruses such as HIV-1. Accessory genes are so named as being dispensable for viral replication under some artificial conditions in tissue culture
- Permissive
a cell line or organism that supports replication of the virus
- Provirus
a DNA copy of the retroviral genome integrated into the host cell genome
- Nucleocapsid (NC)
a proteolytic cleavage product of the Gag precursor protein, containing one or two zinc fingers, required for packaging of the RNA genome into the virion
- Group-specific antigen (gag) gene
the retrovirus gene encoding the precursor protein that is cleaved to give rise to the major structural components of the virion particle
- MLV
murine leukemia virus
- LTR
long terminal repeat
- PIC
preintegration complex
- Primer binding site (PBS)
the region of a retroviral genome complementary to the 3′ end of a cellular tRNA sequence. During reverse transcription a cellular tRNA annealed to this sequence is used to prime the first strand of DNA synthesis
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
References
- 1.Abudu A, Takaori-Kondo A, Izumi T, Shirakawa K, Kobayashi M, et al. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr. Biol. 2006;16:1565–1570. doi: 10.1016/j.cub.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 2.Akgun E, Ziegler M, Grez M. Determinants of retrovirus gene expression in embryonal carcinoma cells. J. Virol. 1991;65:382–388. doi: 10.1128/jvi.65.1.382-388.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 4.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, Hope TJ. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol. 2006;80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barklis E, Mulligan RC, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–399. doi: 10.1016/0092-8674(86)90596-9. The first identification of the PBS of MLV as the target site for transcriptional silencing in EC cells.
- 7.Bartee E, McCormack A, Fruh K. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2006;2:e107. doi: 10.1371/journal.ppat.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassin RH, Duran-Troise G, Gerwin BI, Rein A. Abrogation of Fv-1b restriction with murine leukemia viruses inactivated by heat or by gamma irradiation. J. Virol. 1978;26:306–315. doi: 10.1128/jvi.26.2.306-315.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkhout B, van Hemert FJ. The unusual nucleotide content of the HIV RNA genome results in a biased amino acid composition of HIV proteins. Nucleic Acids Res. 1994;22:1705–1711. doi: 10.1093/nar/22.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkowitz RD, Ohagen A, Hoglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besnier C, Ylinen L, Strange B, Lister A, Takeuchi Y, et al. Characterization of murine leukemia virus restriction in mammals. J. Virol. 2003;77:13403–13406. doi: 10.1128/JVI.77.24.13403-13406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. Describes the initial molecular cloning of the fv1 locus.
- 13.Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishop KN, Bock M, Towers G, Stoye JP. Identification of the regions of Fv1 necessary for murine leukemia virus restriction. J. Virol. 2001;75:5182–5188. doi: 10.1128/JVI.75.11.5182-5188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 17.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 21.Boone LR, Glover PL, Innes CL, Niver LA, Bondurant MC, Yang WK. Fv-1 N- and B-tropismspecific sequences in murine leukemia virus and related endogenous proviral genomes. J. Virol. 1988;62:2644–2650. doi: 10.1128/jvi.62.8.2644-2650.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boone LR, Myer FE, Yang DM, Ou CY, Koh CK, et al. Reversal of Fv-1 host range by in vitro restriction endonuclease fragment exchange between molecular clones of N-tropic and B-tropic murine leukemia virus genomes. J. Virol. 1983;48:110–119. doi: 10.1128/jvi.48.1.110-119.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borman AM, Quillent C, Charneau P, Kean KM, Clavel F. A highly defective HIV-1 group O provirus: evidence for the role of local sequence determinants in G → A hypermutation during negative-strand viral DNA synthesis. Virology. 1995;208:601–609. doi: 10.1006/viro.1995.1191. [DOI] [PubMed] [Google Scholar]
- 24.Bourara K, Liegler TJ, Grant RM. Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1. PLoS Pathog. 2007;3:1477–1485. doi: 10.1371/journal.ppat.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan G, Kozyrev Y, Hu SL. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis . Proc. Natl. Acad. Sci. USA. 2008;105:3569–3574. doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 27.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 28.Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- 29.Chatterji U, Bobardt MD, Gaskill P, Sheeter D, Fox H, Gallay PA. Trim5alpha accelerates degradation of cytosolic capsid associated with productive HIV-1 entry. J. Biol. Chem. 2006;281:37025–37033. doi: 10.1074/jbc.M606066200. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, et al. APOBEC3A is a potent inhibitor of adenoassociated virus and retrotransposons. Curr. Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 31.Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 32.Cowan S, Hatziioannou T, Cunningham T, Muesing MA, Gottlinger HG, Bieniasz PD. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DesGroseillers L, Jolicoeur P. Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia viruses. J. Virol. 1983;48:685–696. doi: 10.1128/jvi.48.3.685-696.