Abstract
Background
Ten generations of domestication selection has caused farmed Atlantic salmon Salmo salar L. to deviate from wild salmon in a range of traits. Each year hundreds of thousands of farmed salmon escape into the wild. Thus, interbreeding between farmed escapees and wild conspecifics represents a significant threat to the genetic integrity of wild salmon populations. In a previous study we demonstrated how domestication has inadvertently selected for reduced responsiveness to stress in farmed salmon. To complement that study, we have evaluated the expression of seven stress-related genes in head kidney of salmon of farmed, hybrid and wild origin exposed to environmentally induced stress.
Results
In general, the crowding stressor used to induce environmental stress did not have a strong impact on mRNA expression levels of the seven genes, except for insulin-like growth factor-1 (IGF-1) that was downregulated in the stress treatment relative to the control treatment. mRNA expression levels of glutathione reductase (GR), Cu/Zn superoxide dismutase (Cu/Zn SOD), Mn superoxide dismutase (Mn SOD), glutathione peroxidase (GP) and IGF-1 were affected by genetic origin, thus expressed significantly different between the salmon of farmed, hybrid or wild origin. A positive relationship was detected between body size of wild salmon and mRNA expression level of the IGF-1 gene, in both environments. No such relationship was observed for the hybrid or farmed salmon.
Conclusion
Farmed salmon in this study displayed significantly elevated mRNA levels of the IGF-1 gene relative to the wild salmon, in both treatments, while hybrids displayed a non additive pattern of inheritance. As IGF-1 mRNA levels are positively correlated to growth rate, the observed positive relationship between body size and IGF-1 mRNA levels detected in the wild but neither in the farmed nor the hybrid salmon, could indicate that growth selection has increased IGF-1 levels in farmed salmon to the extent that they may not be limiting growth rate.
Keywords: Atlantic salmon, Farmed escapees, Introgression, Hybrid, Common garden, mRNA level, Insulin-like growth factor −1, Oxidative stress, Non additive inheritance
Background
The commercial production of Atlantic salmon Salmo salar L. was established in Norway in the 1970's [1], and each year hundreds of thousands of farmed salmon escape into the wild [2], possibly exceeding the number of wild salmon in the natural habitat. Farmed escapees have been documented to enter freshwater, and in some rivers, in some years, represent more than 80% of the total number of spawners [3]. As a consequence, genetic introgression between farmed and wild conspecifics has been documented in several rivers [4-10]. Hence, farmed escaped Atlantic salmon represents one of the largest threats to the genetic integrity of wild salmon populations.
Both directional and inadvertent selective breeding causes farmed salmon to deviate from wild populations in a range of traits, e.g., body size [11-13], body proportions [14], fat reserves [15], time of sexual maturation [16], survival [17], aggressiveness [14,18,19], predator awareness [20], neutral genetic markers [21,22], allele frequencies [23] and gene expressions [24-26]. By comparing the growth reaction norms of farmed, hybrid and wild salmon exposed to an environmentally induced stressor, we have recently demonstrated how domestication selection over approximately ten generations has inadvertently selected for reduced responsiveness to stress in farmed Atlantic salmon [27]. A number of genes have been associated with stress in salmonids and here we have evaluated the expression of seven commonly studied genes in Atlantic salmon, in salmon of farmed, hybrid and wild origin exposed to environmentally induced stress.
Five of the genes investigated in the present study are known to be regulated by oxidative stress [28,29]. These are the four antioxidant genes, glutathione reductase (GR), Cu/Zn superoxide dismutase (Cu/Zn SOD), Mn superoxide dismutase (Mn SOD), glutathione peroxidase (GP) and the heat-shock protein 70 (HSP70). Insulin-like growth factor-1 (IGF-1), a protein important in the regulation of most physiological processes in fish, including somatic growth and metabolism, is downregulated by starvation and nutritional stress and activates the insulin-like growth factor-1 receptor (IGF-1R) [30-33]. The reference gene, eukaryotic elongation factor 1 alpha (EF1AA) is involved in protein synthesis and has been thoroughly validated as a reliable reference gene in quantitative real time PCR examination of gene expressions in Atlantic salmon [34,35], as well as in a broad range of other organism, e.g., plants [36], copepods [37], fish [38] and humans [39].
A total of 29 families were mixed together in a common garden experiment, exposed to standard hatchery conditions or in addition environmentally induced stress, i.e., reduction of water level, twice a day for 30 minutes. Thus, our objectives were to determine the effect of environmentally induced stress upon regulation of the selected genes and further examine whether the process of domestication has caused alterations in the mRNA expression levels. Based upon the results from our previous growth study, documenting reduced responsiveness to stress in the farmed salmon studied here, we hypothesised that the farmed salmon would display attenuated regulations of the genes investigated in this study in comparison to their wild counterparts. Although the crowding stressor used in this study did not inflict a strong regulation in the mRNA expression level of the genes studier here, except for the IGF-1 gene that was downregulated in the stress treatment, genetic origin had an impact on expression of five of the genes. Here we report of significant differences in mRNA levels of GR, Cu/Zn SOD, Mn SOD, GP and IGF-1 between farmed, hybrid or wild Atlantic salmon. In the wild salmon a positive relationship was detected between IGF-1 mRNA levels and body size, in both treatments, while no relationship was detected in the hybrid and farmed salmon where IGF-1 levels were significantly elevated.
