Abstract
Epigenetic mechanisms integrate genetic and environmental causes of disease. Comprehensive genome-wide analyses of epigenetic modifications have not demonstrated robust association with common diseases. Using Illumina HumanMethylation450 arrays on 354 ACPA positive rheumatoid arthritis (RA) cases and 337 controls, we identified two clusters within the MHC region whose differential methylation potentially mediates genetic risk for RA. To reduce confounding hampering previous epigenome-wide studies, we corrected for cellular heterogeneity by estimating and adjusting for cell-type proportions and used mediation analysis to filter out associations likely consequential to disease. Four CpGs also showed association between genotype and variance of methylation in addition to mean. The associations for both clusters replicated at least one CpG (p<0.01), with the rest showing suggestive association, in monocytes in an independent 12 cases and 12 controls. Thus, DNA methylation is a potential mediator of genetic risk.
Epigenetic mechanisms can cause durable changes of gene expression that are heritable during cell division by covalent modifications of DNA bases and potentially other chromatin alterations. They might influence disease development in a manner complementary to direct mutations of the DNA sequence.
The role of epigenetic modifications in cancer etiology and progression is well established1, and a number of small surveys of DNA methylation in common disease have been carried out2-5. We and others have suggested that genetic and epigenetic modifications could interact biologically6-8, and that methylation analysis might uncover heritable genetic variants contributing to disease that are invisible to conventional GWAS. A comprehensive genome-wide methylation analysis has not yet demonstrated robust association of specific methylation alterations with a common disease, however. This may be due in part to several limitation to such studies including (1) the cellular heterogeneity of the sample material, and (2) the potential for methylation changes that are a consequence of disease rather than part of the etiology Here, we apply a series of ad hoc filtering steps that address these issues to identify CpG methylation that likely mediates genetic risk for rheumatoid arthritis (RA) from genome-wide epigenetic and genetic data. This process may serve as a guidepost for epigenetic epidemiological studies generally.
RA is a complex and heterogeneous disease, where onset as well as disease course is dependent on interactions between different genetic and environmental or life style factors9,10. Several meta-analyses of genome-wide association studies (GWAS) have identified close to 40 genetic variants that confer risk for the citrullinated protein antibody-associated (ACPA) subtype of RA11-14. However, the fact that these discoveries can only explain less than 20% of disease variance suggests that other factors are likely involved in the disease13.
Two additional factors make RA an ideal test case for analyzing the relationships between genes, methylation and disease pathogenesis. In RA, one of the main classes of cells involved in the disease, leucocytes, is readily available for DNA methylation analysis and disease state can be reproducibly determined by the presence of antibodies to citrullinated protein antigens (ACPA)
In our present study, 354 RA patients (cases) with citrullinated protein antigens(ACPA) antibodies 15 and 337 healthy individuals (controls) were selected from the Epidemiological Investigation of RA (EIRA)16,17, a Swedish population-based case-control study. Cases were recruited at the first visit to a rheumatology clinic before initiation of treatment with disease-modifying small molecule or biological agents. At this first visit, blood samples were collected for DNA analysis and serology16,18. Control subjects were selected from the same study to match RA cases in terms of age, gender, smoking status and residential area at the time of diagnosis (Supplementary Table 1). An additional advantage of these samples is that genome-wide SNP data were available on the same individuals, enabling us to determine the relationship between genotype, epigenotype and phenotype. On these 691 samples, we first performed genome-wide DNA methylation analysis using the Illumina 450K methylation array, to identify RA-associated epigenetic differences. After excluding two samples with poor quality and 187,468 probes containing SNPs, which might affect the measurement of DNA methylation, the final dataset used for downstream analysis comprised 354 cases and 335 controls for 298,109 CpG positions [see online methods for details].
Correcting for cellular heterogeneity
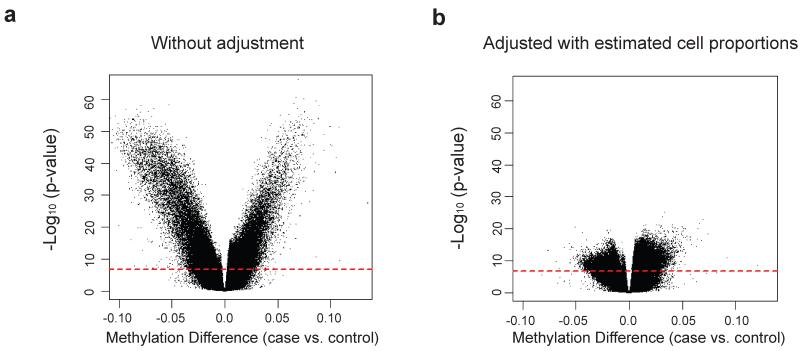
Our first challenge is that the DNA samples available for methylation analysis are generally derived from heterogeneous cell populations. For example, the DNA samples most readily available from large numbers of individuals are from whole blood, which consists of many distinct populations in varying proportions. It has been shown that these functionally distinct populations have unique DNA methylation signatures19, thus cell heterogeneity may act as a potential confounder when investigating DNA methylation differences between cases and controls, if cell distribution itself differs by disease status. To address this, we attempted to adjust for cell proportion using linear regression models in our epigenetic association analysis. To obtain sample-specific estimates of cell proportion for the major cell types in blood, we applied a statistical algorithm developed by Houseman et. al.20 that uses reference information on cell-specific methylation signatures to estimate cell proportions from genome-scale methylation data. The estimated cell type distribution from this algorithm was consistent with our experimental results from flow cytometry and did show distinct patterns between RA cases and controls (Supplementary Table 2), suggesting that it is critical to adjust for cell type distribution in the downstream analyses. For example, Figure 1 shows epigenome-wide association results before and after adjustment using estimate cell proportions, showing a notable reduction in association signals after adjustment.