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz-Griffero F, Li X, Javanbakht H, Song B, Welikala S, et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 36.Doehle BP, Schafer A, Cullen BR. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Doehle BP, Schafer A, Wiegand HL, Bogerd HP, Cullen BR. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 2005;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Douaisi M, Dussart S, Courcoul M, Bessou G, Vigne R, Decroly E. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem. Biophys. Res. Commun. 2004;321:566–573. doi: 10.1016/j.bbrc.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Duran-Troise G, Bassin RH, Rein A, Gerwin BI. Loss of Fv-1 restriction in Balb/3T3 cells following infection with a single N tropic murine leukemia virus particle. Cell. 1977;10:479–488. doi: 10.1016/0092-8674(77)90035-6. [DOI] [PubMed] [Google Scholar]
- 40.Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, et al. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 41.Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 2006;34:1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Estevez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei . EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feuer G, Taketo M, Hanecak RC, Fan H. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J. Virol. 1989;63:2317–2324. doi: 10.1128/jvi.63.5.2317-2324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher AG, Ensoli B, Ivanoff L, Chamberlain M, Petteway S, et al. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- 45.Fitzgibbon JE, Mazar S, Dubin DT. A new type of G → A hypermutation affecting human immunodeficiency virus. AIDS Res. Hum. Retroviruses. 1993;9:833–838. doi: 10.1089/aid.1993.9.833. [DOI] [PubMed] [Google Scholar]
- 46.Flanagan JR, Krieg AM, Max EE, Khan AS. Negative control region at the 5′ end of murine leukemia virus long terminal repeats. Mol. Cell Biol. 1989;9:739–746. doi: 10.1128/mcb.9.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 48.Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, et al. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. Identifies the ZAP restriction factor.
- 50.Gautsch JW, Elder JH, Schindler J, Jensen FC, Lerner RA. Structural markers on core protein p30 of murine leukemia virus: functional correlation with Fv-1 tropism. Proc. Natl. Acad. Sci. USA. 1978;75:4170–4174. doi: 10.1073/pnas.75.9.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geraghty RJ, Talbot KJ, Callahan M, Harper W, Panganiban AT. Cell type-dependence for Vpu function. J. Med. Primatol. 1994;23:146–150. doi: 10.1111/j.1600-0684.1994.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 52.Gorman CM, Rigby PW, Lane DP. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell. 1985;42:519–526. doi: 10.1016/0092-8674(85)90109-6. [DOI] [PubMed] [Google Scholar]
- 53.Goto T, Kennel SJ, Abe M, Takishita M, Kosaka M, et al. A novel membrane antigen selectively expressed on terminally differentiated human B cells. Blood. 1994;84:1922–1930. [PubMed] [Google Scholar]
- 54.Gottlinger HG, Dorfman T, Cohen EA, Haseltine WA. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc. Natl. Acad. Sci. USA. 1993;90:7381–7385. doi: 10.1073/pnas.90.15.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grez M, Akgun E, Hilberg F, Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1990;87:9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 2004;78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. USA. 2007;104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haas DL, Lutzko C, Logan AC, Cho GJ, Skelton D, et al. The Moloney murine leukemia virus repressor binding site represses expression in murine and human hematopoietic stem cells. J. Virol. 2003;77:9439–9450. doi: 10.1128/JVI.77.17.9439-9450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harada F, Peters GG, Dahlberg JE. The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J. Biol. Chem. 1979;254:10979–10985. [PubMed] [Google Scholar]
- 60.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 61.Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 2003;22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. USA. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hatziioannou T, Princiotta M, Piatak M, Jr, Yuan F, Zhang F, et al. Generation of simian-tropic HIV-1 by restriction factor evasion. Science. 2006;314:95. doi: 10.1126/science.1130994. [DOI] [PubMed] [Google Scholar]
- 64.Hilberg F, Stocking C, Ostertag W, Grez M. Functional analysis of a retroviral host-range mutant: altered long terminal repeat sequences allow expression in embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA. 1987;84:5232–5236. doi: 10.1073/pnas.84.15.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Himathongkham S, Luciw PA. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 66.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, et al. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 68.Hopkins N, Schindler J, Hynes R. Six-NB-tropic murine leukemia viruses derived from a B-tropic virus of BALB/c have altered p30. J. Virol. 1977;21:309–318. doi: 10.1128/jvi.21.1.309-318.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 72.Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, et al. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 73.Johansson E, Mejlhede N, Neuhard J, Larsen S. Crystal structure of the tetrameric cytidine deaminase from Bacillus subtilis at 2.0 Å resolution. Biochemistry. 2002;41:2563–2570. doi: 10.1021/bi011849a. [DOI] [PubMed] [Google Scholar]