Methods
Ten pure wild Atlantic salmon families, ten pure farmed families and nine F1 hybrid families were generated for this experiment in November 2009. Farmed parental salmon originated from the Norwegian Mowi strain, while wild parental salmon were caught by rod in the river Etne (59°40´N, 5°56´E). Hybrids were created by crossing farmed females with wild males. All families were created in the hatchery, located on the river Etne. Fertilized eggs (50 eggs/family) were mixed in four replicated tanks (n = 1450), and transported to Matre Research Station at the eyed-egg stage before hatching.
Two tanks were reared under standard hatchery conditions throughout the entire experiment running from June 3 - September 23–24, 2010. The two remaining tanks were subjected to a stressor, twice a day five days a week, in the same period. Stress was induced by a dramatic lowering of the water level for 30 minutes, hence the fish density increased although water circulation was maintained. Panic behaviour was observed as rapid movement within the tank. A stop watch was initiated when the water level was stabilized at the reduced level (3 cm). Water level during the stress treatments was adjusted throughout the experimental period in order to control for the increasing size of the fish during the experiment (5 cm depth at the time of termination). In all other aspects, the two treatments were given identical conditions throughout the experiment. These two treatments we hereon refer to as the control and stress treatments.
The experimental protocol (permit number 2648) was approved May 3, 2010, by the Norwegian Animal Research Authority (NARA).
Sampling
The experiment was terminated after 16 weeks of treatment. Two weeks prior to termination (i.e., week 14), 750 individuals had been removed from each of the four tanks for phenotypic growth comparisons [27]. The treatments (i.e., stress and control) were maintained in weeks 14–16. At the time of terminal sampling in week 16, there were 700 individuals within each tank, minus mortality (124, 125, 77 and 105 individuals, from hatching throughout the experimental period, in tank 1, 2, 3 and 4 respectively). The terminal sample consisted of removing at random 75 individuals from each tank. This was conducted over a period of 2 days.
On the first day of the terminal sampling, a control treatment tank was sampled first, followed by a stress treatment tank, and vice versa the second day. All sampled individuals were euthanized with an overdose of benzocain (160 mg/L) (Benzoak® Vet, A.C.D Pharmaceuticals, Leknes, Norway) in a combination with metodmidat hydrochloride (10 mg/L) (Aquacalm® Vet, ScanVacc, Årnes, Norway), to inhibit the acute cortisol stress response [40]. The concentrated euthanizing agents were added to a mixture of water and ice (7:3), and the individuals were left in the solution for a maximum of 27 minutes. 25 individuals were sampled at once, leaving the experimental tanks subject to only three strokes by the landing net, 3 sampling periods, and 1 h from the first to the last stroke. Fork length and weight were measured, before the individuals were caudal fin clipped and head kidney was sampled. Fins were placed on 95% ethanol, and samples for qPCR analyses were preserved on RNAlater™. To allow the RNAlater™ to protrude into the biological tissue, the samples were stored at < 4°C for 24 h, before being transferred to −20°C.
Microsatellite genotyping and parentage testing
280 of the individuals as sampled above were assigned to family using DNA microsatellite markers (70 individuals randomly selected per tank). Following procedures recommended by the manufacturer, DNA was extracted in 96 well plates using a Qiagen DNeasy®96 Blood & Tissue Kit. To ensure correct genotyping, parental DNA was extracted twice. On each 96-well plate, two randomly assigned blank wells were included, thus to ensure a unique identification of the plate. Six microsatellite loci were amplified in one multiplex PCR; SsaF43 [GenBank: U37494] [41], Ssa197 [GenBank: U43694.1] [42], SSsp3016 [GenBank: AY372820], MHCI[43], MHCII[44] and SsOSL85 [GenBank: Z48596.1] [45]. PCR products were run on an ABI Applied Biosystems ABI 3730 Genetic Analyser and sized-called according to the 500LIZ™ standard. Genotypes were identified using GeneMapper V4.0., with manual control of scored alleles. Assignment to family were performed by FAP Family Assignment Program v3.6 [46], using an exclusion-based approach to unambiguously identify parental origin. This program has successfully been used on several occasions for parentage testing common garden studies using these facilities [47,48]. The genetic markers analysed here have revealed very low genotyping errors in this laboratory [49] and are routinely used in association with a genotyping service for the Norwegian legal authorities to identify the farm of origin for escapees [50,51].
After DNA identification, 15 farmed, hybrid and wild individuals, respectively, within each tank were selected for the gene expression profiling. Individuals were selected by family, representing all 29 families if possible and in an even number (range 0 – 3 fish per family per tank). Choice of individuals within families on which to conduct qPCR was first based upon sampling period, then upon time in the euthanizing solution. Individuals from sampling period 1 were preferred over individuals from the subsequent periods, and within each sampling period individuals with the fewest minutes in the euthanizing solution were selected first (range 1–27 minutes). The 100 excess individuals were excluded from any further studies, thus leaving the total data set consisting of 180 individuals (45 individuals per tank).