Figure 1.
Differential cell counts in identifying RA-associated differentially methylated positions (DMPs). Volcano plot of −log10 (p-value) against beta, representing the methylation difference between ACPA positive RA cases and controls, without a) or with b) adjusting for differential cell counts, in addition to control for age, sex and smoking status. The dashed red lines represent the threshold used for statistical cutoff (Bonferroni-adjusted p-value = 0.05).
Establishing epigenetic mediation of genetic risk
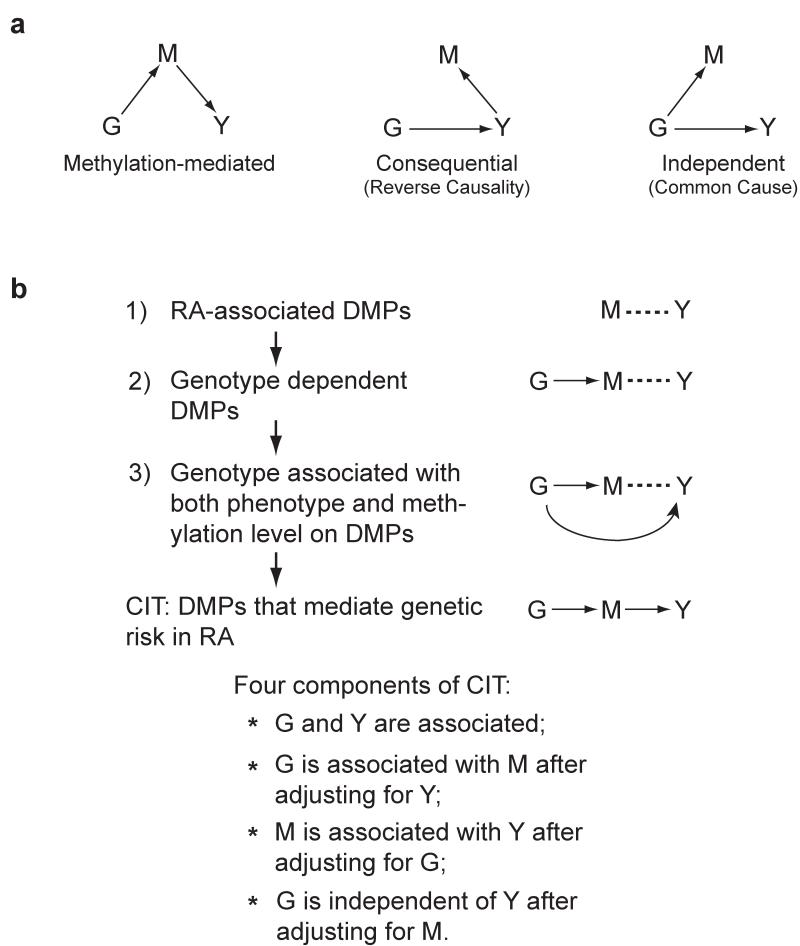
Our second challenge is that many methylation differences are likely a consequence of RA. To filter these out, and reveal biology related to the cause, we applied methods from the causal inference literature21-24. In this approach, which employs a series of conditional correlation analyses, one considers the possible directed relationships between a causal factor, a potential mediator and an outcome (Fig.2a).
Figure 2.
Identification of epigenetically mediated genetic risk factors for RA disease. (a) Possible relationships between a causal factor (G), a possible mediator (M) and an outcome (Y). The diagram on the left represents the methylation-mediated relationship, in which genotype (G) acts on phenotype (Y) through methylation (M). The diagram in the middle represents the consequential methylation model, in which DNA methylation (M) changes are the consequence of phenotype (Y). The diagram on the right represents the independent model, in which the genotype (G) acts on DNA methylation (M) and phenotype (Y) independently. (b) Summary workflow for identifying epigenetically mediated genetic risk factors for RA. The diagrams on the right represent the relationships between genotype (G), DNA methylation (M) and RA phenotype (Y). The dashed lines represent the association relationship, whereas the arrowed lines represent the causal relationship.
As we were particularly interested in epigenetic marks that may mediate the genetic risk for RA, we applied this method with genotype as a causal factor (G, Fig. 2a), DNA methylation as a potential mediator (M, Fig. 2a) and rheumatoid arthritis as the outcome (Y, Fig. 2a). We developed a 3-step filtering process followed by the application of the Causal Inference Test (CIT)24 to identify the RA-associated DMPs that are most likely to be acting as mediators of genetic risk rather than a consequence of RA. These three filtering steps are (Fig. 2b): 1) establish the relationship between the potential mediator (M) and outcome (Y); 2) from these, establish the relationship between the primary cause (G) and the mediator (M); and 3) from these, establish the relationship between the cause (G) and outcome (Y). We then applied the Causal Inference Test to establish that methylation (M) is the mediator between the cause (G) and outcome (Y). Our approach results in a candidate set of mediators, but it should be noted that, as in all epidemiological studies, it is impossible to conclusively prove causal relationships on the basis of observational data alone.
Briefly, the CIT requires the following criteria24: G and Y are associated; G is associated with M after adjusting for Y; M is associated with Y after adjusting for G; and G is independent of Y after adjusting for M. If methylation is a consequence of Y (Fig. 2a, middle panel) or independently controlled by G (Fig. 2a, right panel), rather than a mediator in the path from G to Y (Fig. 2a, left panel), the estimated effect of G on Y should not be impacted by conditioning on M. However, if methylation is indeed a mediator, this conditioning should drastically reduce the observed effect of G on Y (Fig. 2a, left panel).