- 74.Jolicoeur P, Baltimore D. Effect of Fv-1 gene product on proviral DNA formation and integration in cells infected with murine leukemia viruses. Proc. Natl. Acad. Sci. USA. 1976;73:2236–2240. doi: 10.1073/pnas.73.7.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. The human immunodeficiency virus type 1Vif protein reduces intracellular expression and inhibits packaging ofAPOBEC3G(CEM15), a cellular inhibitor of virus infectivity. J. Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kerns JA, Emerman M, Malik HS. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4:e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ko TP, Lin JJ, Hu CY, Hsu YH, Wang AH, Liaw SH. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J. Biol. Chem. 2003;278:19111–19117. doi: 10.1074/jbc.M300874200. [DOI] [PubMed] [Google Scholar]
- 80.Kozak CA, Chakraborti A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 81.Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. Bst-2/HM1.24 is a raftassociated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 82.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 84.Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G antiretroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, et al. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 86.Li J, Potash MJ, Volsky DJ. Functional domains of APOBEC3G required for antiviral activity. J. Cell Biochem. 2004;92:560–572. doi: 10.1002/jcb.20082. [DOI] [PubMed] [Google Scholar]
- 87.Liao CH, Kuang YQ, Liu HL, Zheng YT, Su B. A novel fusion gene, TRIM5-Cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. Aids. 2007;21(Suppl. 8):S19–S26. doi: 10.1097/01.aids.0000304692.09143.1b. [DOI] [PubMed] [Google Scholar]
- 88.Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 89.Lilly F. Susceptibility to two strains of Friend leukemia virus in mice. Science. 1967;155:461–462. doi: 10.1126/science.155.3761.461. [DOI] [PubMed] [Google Scholar]
- 90.Linney E, Davis B, Overhauser J, Chao E, Fan H. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. Nature. 1984;308:470–472. doi: 10.1038/308470a0. [DOI] [PubMed] [Google Scholar]
- 91.Liu B, Yu X, Luo K, Yu Y, Yu XF. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 2004;78:2072–2081. doi: 10.1128/JVI.78.4.2072-2081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loh TP, Sievert LL, Scott RW. Proviral sequences that restrict retroviral expression in mouse embryonal carcinoma cells. Mol. Cell Biol. 1987;7:3775–3784. doi: 10.1128/mcb.7.10.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Loh TP, Sievert LL, Scott RW. Negative regulation of retrovirus expression in embryonal carcinoma cells mediated by an intragenic domain. J. Virol. 1988;62:4086–4095. doi: 10.1128/jvi.62.11.4086-4095.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Loh TP, Sievert LL, Scott RW. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonal carcinoma cells. Mol. Cell Biol. 1990;10:4045–4057. doi: 10.1128/mcb.10.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 96.Madani N, Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 1998;72:10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 98.Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 99.Margottin F, Bour SP, Durand H, Selig L, Benichou S, et al. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 100.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 101.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 102.Martin A, Bardwell PD, Woo CJ, Fan M, Shulman MJ, Scharff MD. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 2002;415:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- 103.Massiah MA, Matts JA, Short KM, Simmons BN, Singireddy S, et al. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold. J. Mol. Biol. 2007;369:1–10. doi: 10.1016/j.jmb.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 104.Massiah MA, Simmons BN, Short KM, Cox TC. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J. Mol. Biol. 2006;358:532–545. doi: 10.1016/j.jmb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 105.Maurer B, Bannert H, Darai G, Flugel RM. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J. Virol. 1988;62:1590–1597. doi: 10.1128/jvi.62.5.1590-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity ofAPOBEC3Gby promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 107.Mische CC, Javanbakht H, Song B, Diaz-Griffero F, Stremlau M, et al. Retroviral restriction factor TRIM5alpha is a trimer. J. Virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 109.Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, et al. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Modin C, Lund AH, Schmitz A, Duch M, Pedersen FS. Alleviation of murine leukemia virus repression in embryonic carcinoma cells by genetically engineered primer binding sites and artificial tRNA primers. Virology. 2000;278:368–379. doi: 10.1006/viro.2000.0683. [DOI] [PubMed] [Google Scholar]
- 111.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 112.Muller S, Moller P, Bick MJ, Wurr S, Becker S, et al. Inhibition of filovirus replication by the zinc finger antiviral protein. J. Virol. 2007;81:2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nakayama EE, Miyoshi H, Nagai Y, Shioda T. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 2005;79:8870–8877. doi: 10.1128/JVI.79.14.8870-8877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. Identifies the tetherin restriction factor.