RNA extraction
Total RNA was extracted in situ from the macrodissected head kidney samples. The 180 selected individuals were randomized into 15 batches and isolated over a period of 3 days. Up to 50 mg tissue was homogenized in 1 mL TRIzol using a FastPrep homogenisator (Thermo Electron) and Lysing Matrix D ceramic beads (MP Biomedical). Following homogenization, 400 μL chloroform was added and the sample vortexed for 1 min, phase separated by centrifuge and the aqueous phase were collected using iPrep™ Purification Instrument (Invitrogen) with the iPrep™ TRIzol ® Plus RNA Kits, according to the manufacturers protocol. The RNA was eluted in 50 μL. Quantity of the isolated RNA was assessed by Nanodrop® spectrophometer (NanoDrop Technologies, Wilmington, DE). 260/280 absorbance ratio ranged from 1.61 – 2.16 with a mean average value of 2.03, while the 260/230 absorbance ratio ranged from 1.86 – 2.46 with a mean average value of 2.29. From each isolation batch minimum three samples were randomly selected, 48 samples in total, and the RNA integrity was evaluated by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA), using RNA 6000 Nano LabChip® (Agilent Technologies, Palo Alto, CA). With a mean RNA integrity number (RIN) of 9.5 (range 8.0 – 10.0), no samples showed any sign of RNA degradation. Total RNA samples were randomized again before normalized with distilled water (dH2O) to a final concentration of 100 ng/μL and stored in 2 x 96 Well Plates at - 80°C.
cDNA synthesis
For each sample cDNA synthesis were carried out in triplicate from 200 ng total RNA in 10 μL reaction volume using qScript™ cDNA Synthesis Kit (Quanta, Biosciences) in accordance to suppliers protocols. cDNA was subsequent diluted 1:10 in dH2O and stored at - 20°C.
Twenty-eight samples were distributed in triplicate on each cDNA 96 Well Plate. Thus, 16 samples were included a second time on one of the plates to secure full plates at all times. Negative Reverse Transcriptase Controls (nRT, a minus enzyme control) to control for genomic DNA contamination, was made from 200 ng total RNA from the first and the last sample on each RNA tray, and from two random samples in the middle of the RNA tray. nRTs were made from the qScript™ cDNA Synthesis Kit (Quanta, Biosciences) and contained all the reaction components except the reverse transcriptase enzyme, and were diluted and stored in the same manner as the cDNA.
Positive control (PK) was made by a mix of total RNA from all 180 samples. For this purpose 100 ng RNA per sample, in 5 μL reaction volume, were converted into cDNA in the same manner as described above, then all samples were mixed together as one PK. The Positive Control was diluted 1:20 in dH2O and stored at - 20°C.
Genes and primers
Quantitative PCR primers and probes for the genes to be analyzed were obtained from published literature of earlier gene expression studies in Atlantic salmon [34,52-55]. The chosen target genes were heat shock protein 70 HSP70 [GenBank: BG933934] [52], glutathione reductase GR [GenBank: BG934480] [52], Cu/Zn superoxide dismutase Cu/Zn SOD [GenBank: BG936553] [55], Mn superoxide dismutase Mn SOD [GenBank: DY718412] [53], glutathione peroxidase GP [GenBank: BG934453] [55], insulin-like growth factor-1 IGF-1 [GenBank: M81904][54] and the insulin-like growth factor-1 receptor IGF-1R [GenBank: AY049954] [54]. Normalization of target genes was performed against the reference gene eukaryotic elongation factor 1 alpha, EF1AA [GenBank: AF321836] [34]. This gene has been documented to be one of the most reliable reference genes in Atlantic salmon [34,35] and is often used as the sole reference gene in qPCR examination of gene expressions in this species. In our study amount of total RNA was equalized between samples prior to cDNA synthesis, which allowed us to statistically demonstrate that this gene was stable between all three genetic origins and between treatments (see Results). The qPCR primers and hydrolysis probe sequences are presented in Table 1.
Table 1.
Primer and probe sequences for qPCR used in the present study
| Gene | GenBank # | Primer forward 5'-3' | Primer reverse 5'-3' | Hydrolysis probe 5'-3' | Amplicon size (bp)* | Reference |
|---|---|---|---|---|---|---|
|
EF1AA |
AF321836 |
CCCCTCCAGGACGTTTACAAA |
CACACGGCCCACAGGTACA |
6-FAM-ATCGGTGGTATTGGAAC-MGB |
57 |
[34] |
|
HSP70 |
BG933934 |
CCCCTGTCCCTGGGTATTG |
CACCAGGCTGGTTGTCTGAGT |
6-FAM-CGCTGGAGGTGTCATG-MGB |
121 |
[52] |
|
GR |
BG934480 |
CCAGTGATGGCTTTTTTGAACTT |
CCGGCCCCCACTATGAC |
6-FAM-AGCCTTCCTAAGCGCAG-MGB |
61 |
[52] |
|
Cu/Zn SOD |
BG936553 |
CCACGTCCATGCCTTTGG |
TCAGCTGCTGCAGTCACGTT |
6-FAM-ACAACACCAACGGCT-MGB |
140 |
[55] |
|
Mn SOD |
DY718412 |
GTTTCTCTCCAGCCTGCTCTAAG |
CCGCTCTCCTTGTCGAAGC |
6-FAM-CACATCAACCACACCATCTTCTGGACAAAC-TAMRA |
209 |
[53] |
|
GP |
BG934453 |
GATTCGTTCCAAACTTCCTGCTA |
GCTCCCAGAACAGCCTGTTG |
6-FAM-TGAATGGAGACACAGAAC-MGB |
140 |
[55] |
|
IGF-1 |
M81904 |
GTGTGCGGAGAGAGAGGCTTT |
TGTGACCGCCGTGAACTG |
6-FAM-TTTCAGTAAACCAACGGGCTATGG-TAMRA |
68 |
[54] |
| IGF-1R | AY049954 | TGAAGAGCCACCTGAGGTCACT | TCAGAGGTGGGAGGTTGAGACT | 6-FAM-CGGGCTAAAGACCCGTCCCAGTCC-TAMRA | 72 | [54] |
*bp = base pair.