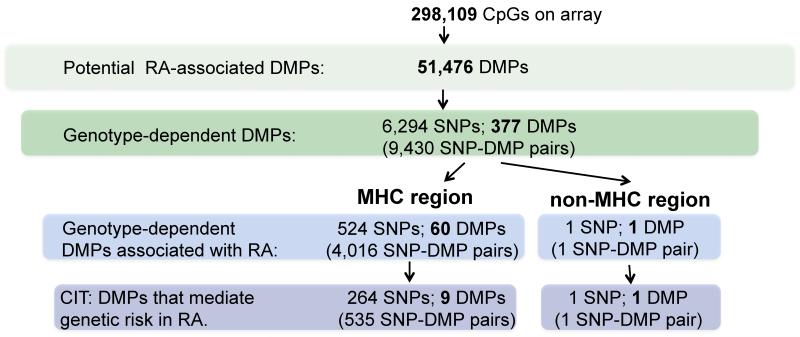
In our first filtering step, we performed epigenome-wide association analysis using adjustment for estimated cell proportions as well as age, sex, and smoking status in the context of a linear model. We took all putatively RA-associated DMPs that achieved a p<0.05 after Bonferroni correction, 51,476, into the next step (step 1 in Fig. 3). We then performed genome-wide SNP association analysis for each of these DMPs (step 2 in Fig. 3) to identify the subset where methylation level appears to be under genetic control. We fit an allelic dosage model for each DMP and each of 1,196,263 SNPs (300,987 genotyped SNPs and 895,276 SNPs imputed from the HapMap 3 panel) and identified 9,430 SNP-DMP pairs (Supplementary Table 4) with genome-wide significance (Bonferroni-adjusted p < 0.05). These SNP-DMP pairs comprised 6,294 unique SNPs and 377 unique DMPs (step 2 in Fig.3). More than half of SNP-DMP pairs are spread over a 5Mb region covering the MHC cluster, which is known to harbor several RA-risk loci14. Given that the MHC cluster was heavily overrepresented, we decided to analyze the MHC region and non-MHC region separately.
Figure 3.
Summary workflow and results for identifying epigenetically mediated genetic risk factors for RA disease.
In-depth analysis of the MHC region
It has been shown that the major genetic risk loci for seropositive rheumatoid arthritis are located within the MHC region, which accounts for more than 10% of the phenotypic variance14. Given that the MHC cluster was heavily overrepresented in the results from the genome-wide SNP-DMP scan, we decided to explore this region in detail using increased density genotype data imputed based on a large reference panel14. We again fit an allelic dosage model for each RA-associated DMP and each of 5,009 imputed SNPs within the MHC region and identified 7,242 significantly associated SNP-DMP pairs (Supplementary Table 5) (Bonferroni-adjusted p < 0.05) in MHC. These SNP-DMP pairs comprised 1,952 unique SNPs and 76 unique DMPs. We then performed pre-filtering step 3 and the causal inference procedure, outlined above, to distinguish between methylation that is a result of disease and methylation that may be in the causal path to disease.
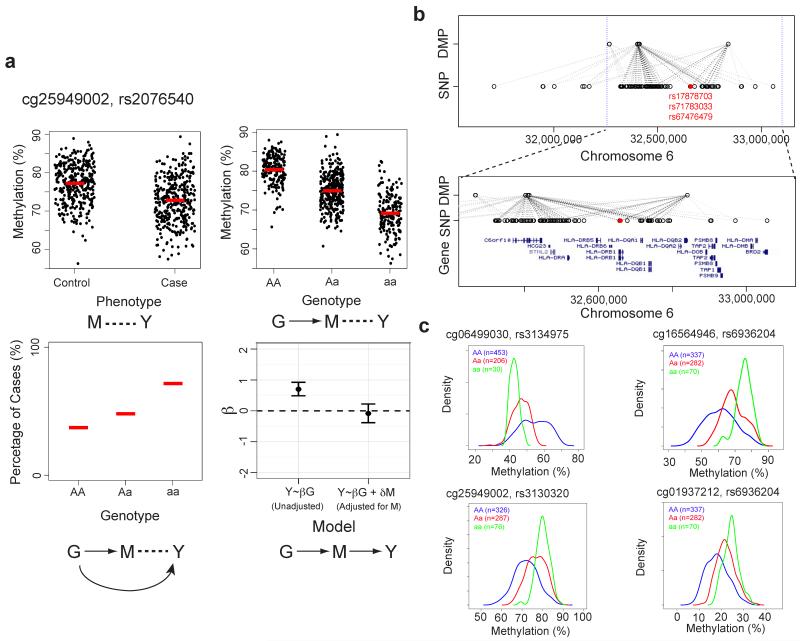
We tested the association between each of 1,952 SNPs and RA using an allelic dosage model. As all these SNPs are located within the MHC region and many are correlated, we permuted adjusted p-values using the step-down maxT multiple testing procedure to control the family wise Type I error rate (FWER)25. Of the 1,952 SNPs, we identified 524 that are significantly associated with RA at an adjusted p-value < 0.05. These 524 SNPs form 4,016 SNP-DMP pairs with 60 unique DMPs (step 3 in Fig.3, Supplementary Table 6). By performing the CIT test24, we identified that for 535 of the 4,016 SNP-DMP pairs, the SNP effect on RA is reduced when conditioning on methylation (Bonferroni adjusted CIT p-values < 0.05, see methods for further detail), suggesting mediation (Fig. 4a). These 535 MHC SNP-DMP pairs comprised 264 unique SNPs and 9 unique DMPs (CIT in Fig. 3, Table 1, Supplementary Table 7) and represent potential methylation-mediated relationships between SNPs and RA disease risk.