- 116.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 117.Newman RM, Hall L, Kirmaier A, Pozzi L-A, Pery E, et al. Evolution of a TRIM5-CypA splice isoform in Old World monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 119.Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- 120.Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, et al. Dual effect of APOBEC3G on Hepatitis B virus. J. Gen. Virol. 2007;88:432–440. doi: 10.1099/vir.0.82319-0. [DOI] [PubMed] [Google Scholar]
- 121.Noguchi C, Ishino H, Tsuge M, Fujimoto Y, Imamura M, et al. G to A hypermutation of hepatitis B virus. Hepatology. 2005;41:626–633. doi: 10.1002/hep.20580. [DOI] [PubMed] [Google Scholar]
- 122.Owens CM, Yang PC, Gottlinger H, Sodroski J. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 2003;77:726–731. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, et al. Reconstitution of the KRAB-KAP1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- 124.Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J. Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Perron MJ, Stremlau M, Song B, Ulm W, Mulligan RC, Sodroski J. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Petersen R, Kempler G, Barklis E. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol. Cell Biol. 1991;11:1214–1221. doi: 10.1128/mcb.11.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pincus T, Hartley JW, Rowe WP. A major genetic locus affecting resistance to infection with murine leukemia viruses. IV. Dose-response relationships in Fv-1-sensitive and resistant cell cultures. Virology. 1975;65:333–342. doi: 10.1016/0042-6822(75)90039-2. [DOI] [PubMed] [Google Scholar]
- 128. Pincus T, Rowe WP, Lilly F. A major genetic locus affecting resistance to infection with murine leukemia viruses. II. Apparent identity to a major locus described for resistance to friend murine leukemia virus. J. Exp. Med. 1971;133:1234–1241. doi: 10.1084/jem.133.6.1234. Describes the initial discovery of the Fv1 locus.
- 129.Pryciak PM, Varmus HE. Fv-1 restriction and its effects on murine leukemia virus integration in vivo and in vitro. J. Virol. 1992;66:5959–5966. doi: 10.1128/jvi.66.10.5959-5966.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rollason R, Korolchuk V, Hamilton C, Schu P, Banting G. Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J. Cell Sci. 2007;120:3850–3858. doi: 10.1242/jcs.003343. [DOI] [PubMed] [Google Scholar]
- 132.Rose KM, Marin M, Kozak SL, Kabat D. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res. Hum. Retroviruses. 2005;21:611–619. doi: 10.1089/aid.2005.21.611. [DOI] [PubMed] [Google Scholar]
- 133.Rosler C, Kock J, Kann M, Malim MH, Blum HE, et al. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42:301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- 134.Sakai H, Shibata R, Sakuragi J, Sakuragi S, Kawamura M, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1Vif protein for maturation of virus particles. J. Virol. 1993;67:1663–1666. doi: 10.1128/jvi.67.3.1663-1666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. Describes and characterizes the TRIMCyp fusion protein.
- 137.Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 138.Schaller T, Hue S, Towers GJ. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J. Virol. 2007;81:11713–11721. doi: 10.1128/JVI.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc. Natl. Acad. Sci. USA. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1- associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD and HIV-1 restriction. J. Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. Identifies APOBEC3G as the restriction factor suppressed by the Vif protein.