Quantitative real-time PCR (qPCR)
qPCR was performed in triplicates in 14 × 384 Well Plates on ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) in 5 μL reaction volume with 1.5 μL cDNA template and Briliant III Ultra-Fast QPCR Master Mix. Primers and probes had a final concentration of 900 μM and 200 μM, respectively. A passive reference dye, ROX™, was included in the reaction mix. On each 384 Well Plate all 8 genes were run with 14 samples in triplicate. For each plate one No Template Control (NTC), two different nRTs and three PKs were included for each gene. NTCs contained all reaction components besides template (cDNA substituted by dH2O) and were added to monitor possible PCR contaminations and primer dimer formations. All genes had previously been validated, thus their efficiency documented to be approximately the same [34,52-55].
Quantification cycle values (Cq) were obtained from the qPCR instrument using SDS (2.4) and RQ Manager (1.2.1) (Applied Biosystems). Baseline and threshold for Cq values were set manually for each gene and kept identical for all plates. One Cq equals a doubling (2^Cq) of the mRNA level.
The experiment was performed in accordance to the general guidelines for qPCR experiments, Minimum Information for Publication of Quantitative Real-Time PCR Experiments “MIQE” [56,57].
Statistical analysis
The comparative quantification cycle (Cq) method [58], were used to analyse the relative gene expression of the target genes. The median ΔCq value of the wild salmon in the control treatment was used as calibrator when calculating ΔΔCq values. ΔΔCq values were multiplied by −1, so that upregulated mRNA levels were displayed as positive values, while downregulated mRNA levels were displayed as negative values.
Quantification cycle values, Cq, were quality checked, first by manually removing non-amplified samples, samples displaying extreme Cq values (Cq <15 and > 39) and aberrant Cq values caused by documented sampling errors. Outliers defined as values more than 1.5 times the interquartile range (IQR) above the 3rd quartile and below the 1st quartile [59] were excluded from the data set. Possible outliers were identified based upon several calculated interquartile ranges; IQR of the Cq values of each target gene and the reference gene, IQR of the standard deviation (SD) of the Cq values and IQR of the Cq, ΔCq and the ΔΔCq values, of each target gene in each treatment. Samples had to pass all the selected criterions to be included in the statistical analysis. For passed samples, the median of the three replicates were used as the sample’s Cq value. For the 16 samples that were run twice, on two different plates, the mean of the two collapsed triplicates were used as the samples Cq value.
Linear mixed effect models (LME), testing for difference in continuous response variables, were used to model variation in weight at termination and mRNA expression levels between treatments and genetic origin. LMEs were fitted for Cq values of the reference gene and ΔΔCq values of the target genes. Model selection was performed based upon Akaike information criterion (AIC) values, calculated using the restricted maximum likelihood (REML) [60]. Models displaying less than 2 AIC values in distance were considered equally good. Thus, by the principle of parsimony, the simplest model that performed best was applied. The initial model fitted for weight included treatment and type as fixed effects, as well as the interaction between them. By forward selection the random effect of tank nested within treatment, as well as a genetic (co)variance matrix across treatments were incorporated if this improved the fit of the model. Due to differences in growth rate between salmon of farmed, hybrid and wild origin and to achieve normality, the response variable, weight at termination, was log transformed (log10) [61-63]. A similar model with treatment and type as fixed effects (and the interaction between them) were fitted for the expression of each of the eight genes (Cq and ΔΔCq values). By forward selection the random effect of tank nested within treatment, a genetic (co)variance matrix, log-weight of fish, sampling period, and minutes in anaesthesia were incorporated if this improved the fit of the model. To satisfy homogeneity and normality in the model, Cq and ΔΔCq values were log transformed. Prior to transformation, ΔΔCq values were added a constant so that all values were above 1. For AIC comparisons of LME models, see Additional file 1. Gene expression in farmed versus hybrid salmon, hybrid versus wild salmon and wild versus farmed salmon were compared by re-running the models while excluding one of the three genetic origins at a time. For the re-runs, multiple comparisons were counteracted by the Bonferroni correction, giving an adjusted significance level of P < 0.017. P-values are given from F-statistics of the simplest model. Numerator degrees of freedom were given as k – 1, where k is the number of factor levels. Denominator degrees of freedom were calculated as N – k, where N was set to the smallest sample size detected in any of the three genetic origins in any of the two treatments. Linear regressions between ΔΔCq values on the y-axis, and weight of fish (g), sampling period (1–3) and time in anaesthesia (minutes) on the x-axis, were performed with a 95% confidence interval. The goodness of fit of the linear regression was validated by the R-square values and by the P-values of the slopes. As we measured 7 genes, multiple comparisons was counteracted by the Bonferroni correction, which in this case gave an adjusted P-value of 0.007.
All statistical analysis was performed using R ver. 2.15.2 [64] with critical P-values set to 0.05, unless otherwise stated. LME’s were fitted using the lmer function in the lme4 package [65].
Results
Growth of experimental fish
The mean weight, length and condition factor of farmed, hybrid and wild salmon in all four tanks is shown in Table 2. Salmon in the control treatment were significantly larger than salmon in the stress treatment and farmed salmon were significantly larger than the hybrid and wild salmon, in both treatments (Table 3; Additional file 2). At the time of sampling the effect of the stress treatment was similar in all groups, as they displayed similar growth reaction norm slopes (Figure 1, solid lines; Table 2), thus the interaction between treatment and type were not included in the final LME model (Table 3).
Table 2.