Figure 4.
Genotype-dependent candidate DMPs that mediate genetic risk within the MHC region. (a) Top panels: association between DNA methylation level at a DMP that mediates genetic risk in RA and phenotype (top left) or genotype (top right). Red lines mark average DNA methylation levels. Bottom left panel: association between genotype and RA phenotype. Red lines mark percentage of cases for each genotype. Bottom right panel: Coefficient (β) represents the dependence of RA phenotype (Y) on genotype (G), with or without adjusting for DNA methylation (M). The error bars represent the 95% confidence interval for the coefficient, β. In the case of the methylation-mediated model, the absolute value of the observed G:Y relationship strength reduces towards zero when adjusting for methylation (M). (b) Association between candidate genetic risk-mediating DMPs and genotype within a 1.5Mb section of the MHC region. Each dashed line represents a potential mediation relationship between a SNP and a DMP as determined by the Causal Inference Test (CIT). The color of the line indicates significance of the p-value for the statistical mediation test. Genotype data is imputed based on a large reference panel. Three previously identified RA-associated genetic variants are illustrated in red. Bottom panel: zoomed-in images of top panel with gene annotations. (c) Examples of RA-associated DMP in which genotype is associated with the change in both mean and variance of DNA methylation. The methylation density plot is color coded by genotype. The number of individuals in each genotype group is shown on the top left corner.
Table 1. DMPs that mediate genetic risk in RA.
| RA-associated CpGs (DMPs) |
SNPs associated with CpG methylation |
|||||||
|---|---|---|---|---|---|---|---|---|
| DMP | Beta* | p-value (meth vs. pheno) |
Gene Name | SNP† | p-value (geno vs. meth) |
Adjusted p-value (geno vs. pheno) |
p-value (CIT) | |
| MHC | cg21325723 | −0.028 | 1.49E-09 | C6orf10 | rs2395163 | <2E-16 | 1.00E-04 | 2.96E-19 |
| cg16609995 | 0.015 | 2.88E-09 | PBX2 | DRB1_AA104_E2_32659926_AE | 1.33E-15 | 3.00E-04 | 5.95E-07 | |
| cg06499030 | 0.038 | 4.01E-09 | HLA-DQB2 | CHR6_POS32657567 | <2E-16 | 1.00E-04 | 6.62E-15 | |
| cg16564946 | −0.046 | 9.74E-09 | C6orf10 | rs9267954 | <2E-16 | 1.00E-04 | 7.30E-08 | |
| cg25949002 | −0.026 | 2.57E-08 | C6orf10 | rs2076540 | <2E-16 | 1.00E-04 | 1.24E-10 | |
| cg14704780 | −0.066 | 2.87E-08 | C6orf10 | rs3916765 | <2E-16 | 1.00E-04 | 8.72E-10 | |
| cg19555708 | 0.012 | 5.11E-08 | GPSM3 | DRB1_AA104_E2_32659926_AE | 8.88E-16 | 3.00E-04 | 5.95E-07 | |
| cg01937212 | −0.021 | 6.00E-08 | C6orf10 | rs477005 | <2E-16 | 1.00E-04 | 5.39E-10 | |
| cg19321684 | 0.023 | 8.26E-08 | GPSM3 | DRB1_AA104_E2_32659926_AE | <2E-16 | 3.00E-04 | 5.95E-07 | |
| Non-MHC | cg00462104 | −0.023 | 8.84E-08 | GSTA2 | rs3996993 | <2E-16 | 0.038 | 0.0016 |
Adjusted methylation difference between cases and controls
For each DMP, only SNP with smallest CIT p-value is shown here. For the full SNP list, see Supplementary Table 7.
Methylation mean and variance mediating genotype-phenotype relationship
In recent work, Raychaudhuri et al.14 have demonstrated that variations in genes coding for five different amino acid positions in the binding grooves of HLA-DR, HLA-DP and HLA-B account for most hitherto described associations between genetic differences in the MHC region and ACPA-positive RA. Three of these five genetic variants are located within the HLA-DRB1 gene and show significant (Bonferroni-adjusted p<0.05) evidence of association with DNA methylation loci that appear to at least partially mediate the genetic risk effect (Fig. 4b). Some of these RA-associated DMPs are located over 100kb from the associated genetic risk variants, possibly as a consequence of the relative sparseness in CpG coverage on the Illumina 450K methylation array.
We proposed recently that genetic variants might regulate phenotypic variability in addition to mean phenotype and that this connection between genotype and phenotypic plasticity would be mediated epigenetically26. Such a mechanism would provide a non-Lamarckian basis for an epigenetic role in natural selection, because the variants themselves would be transmitted genetically, but they would also allow increased phenotypic plasticity in response to a varying environment26. DNA methylation represents a promising candidate for mediating such plasticity, since methylation levels are measured as proportions where variance changes are intrinsically related to mean shifts. Notably, five out of nine DMPs identified here also showed a significant association between genotype and variance of methylation, as suggested by the model (Fig. 4c, Supplementary Table 8).