- 145.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 146.Shibata R, Sakai H, Kawamura M, Tokunaga K, Adachi A. Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 1995;76(Part 11):2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 147.Simon JH, Gaddis NC, Fouchier RA, Malim MH. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 148.Song B, Javanbakht H, Perron M, Park DH, Stremlau M, Sodroski J. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J. Virol. 2005;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sonigo P, Alizon M, Staskus K, Klatzmann D, Cole S, et al. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985;42:369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- 150.Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive d-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 151.Sova P, Volsky DJ. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Speck NA, Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol. Cell Biol. 1987;7:1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 154.Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 155.Strebel K, Klimkait T, Maldarelli F, Martin MA. Molecular and biochemical analyses of human immunodeficiency virus type 1 vpu protein. J. Virol. 1989;63:3784–3791. doi: 10.1128/jvi.63.9.3784-3791.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Strebel K, Klimkait T, Martin MA. A novel gene of HIV-1, vpu, and its 16-kDa product. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- 157. Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. Describes the first identification of the TRIM5α restriction factor.
- 158.Stremlau M, Perron M, Lee M, Li Y, Song B, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J. Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Suspene R, Guetard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, et al. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 162.Teich NM, Weiss RA, Martin GR, Lowy DR. Virus infection of murine teratocarcinoma stem cell lines. Cell. 1977;12:973–982. doi: 10.1016/0092-8674(77)90162-3. [DOI] [PubMed] [Google Scholar]
- 163.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 164.Towers G, Bock M, Martin S, Takeuchi Y, Stoye JP, Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Towers G, Collins M, Takeuchi Y. Abrogation of Ref1 retrovirus restriction in human cells. J. Virol. 2002;76:2548–2550. doi: 10.1128/jvi.76.5.2548-2550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 167.Tsukiyama T, Niwa O, Yokoro K. Mechanism of suppression of the long terminal repeat of Moloney leukemia virus in mouse embryonal carcinoma cells. Mol. Cell Biol. 1989;9:4670–4676. doi: 10.1128/mcb.9.11.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 169.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vartanian JP, Meyerhans A, Asjo B, Wain-Hobson S. Selection, recombination, and G → A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 1991;65:1779–1788. doi: 10.1128/jvi.65.4.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Varthakavi V, Smith RM, Bour SP, Strebel K, Spearman P. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. USA. 2003;100:15154–15159. doi: 10.1073/pnas.2433165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T. Independent genesis of chimeric TRIM5- cyclophilin proteins in two primate species. Proc. Natl. Acad. Sci. USA. 2008;105:3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wilson SJ, Webb BL, Ylinen LM, Verschoor E, Heeney JL, Towers GJ. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2008;105:3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wolf D, Cammas F, Losson R, Goff SP. PBS-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J. Virol. 2008;82:4675–4679. doi: 10.1128/JVI.02445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. Identifies TRIM28 as a cellular factor required PBS-mediated repression of MLV in EC and ES cells.
- 179.Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. USA. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan MA, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xu L, Yang L, Moitra PK, Hashimoto K, Rallabhandi P, et al. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp. Cell Res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- 182.Yamauchi M, Freitag B, Khan C, Berwin B, Barklis E. Stem cell factor binding to retrovirus primer binding site silencers. J. Virol. 1995;69:1142–1149. doi: 10.1128/jvi.69.2.1142-1149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yang B, Chen K, Zhang C, Huang S, Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 2007;282:11667–11675. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- 184.Yang WK, Kiggans JO, Yang DM, Ou CY, Tennant RW, et al. Synthesis and circularization of N- and B-tropic retroviral DNA Fv-1 permissive and restrictive mouse cells. Proc. Natl. Acad. Sci. USA. 1980;77:2994–2998. doi: 10.1073/pnas.77.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yao XJ, Gottlinger H, Haseltine WA, Cohen EA. Envelope glycoprotein and CD4 independence of Vpu-facilitated human immunodeficiency virus type 1 capsid export. J. Virol. 1992;66:5119–5126. doi: 10.1128/jvi.66.8.5119-5126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yap MW, Nisole S, Lynch C, Stoye JP. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of humanTrim5alpha leads to HIV-1 restriction. Curr. Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 188.Ylinen LM, Keckesova Z, Webb BL, Gifford RJ, Smith TP, Towers GJ. Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J. Virol. 2006;80:7332–7338. doi: 10.1128/JVI.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 190.Yu X, Yu Y, Liu B, Luo K, Kong W, et al. Induction ofAPOBEC3Gubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 191.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3Gincorporation into human immunodeficiency virus type 1 particles. J. Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353:396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 193.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 2007;81:11246–11255. doi: 10.1128/JVI.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]