Growth measurements of Salmo salar L. of wild, hybrid and farmed origin
| Group | Treatment | Tank |
Measurements at termination (week 16) |
Weight difference |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean W (g) | ± SD | Mean L (cm) | ± SD | Mean K | ± SD | n | Mean W (g) | ± SD | Absolute (g) | Percent (%) | |||
|
Wild |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Control |
1 |
15 |
21.07 |
9.58 |
11.63 |
2.08 |
1.28 |
0.05 |
30 |
18.6 |
8.8 |
4.3 |
23.12 |
|
| 2 |
15 |
16.13 |
7.44 |
10.71 |
1.76 |
1.27 |
0.07 |
|||||||
| Stress |
3 |
15 |
14.47 |
6.77 |
10.34 |
1.74 |
1.24 |
0.05 |
30 |
14.3 |
6.48 |
|||
| 4 |
15 |
14.13 |
6.4 |
10.42 |
1.63 |
1.18 |
0.05 |
|||||||
|
Hybrid |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Control |
1 |
15 |
30.73 |
10.67 |
13.29 |
1.49 |
1.32 |
0.1 |
30 |
30.83 |
9.81 |
8.23 |
26.69 |
|
| 2 |
15 |
30.93 |
9.24 |
13.65 |
0.6 |
1.24 |
0.32 |
|||||||
| Stress |
3 |
15 |
22.8 |
5.82 |
12.23 |
1.15 |
1.23 |
0.06 |
30 |
22.6 |
5.97 |
|||
| 4 |
15 |
22.4 |
6.35 |
12.13 |
1.23 |
1.24 |
0.04 |
|||||||
| Farm | |
|
|
|
|
|
|
|
|
|
|
|
|
|
| Control |
1 |
15 |
46.47 |
8.21 |
15.22 |
0.9 |
1.31 |
0.04 |
30 |
47.57 |
9.58 |
11.9 | 25.02 | |
| 2 |
15 |
48.67 |
10.97 |
15.47 |
1.18 |
1.29 |
0.06 |
|||||||
| Stress | 3 |
15 |
33.67 |
3.96 |
13.93 |
0.5 |
1.28 |
0.05 |
30 | 35.67 | 5.74 | |||
| 4 | 15 | 37.67 | 6.64 | 14.32 | 0.82 | 1.29 | 0.04 | |||||||
Weight (gram), length (cm) and condition factor (K) with standard deviations. Differences in weight between treatments; Absolute (grams); Percent (percent reduction in weight in the stressed environment, compared to in the control environment).
Table 3.
The effect of treatment, type and their interaction on fish weight and gene expressions
| Salmo salar L. | n |
Effects |
||||||
|---|---|---|---|---|---|---|---|---|
| Fixed | Random | DFn | DFd | Sum Sq | F | P | ||
| Weight [log] |
180 |
Treatment |
Treatment:Tank |
1 |
28 |
0.62 |
23.19 |
<0.0001*** |
| |
|
Type |
|
2 |
27 |
6.06 |
113.11 |
<0.0001*** |
| Gene |
n |
|
|
DFn |
DFd |
|
F |
P |
|
EF1A A |
177 |
Type |
Treatment:Tank |
2 |
26 |
0.0002 |
1.17 |
0.3 |
|
HSP70 |
173 |
Treatment |
Treatment:Tank |
1 |
26 |
0.0017 |
0.72 |
0.3 |
|
GR |
173 |
Type |
Treatment:Tank |
2 |
25 |
0.08 |
7.92 |
0.001*** |
|
Cu/Zn SOD |
166 |
Type |
Treatment:Tank |
2 |
24 |
0.06 |
3.49 |
0.04* |
|
Mn SOD |
170 |
Type |
Weight [log] |
2 |
24 |
0.03 |
5.42 |
0.01** |
|
GP |
172 |
Type |
Weight [log] |
2 |
24 |
0.07 |
6.79 |
0.003** |
|
IGF-1 |
168 |
Treatment |
Weight [log] |
1 |
25 |
0.09 |
13.57 |
<0.001*** |
| |
|
Type |
|
2 |
24 |
0.15 |
10.97 |
<0.001*** |
| IGF-1R | 173 | Type | Treatment:Tank | 2 | 23 | 0.01 | 1.37 | 0.24 |
Summary of the best linear mixed effect models testing for difference in fish weight, and mRNA levels (raw Cq values of EF1AA, ΔΔCq values of target genes) between treatments and genetic origins (type). Including the interaction between treatment and type did not improve the fit of any of the models, thus all groups were affected by treatment in the same matter. The random effects were included by forward selection. Treatment:Tank; tank nested within treatment, Weight [log]; log transformed weight measurement of the fish. For more information on model selection based upon AIC values, see Additional file 1. The statistical significance is marked with asterisks, where * = P ≤ 0.05, ** = P ≤ 0.01 and *** = P ≤ 0.001.
Figure 1.
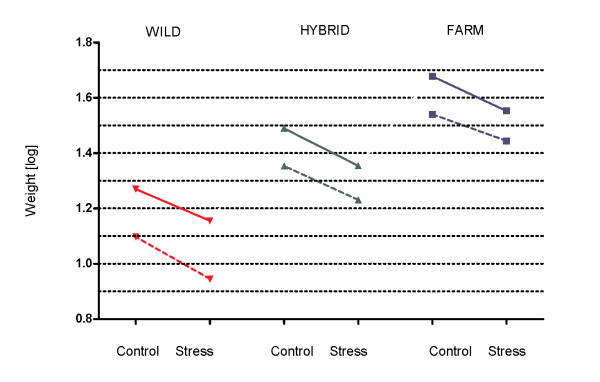
Growth reaction norms of salmon of farmed, hybrid and wild origin. Reaction norms for the log transformed weight measurements of Atlantic salmon of wild , hybrid and farmed origin reared in the control treatment and the stress treatment at week 14 (dotted line) of the experimental period and at termination, week 16 (solid line). Replicated tanks are pooled. The within-tank biomasses were dramatically reduced at week 14, thus the dotted (−−) and the solid (__) lines display the reaction norm before and after the reduction, respectively.