Identification of methylation-mediated genetic risks in non-MHC regions
We further analyzed the 4,619 SNP-DMP pairs (including 4,540 SNPs and 343 DMPs) outside of the MHC region that were identified in the genome-wide scan. Of the 4,540 SNPs, one SNP is significantly associated with RA phenotype (step-down maxT adjusted p-value < 0.05) (step 3 in Fig.3). Using the CIT24 as described before, we concluded that the effect of genotype on RA risk appears to be mediated by a DNA methylation change in the promoter region of a gene called GSTA2 (Supplementary Fig. 1,Table 1). GSTA2 belongs to the glutathione S-transferase supergene family, which is important in the detoxification of electrophilic compounds, including environmental toxins (such as tobacco smoke)27-29. Polymorphisms within the μ (GSTM1), θ (GSTT1) and π (GSTP1) classes of GST previously have been identified and are associated with RA risk and severity30-33, although GSTA2 has not previously been implicated in RA through GWAS13. This underscores the usefulness of our approach. In fact, we failed to identify any RA-associated non-MHC genetic variants by fitting a standard allelic dosage model at each SNP within our own samples (Supplementary Fig. 2).
Replication of methylation differences in flow-sorted cells
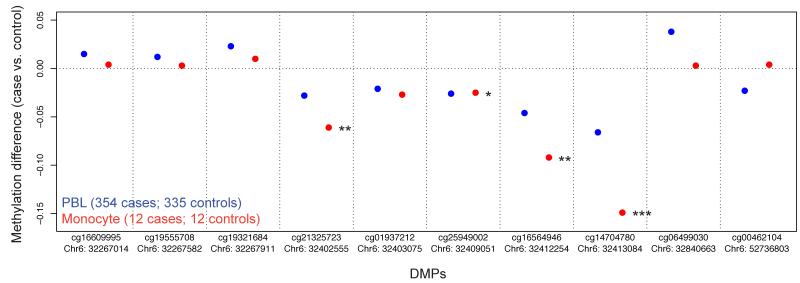
Having identified 10 differentially methylated positions (DMPs) (9 within the MHC region and 1 outside of the MHC region) whose methylation level putatively mediates genetic risk in RA (Table 1), we attempted to replicate these methylation differences in fresh flow sorted cell populations of untreated RA cases and controls. We separated PBLs from 12 case-control pairs into separate cell fractions. For the monocyte cell fraction, nine out of ten DMPs showed methylation changes in the same direction as that seen in the large scale PBL analysis. Of these, three of the CpG sites were significant at p < 0.05, one at p = 0.063, and a fifth at 0.11, even with this small sample number (Supplementary Table 9). The three with greatest significance also showed larger beta values than seen in PBL (Fig. 5), suggesting that monocytes are more proximal to the pathogenic cell type. Given that monocytes represented less than 10% of the PBL fraction, this may explain the smaller effect size seen using total PBLs.
Figure 5.
Replication data for ten candidate DMPs that mediate genetic risk in RA from sorted monocytes. DNA methylation data on CD14+ monocytes from a replication set of 12 ACPA positive RA cases and 12 controls was measured using the Illumina 450K methylation array. Results from PBLs are illustrated in blue while the results from monocytes are in red. For PBLs, all 10 DMPs have p-value less than 10−7. For sorted monocytes: one asterisk, p-value < 0.1; two asterisks, p-value < 0.01; three asterisks, p-value < 0.001.
Discussion
In summary, we have applied an approach that corrects for the confounding influence of cell heterogeneity and filters out signals likely due to the disease itself Using a strategy of three filtering steps followed by the application of mediation analysis using the CIT algorithm, we performed genome-scale methylation and SNP analysis and identified ten putative DMPs that mediate genetic risk for RA, nine in the MHC cluster, and one outside on the same chromosome (6p12.1).
Our approach for adjusting for cell heterogeneity should be applicable for many tissue sources, if cell-specific methylation signatures for the particular mixture in question are available. Even samples from primary affected tissues tend to consist of a mixture of many cell types making an adjustment for cell type proportions a prerequisite for epigenetic association analysis, somewhat analogous to the correction for population stratification using empirically estimated ancestry proportions in GWAS studies34,35. This adjustment for cell proportions does not address the question of whether the chosen tissue is the appropriate surrogate tissue for the disease in question, but simply handles the heterogeneity issue regardless of surrogate or primary status. In this report, we have assumed that blood is the primary tissue for an inflammatory disease.
While we show that our cell type adjustment is a notable improvement over unadjusted analyses and reduces confounding by cell type bias, there may be residual confounding not fully accommodated in the specific proportion estimation and linear adjustment we pursued. Further methodological work to improve this estimation and modeling approach is important. Other sources of confounding in array analyses must also be considered. In addition to age, sex, and other demographic confounders, batch effects such as date and laboratory should be evaluated. For example, while we did not anticipate strong batch effects for 450K methylation data to date, we examined our own data via principle components analysis and do observe a relationship between date of assay and overall methylation signals. While ideally one would design assays runs to have equivalent spread of phenotypes across dates/labs, this is often not practical. In our study, there was an imbalance between the number of cases and controls run per date, and thus batch effects in these 450K data could potentially confound associations36. To address these issues, we re-analyzed the results for our top 10 CpGs using a procedure that simultaneously corrects for batch and cell type composition (see Methods, Supplementary Fig. 3). While the statistical significance of all CpGs is affected by this adjustment, it is notable that the five CpGs in the two regions that were replicated in flow-sorted monocytes retain the strongest effect size (Supplementary Fig. 4, Supplementary Table 10). We would recommend in future studies addressing batch effect issues via principle components analysis or SVA37 as a first step. This may in fact improve cell proportion estimation for subsequent adjustment. We also noted that although our approach to cell proportion adjustment, which is a convenient tool for blood-derived samples, is a considerable improvement over no adjustment, residual confounding due to cell type may remain. This can be dealt with via replication in cell-sorted samples as we have shown, or via advanced estimation and statistical adjustment methods - a call for additional methodological work.