Quality of qPCR
The expression of the reference gene EF1AA (raw Cq values) was stable between farmed, hybrid and wild salmon in both treatments (Table 3). The average mean Cq value of samples before and after quality check, displayed a minor deviation of 0.02, while the median was identical (Table 4). Collapsing samples where all triplicates passed the selected quality criterions resulted in 98% of the reference gene samples and 95% of the target gene samples being included in the statistical analysis.
Table 4.
Variation of raw EF1AA Cq values
| Quality checked | n | Median | ± SD | Mean | Min | 1st Qu | 3rd Qu | Max | NA |
|---|---|---|---|---|---|---|---|---|---|
| No |
576 |
22.13 |
0.53 |
22.19 |
20.90 |
21.85 |
22.48 |
26.16 |
12* |
| Yes | 177 | 22.13 | 0.47 | 22.17 | 21.04 | 21.89 | 22.43 | 23.92 | 3 |
* 6 runs not amplified (2 individuals), 6 runs removed due to a sampling error (1 individual). Triplicates collapsed after quality check. 1st Qu; lower quartile (25 percentile), 3rd Qu; upper quartile (75 percentile).
For gene HSP 70, 64% of the negative Reverse Transcriptase Controls (nRTs) turned out positive due to amplification of genomic DNA. The HSP70 assay is based upon an EST sequence of the gene, hence no information on the exon-exon junctions was used in assay design, which likely explains these results. However, this did not impose a problem in analysing the data, as the difference between the average Cq value for positive nRTs and the samples average Cq value were larger than 9. Ignoring HSP70, nRTs were negative in 98% of the cases, and the difference between average Cq value for positive nRTs and the samples average Cq value were larger than 10 in all positive nRTs (10, 10, 10 and 12, respectively). Positive controls PK’s were amplified in > 99% of the controls, while No Template Controls NTC’s turned out non-amplified in > 97% of the controls. The NTCs that were amplified displayed a Cq value of +10, + 13 and +14, when compared to the adjoining samples average Cq value.
The effect of treatment and tank
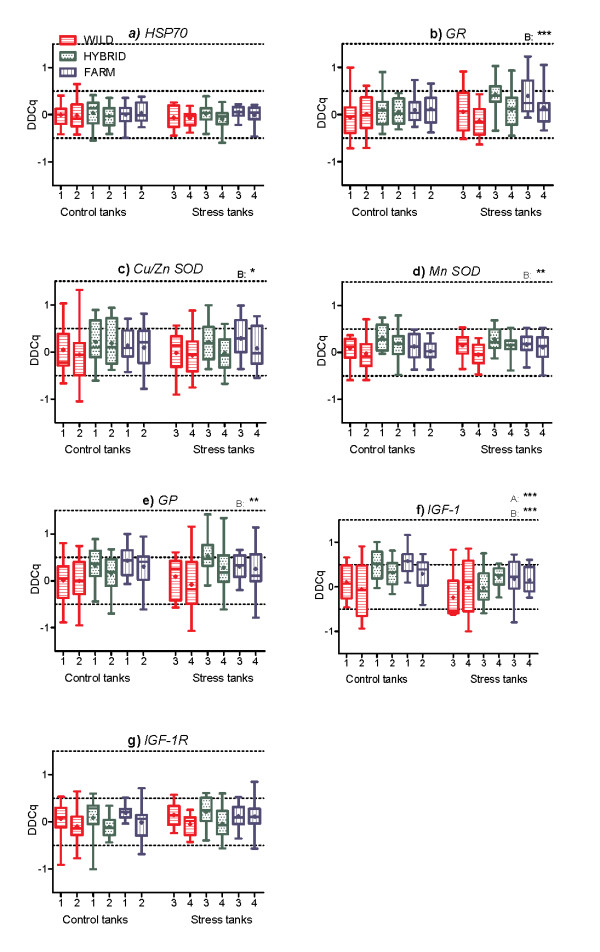
Some tank effects were detected (Figure 2a-c, g). Statistically, this was controlled for by including the random effect of tank nested within treatment in the linear mixed effect models, which significantly improved the fit of the models (Table 3). With the exception of the IGF-1 gene, that was downregulated in the stress treatment, the genes investigated in this study were not upregulated, nor downregulated by the environmentally induced stress (Table 3). Thus, treatment, i.e., control/stress, did not have a significant effect upon mRNA expression levels, except for the IGF-1 gene where the median ΔΔCq value displayed in the control treatment were downregulated by −0.23 in the stress treatment (Figure 2f; Additional file 3).
Figure 2.
qPCR analyses of the seven selected genes in salmon of farmed, hybrid and wild origin. Expressions of a) HSP70, b) GR, c) Cu/Zn SOD, d) Mn SOD, e) GP, f) IGF-1 and g) IGF-1R in Atlantic salmon of wild, hybrid and farmed origin, reared in a standard hatchery environment (tank 1 and 2) or in addition exposed to environmentally induced stress (tank 3 and 4). One quantification cycle (Cq) equals a doubling of the amount of mRNA (2^Cq). All values are relative to the wild salmon in control treatment (pooled), and ΔΔCq values with positive and negative values indicate upregulated and downregulated mRNA levels, respectively. Boxes show the median (thick line), mean (+), 1st and 3rd quartiles (lower and upper boundary) and the lower and upper extreme (whiskers). The statistical significance of the effect of treatment (A) and type (B) on gene expressions is marked with asterisks, where * = P ≤ 0.05, ** = P ≤ 0.01 and *** = P ≤ 0.001.
The effect of genetic origin (farm/hybrid/wild)
In five of the genes, GR, Cu/Zn SOD, Mn SOD, GP and IGF-1, mRNA expression levels were significantly different between the genetic origins (Table 3, Figure 2b-f). Thus, mRNA expression levels of HSP70 and IGF-1R were not significantly different between salmon of farmed, hybrid and wild origin (Table 3, Figure 2a, g).
Farmed salmon displayed elevated mRNA expression levels of GR, CuZn SOD, GP and IGF-1, relative to the wild salmon (Table 5, Additional files 2 and 3). Mn SOD was expressed insignificantly different in the farmed and wild salmon (Table 5, Additional files 2 and 3).
Table 5.
Median ΔΔCq values of the seven genes in farmed and hybrid-, relative to wild salmon
| Group | HSP70 | GR*** | Cu/ ZnSOD * | MnSOD** | GP** | IGF- 1 *** | IGF-1R | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild |
0 |
(a,a) |
0 |
(a,a) |
0 |
(a,a) |
0 |
(a,a) |
0 |
(a,a) |
0 |
(a,a) |
0 |
(a,a) |
| Hybrid |
0.01 |
(a,a) |
0.27 |
(b,b) |
0.13 |
(b,ab) |
0.17 |
(b,b) |
0.32 |
(b,b) |
0.32 |
(b,b) |
0.00 |
(a,a) |
| Farm | 0.03 | (a,a) | 0.18. | (b,b) | 0.15 | (b,b) | 0.09 | (a,a) | 0.35 | (b,b) | 0.36 | (b,b) | 0.07 | (a,a) |
Upregulated mRNA levels are displayed as positive values (multiplied by −1). The statistical significance of the effect of genetic origin on gene expressions, in general, is marked with asterisks, where * = P ≤ 0.05, ** = P ≤ 0.01 and *** = P ≤ 0.001. Letters in brackets represent statistical significance among the three origins, the first with the significance level set at P ≤ 0.05 and the second with the Bonferroni correction for multiple comparisons, P ≤ 0.017.
For the hybrid salmon, mRNA levels of the GR, Cu/Zn SOD, GP and IGF- 1 gene were similar to the mRNA levels detected in the farmed salmon (Table 5, Additional files 2 and 3). When the significance levels were adjusted for multiple comparisons, Cu/Zn SOD was in addition expressed insignificantly different between hybrid and wild salmon, P = 0.023 (Table 5, Additional files 2 and 3). For Mn SOD, hybrids displayed elevated mRNA levels, in comparison to both farmed and wild salmon (Table 5, Additional files 2 and 3). Thus, hybrids displayed three out of five genes similar to farmed salmon, one gene intermediate to farmed and wild salmon, and one gene significantly elevated compared to both farmed and wild salmon (Table 5).
The influence of fish size and sampling
The relationship between mRNA expression levels and fish size, sampling period, and time in anesthesia, for all three groups in both treatments, is shown in Table 6. The influence of fish size was significant for four genes in the control treatment, and for three genes in the stress treatment. Interestingly, these trends were only observed in the wild fish, and all relationships between gene expression and fish size were positive (Table 6; Additional file 4).
Table 6.
Linear regression between gene expression and fish weight, sampling period and time in anaesthesia
|
HSP70 |
GR |
Cu/Zn SOD |
Mn SOD |
GP |
IGF-1 |
IGF-1R |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R square | P | R square | P | R square | P | R square | P | R square | P | R square | P | R square | P | |
| Weight |
||||||||||||||
| Control |
||||||||||||||
| Wild |
0.046 |
0.265 |
0.046 |
0.254 |
0.250 |
0.005 ** |
0.290 |
0.002 ** |
0.154 |
0.036 * |
0.320 |
0.002 ** |
0.000 |
0.917 |
| Hybrid |
0.018 |
0.492 |
0.028 |
0.393 |
0.011 |
0.601 |
0.023 |
0.440 |
0.014 |
0.547 |
0.038 |
0.311 |
0.037 |
0.306 |
| Farm |
0.002 |
0.798 |
0.066 |
0.178 |
0.021 |
0.463 |
0.015 |
0.533 |
0.007 |
0.663 |
0.023 |
0.442 |
0.062 |