Another issue that complicates epigenetic studies over purely genetic analysis is that the primary tissue may harbor DNA methylation changes that are a consequence of the disease, rather than a marker of causal mechanism. To address this, we applied mediation approaches already used in the gene expression and epidemiology literature, but not previously applied to epigenetic studies. We emphasize that our findings, as in all epidemiological studies, are hypotheses that will ultimately require verification in independent and/or mechanistic studies. In particular, there exist conditions, such as the presence of unmeasured confounders, where it may be impossible to distinguish causal from consequential methylation events based on observational data alone38. Although much will need to be worked out over time, just as it was in the development of GWAS, we feel that our approach directly addresses the fundamental question of epigenetic epidemiology i.e. how one can link genetics to epigenetics to phenotype. Similarly, mediation analysis can be applied to the other component of epigenetic epidemiology—the role of the environment—if one assumes that the environmental factors are causal in the disease.
It is notable that our top 10 CpGs represent signals across 5 genomic regions, and the 5 CpGs that replicate most robustly in monocytes cluster in two regions (Supplementary Fig. 4). This supports use of region-based statistical approaches such as “bump hunting”39 to epigenetic association analyses and further suggests that denser coverage than the 450K array will be better in identifying methylation differences moving forward either by a new array design or capture bisulfite sequencing. It is notable that monocyte subfractions showed stronger effect sizes compared to unfractionated PBLs, with statistical significance for 3 of the DMPs and marginal significance for another 2, with only 12 case-control pairs, supporting a role for monocytes in RA pathogenesis, something that is also suggested from many previous cell biologic studies in RA40. In addition, MHC class II gene expression in macrophages, which are derived from monocytes, show a strong relationship to RA progression41.
A byproduct of the analysis presented here was the identification of suggestive evidence for vSNPs for epigenetic modification, i.e. SNPs regulating DNA methylation variation. These vSNPs are predicted by a model we proposed in which genetic variants might increase epigenetic plasticity, providing a non-Lamarckian basis for an epigenetic role in natural selection26. They included 5 of the 9 DMPs identified in the MHC region.
This research also makes a prediction that is beyond the scope of the current experiments. Given that genetic association in the MHC cluster with RA has already been shown to be linked to specific HLA protein epitopes, the methylation mediation we observe implies an additional complementary mechanism for RA, e.g. basal levels of gene expression, expression in response to antigen provocation, or alternative splicing, since both gene expression and splicing are regulated by DNA methylation. Given that there are more than 10 genes whose promoters are within 100kb distance of the identified DMPs (Supplementary Table 11) and over 50 genes within the region defined by the SNP-DMP-phenotype associations reported here, at least one of these genes should show altered regulation related to DNA methylation in RA, in addition to the linear gene-protein relationships already known.
Methods
Sample preparation
Recruitment of RA patients in the EIRA study was described in details previously11. Briefly, only incident cases of RA were invited for the study within the years 1996-2009, from 18 clinics in Middle Sweden. Individuals were examined by rheumatologists and all patients correspond to ACR1987 criteria. The controls from the same population were matched by sex, age, smoking status and residence area. DNA was extracted from EDTA blood and kept at −80 °C until use. The cell purification protocol was described previously42.
Illumina genome-wide genotyping
The genotyping and QC procedures have been described previously11. Briefly, the EIRA samples were genotyped with the Illumina Human Hap300 v1.0 chip, Hap370CNVduo chip or Hap550duo chip. Samples included for analysis had call-rates > 95% and inferred gender consistent with clinical records. SNP filtering was performed based on chip type, eliminating SNPs with call-rates below 95%; monomorphic SNPs; SNPs with a minor allele frequency < 0.005; SNPs with a Hardy-Weinberg equilibrium p-value of < 1.0 × 10−7 in controls, and SNPs mapping to multiple locations and non-autosomal chromosomal SNPs. This resulted in 306,994 autosomal SNPs on 1966 samples in Hap300; 324,981 autosomal SNPs in Hap370CNVduo on 674 samples; 527,434 autosomal SNPs on 520 samples in Hap550duo passing the quality control filters. Closely related individuals were identified by RELPAIR and PLINK. The member of each pair with the lower call rate was dropped from further analysis. To quantify and control for population stratification, we used a principal components approach implemented in the EIGENSTRAT software. EIGENSTRAT identified a total of 141 significant outliers, which were removed from further analysis. This resulted in a dataset of 1934 RA cases and 1079 controls on 297,393 SNPs.
Genomic imputation
Imputation was performed using the MACH algorithm based on HapMap3. The cleaned EIRA GWAS dataset (3000 individuals) was used for imputation. The genotype calls are based on a QC cutoff of 0.9. Amino acids imputation within HLA-DRB1, HLA-DPB1 and HLA-B was performed previously14.
Rheumatoid arthritis genetic risk genome wide association analysis
SNPs (n=1,196,263) were tested for association with RA case-control status using an additive minor-allele dosage model in the cohort of 354 ACPA positive RA cases and 335 population-matched controls selected for Illumina 450K methylation assay. No non-MHC SNPs were significantly associated with RA phenotype after adjusting for multiple testing using a Bonferroni-adjusted α=0.05 significance level.
Illumina 450K methylation assay
For each sample, 1 μg of genomic DNA was bisulfite-converted using an EZ DNA methylation Kit (ZYMO research) according to the manufacturer’s recommendations. Converted genomic DNA was eluted in 22 μl of elution buffer. DNA methylation level was measured using the Illumina Infinium HD Methylation Assay (Illumina) according to the manufacturer’s instructions. Briefly, 4 μl of bisulfite-converted DNA was isothermally amplified overnight (20-24 hours) and fragmented enzymatically. Precipitated DNA was resuspended in hybridization buffer and dispensed onto the Infinium HumanMethylation450 BeadChips (12 samples/chip) using a Freedom EVO robot (Tecan). The hybridization procedure was performed at 48 °C overnight (16-20 hours) using an Illumina Hybridization oven. After hybridization, free DNA was washed away and the BeadChips were processed through a single nucleotide extension followed by immunohistochemistry staining using a Freedom EVO robot (Tecan). Finally, the BeadChips were imaged using an Illumina iScan.
Illumina 450K microarray data preprocessing
Detection p-values were calculated to identify failed probes as per Illumina’s recommendations. No arrays exceeded our quality threshold of >5% failed probes. Probes on sex chromosomes or containing SNPs (dbSNP v132) in the probe sequence were excluded. Raw data was normalized using Illumina’s control probe scaling procedure and converted to methylation values on the 0-1 scale (M/(M + U + 100), where M and U represent the methylated and unmethylated signal intensities respectively). The Illumina 450K array data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), under accession number GSE42861.
Estimate differential cell counts
Differential cell counts for each individual were estimated using the algorithm developed by Houseman et. al.20 with a slight modification. Briefly, the distribution of cell types for each sample was inferred based on DNA methylation signatures of the constituent cell types. A total of five different cell types, including T cells, NK cells, B cells, monocytes and granulocytes, were included in the estimation. DNA methylation signatures on sorted human cells from the Illumina 450K arrays19 were used as validation data. Among the 500 most informative CpG probes for distinguishing cell types chosen from the Illumina Infinium 27K array20, all 473 probes also present in the Illumina 450K array are included in the analysis.
Identify RA-associated DMPs
To identify the differentially methylation positions (DMPs) associated with RA phenotype, we fit a linear regression model predicting methylation at each CpG sites as a function of RA status, adjusted for age, sex, smoking status and estimated differential cell counts. RA-DMP associations were corrected for multiple testing using a stringent Bonferroni-adjusted threshold of 0.05/(298,109 CpGs) = 1.68×10−7.
Identify genotype dependent DMPs
All genome-wide significant (Bonferroni-adjusted p < 0.05) RA-associated DMPs were subsequently tested for association with genotype (1,196,263 SNPs) using an additive minor-allele dosage model. Genotype-DMP associations were corrected for multiple testing using a stringent Bonferroni-adjusted threshold of 0.05/(51,476 DMPs × 1,196,263 SNPs) = 8.12×10−13. SNPs associated with methylation variance were identified by fitting an additive minor-allele dosage model to absolute methylation residuals, calculated as the difference between a subject’s methylation value and the genotype-specific mean. A Bonferroni-adjusted α=0.05 cutoff was used to determine significance.
Causal Inference Test
Each of the genotype (G) - methylation (M) - phenotype (Y) relationships were assessed using the Causal Inference Test (CIT)24 to classify them as methylation-mediated, methylation consequential and independent. Note that the corresponding terms originally used by Schadt et al. in describing the CIT are causal, reactive and independent21. Briefly, the CIT performs statistical tests for four conditions, all of which must be met for the methylation-mediated (causal) classification: 1) G and Y are associated, 2) G is associated with M after adjusting for Y, 3) M is associated with Y after adjusting for G, and 4) G is independent of Y after adjusting for M. Because all component conditions must be satisfied, the CIT p-value is defined using the intersection-union test framework43 as the maximum of the four component test p-values. Since the CIT was designed for continuous phenotypes rather than case/control studies, we developed a modified version based on logistic regression and confirmed that all 10 reported SNP-CpG pairs retained significant causal p-values.
Post hoc statistical analysis
Within each batch, we calculated the first principal component of cell type composition using a balanced sample of cases and controls. We then used a linear model to fit methylation values to this cell type composition proxy calculated residuals representing batch and cell type corrected estimates, and used these in epigenotype-RA association analyses.
DNA methylation analysis from flow sorted cells
30-50 mL of whole blood with heparin as a conservative was collected by a clinician from RA patients and controls with consent, and separated with Ficoll within 24 hours of collection. Cells were sorted using AutoMACS (Miltenyi Biotech) into four populations: CD4+, CD8+, CD14+, CD19+ and frozen as cell pellets in PBS at −80 °C. Genomic DNA was extracted with salting-out method. DNA methylation level was measured using the Illumina 450K methylation array as described before.
Analysis software
All analysis was performed in R 2.14 and Bioconductor 2.9. Illumina 450K microarray data was analyzed with the minfi package.
Supplementary Material
Acknowledgements
We thank Michael Rosenblum for helpful discussions on the application of statistical mediation methodology. We thank Eugene Andres Houseman for codes used for estimating cell proportions. The authors thank Rafa Irizarry for his contributions to the concepts in this work, his statistical insights on batch effects, and helpful comments. We also thank the EIRA study group18 for contributing invaluable clinical information and biological samples. This work was supported by the US National Institutes of Health Centers of Excellence in Genomic Science, 5P50HG003233 to A.P.F., and by the Swedish Research Council, the Swedish Combine project, the Swedish Strategic Foundations, the AFA Insurance, and the European Research Council (ERC).
Footnotes
Author Contributions
Y.L. performed the experiments. Y.L. and M.J.A. analyzed data. L.P. and L.A. performed epidemiological data collection and evaluation. L.P. did sample genotyping and genotype imputation. M.D.F performed epidemiology analysis and data interpretation and assisted in experimental design. L.P., M.R.,K.S., and E.H. prepared nucleic acids and/or cell sorting. A.R. performed the 450K arrays. M.T. assisted in statistical analysis. J.K., L.R., N.A., and A.S. provided reference normal 450K data from sorted cells for estimating cell proportions. Y.L., M.J.A., L.P., M.D.F. L.K., T.J.E., and A.P.F. conceived the experiments and wrote the paper.
References
- 1.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 2.Kaminsky ZA, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–5. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med. 2010;2:49ra67. doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Javierre BM, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–9. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakyan VK, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7:e1002300. doi: 10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–8. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Bjornsson HT, et al. Intra-individual change over time in DNA methylation with familial clustering. J Amer Med Assoc. 2008;299:2877–83. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–41. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klareskog L, Catrina AI, Paget S. Rheumatoid arthritis. Lancet. 2009;373:659–72. doi: 10.1016/S0140-6736(09)60008-8. [DOI] [PubMed] [Google Scholar]
- 10.Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. Lancet. 2010;376:1094–108. doi: 10.1016/S0140-6736(10)60826-4. [DOI] [PubMed] [Google Scholar]
- 11.Padyukov L, et al. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann Rheum Dis. 2011;70:259–65. doi: 10.1136/ard.2009.126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raychaudhuri S, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–8. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–6. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klareskog L, Ronnelid J, Lundberg K, Padyukov L, Alfredsson L. Immunity to citrullinated proteins in rheumatoid arthritis. Annu Rev Immunol. 2008;26:651–75. doi: 10.1146/annurev.immunol.26.021607.090244. [DOI] [PubMed] [Google Scholar]
- 16.Padyukov L, Silva C, Stolt P, Alfredsson L, Klareskog L. A gene-environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum. 2004;50:3085–92. doi: 10.1002/art.20553. [DOI] [PubMed] [Google Scholar]
- 17.Mahdi H, et al. Specific interaction between genotype, smoking and autoimmunity to citrullinated alpha-enolase in the etiology of rheumatoid arthritis. Nat Genet. 2009;41:1319–24. doi: 10.1038/ng.480. [DOI] [PubMed] [Google Scholar]
- 18.Klareskog L, et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 19.Reinius LE, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–7. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen LS, Emmert-Streib F, Storey JD. Harnessing naturally randomized transcription to infer regulatory relationships among genes. Genome Biol. 2007;8:R219. doi: 10.1186/gb-2007-8-10-r219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacKinnon DP, MacKinnon D. Introduction to Statistical Mediation Analysis. Routledge Academic; 2008. [Google Scholar]
- 24.Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23. doi: 10.1186/1471-2156-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Laan MJ, Dudoit S, Pollard KS. Multiple testing. Part II. Step-down procedures for control of the family-wise error rate. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1041. Article14. [DOI] [PubMed] [Google Scholar]
- 26.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1757–64. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology. 2000;61:154–66. doi: 10.1159/000028396. [DOI] [PubMed] [Google Scholar]
- 28.Strange RC, Jones PW, Fryer AA. Glutathione S-transferase: genetics and role in toxicology. Toxicol Lett. 2000:112–113. 357–63. doi: 10.1016/s0378-4274(99)00230-1. [DOI] [PubMed] [Google Scholar]
- 29.Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482:21–6. doi: 10.1016/s0027-5107(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 30.Bohanec Grabar P, Logar D, Tomsic M, Rozman B, Dolzan V. Genetic polymorphisms of glutathione S-transferases and disease activity of rheumatoid arthritis. Clin Exp Rheumatol. 2009;27:229–36. [PubMed] [Google Scholar]
- 31.Yun BR, El-Sohemy A, Cornelis MC, Bae SC. Glutathione S-transferase M1, T1, and P1 genotypes and rheumatoid arthritis. J Rheumatol. 2005;32:992–7. [PubMed] [Google Scholar]
- 32.Keenan BT, et al. Effect of interactions of glutathione S-transferase T1, M1, and P1 and HMOX1 gene promoter polymorphisms with heavy smoking on the risk of rheumatoid arthritis. Arthritis Rheum. 2010;62:3196–210. doi: 10.1002/art.27639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundstrom E, et al. Effects of GSTM1 in rheumatoid arthritis; results from the Swedish EIRA study. PLoS One. 2011;6:e17880. doi: 10.1371/journal.pone.0017880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao K, Chudin E, Greenawalt D, Schadt EE. Magnitude of stratification in human populations and impacts on genome wide association studies. PloS one. 2010;5:e8695. doi: 10.1371/journal.pone.0008695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–9. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–35. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang EY, Ye C, Shpitser I, Eskin E. Detecting the presence and absence of causal relationships between expression of yeast genes with very few samples. J Comput Biol. 2010;17:533–46. doi: 10.1089/cmb.2009.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaffe AE, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41:200–9. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thurlings RM, et al. Monocyte scintigraphy in rheumatoid arthritis: the dynamics of monocyte migration in immune-mediated inflammatory disease. PloS one. 2009;4:e7865. doi: 10.1371/journal.pone.0007865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller RB, et al. MHC class II expression on myeloid cells inversely correlates with disease progression in early rheumatoid arthritis. Rheumatology (Oxford) 2007;46:931–3. doi: 10.1093/rheumatology/kem025. [DOI] [PubMed] [Google Scholar]
- 42.Ronninger M, et al. The balance of expression of PTPN22 splice forms is significantly different in rheumatoid arthritis patients compared with controls. Genome Med. 2012;4:2. doi: 10.1186/gm301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casella G, Berger RL. Statistical Inference. Duxbury Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.