0.201 |
| Stress |
||||||||||||||
| Wild |
0.095 |
0.111 |
0.161 |
0.034 * |
0.002 |
0.809 |
0.004 |
0.746 |
0.190 |
0.023 * |
0.324 |
0.001 ** |
0.012 |
0.602 |
| Hybrid |
0.060 |
0.192 |
0.029 |
0.379 |
0.017 |
0.519 |
0.018 |
0.500 |
0.090 |
0.108 |
0.034 |
0.360 |
0.058 |
0.208 |
| Farm |
0.007 |
0.671 |
0.002 |
0.821 |
0.028 |
0.405 |
0.027 |
0.407 |
0.001 |
0.898 |
0.000 |
0.986 |
0.010 |
0.607 |
| Period |
||||||||||||||
| Control |
||||||||||||||
| Wild |
0.015 |
0.524 |
0.039 |
0.293 |
0.022 |
0.438 |
0.002 |
0.836 |
0.049 |
0.250 |
0.005 |
0.731 |
0.010 |
0.607 |
| Hybrid |
0.090 |
0.120 |
0.018 |
0.498 |
0.031 |
0.383 |
0.002 |
0.814 |
0.022 |
0.448 |
0.067 |
0.176 |
0.022 |
0.439 |
| Farm |
0.178 |
0.023 * |
0.021 |
0.455 |
0.007 |
0.678 |
0.033 |
0.343 |
0.037 |
0.325 |
0.053 |
0.238 |
0.070 |
0.173 |
| Stress |
||||||||||||||
| Wild |
0.033 |
0.356 |
0.019 |
0.488 |
0.020 |
0.483 |
0.000 |
0.951 |
0.053 |
0.249 |
0.047 |
0.257 |
0.000 |
0.993 |
| Hybrid |
0.057 |
0.204 |
0.058 |
0.209 |
0.001 |
0.862 |
0.000 |
0.961 |
0.001 |
0.847 |
0.036 |
0.346 |
0.139 |
0.047* |
| Farm |
0.070 |
0.165 |
0.006 |
0.678 |
0.054 |
0.244 |
0.023 |
0.439 |
0.009 |
0.634 |
0.028 |
0.407 |
0.012 |
0.566 |
| Minutes |
||||||||||||||
| Control |
||||||||||||||
| Wild |
0.217 |
0.011 * |
0.011 |
0.587 |
0.051 |
0.231 |
0.083 |
0.123 |
0.113 |
0.074 |
0.050 |
0.252 |
0.130 |
0.050 * |
| Hybrid |
0.001 |
0.859 |
0.006 |
0.695 |
0.002 |
0.836 |
0.010 |
0.614 |
0.032 |
0.354 |
0.008 |
0.643 |
0.005 |
0.721 |
| Farm |
0.007 |
0.666 |
0.002 |
0.806 |
0.007 |
0.668 |
0.056 |
0.215 |
0.005 |
0.714 |
0.158 |
0.036 * |
0.030 |
0.382 |
| Stress |
||||||||||||||
| Wild |
0.008 |
0.643 |
0.025 |
0.424 |
0.006 |
0.706 |
0.002 |
0.816 |
0.014 |
|
0.001 |
0.891 |
0.023 |
0.462 |
| Hybrid |
0.006 |
0.682 |
0.008 |
0.650 |
0.002 |
0.830 |
0.050 |
0.252 |
0.026 |
0.395 |
0.013 |
0.570 |
0.079 |
0.140 |
| Farm | 0.000 | 0.979 | 0.012 | 0.570 | 0.099 | 0.110 | 0.002 | 0.821 | 0.002 | 0.819 | 0.070 | 0.183 | 0.001 | 0.890 |
* P < 0.05; ** P < 0.007; Weight (gram); Period (1–3); Time (1–27 minutes).
A significant positive relationship was detected between mRNA expression levels and sampling period in two of the genes, although only for one group, in one treatment (Table 6; Additional file 5). The effect of time in anaesthesia was significant in three genes, although for all genes the negative relationship between gene expression and sampling period was only displayed in one group, and only in one of the treatments (Table 6; Additional file 6).
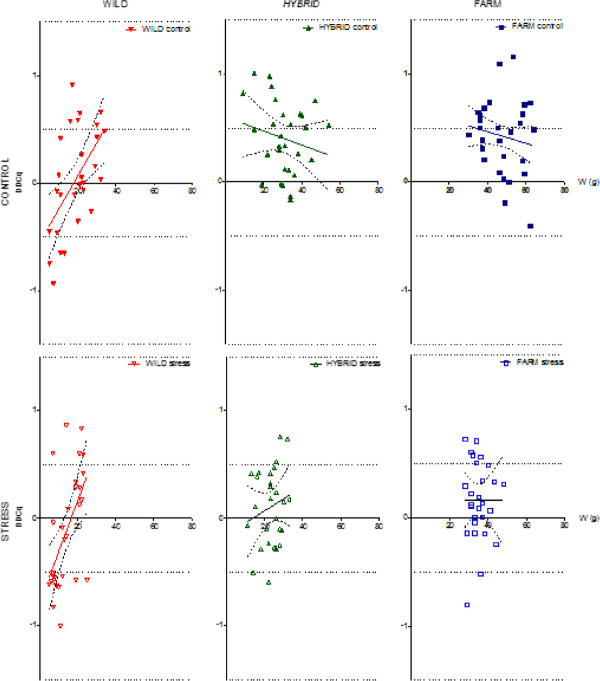
Overall, 9 out of the 12 significant regressions detected here, and all of the regressions detected between mRNA level and fish weight, were displayed in the wild salmon. When adjusted for multiple testing, the adjusted P-values displayed only significant relationships between the expression of three genes and fish size in the wild salmon. For Cu/Zn SOD and Mn SOD the significant positive relationship were only detected in the control treatment. However, the relationship between fish size and IGF-1 was significant for wild salmon in both treatments (r2 = 0.320 and 0.324, in the control and stress treatment, respectively) and as the significant regression between gene expression and fish size was positive, the largest wild salmon displayed the highest mRNA expression levels of the insulin-like growth factor-I IGF-1gene (Figure 3).
Figure 3.
Linear regressions between expression of the IGF-1 gene and fish weight in Atlantic salmon. ΔΔCq value of the IGF-1 gene plotted against fish weight, for salmon of wild, hybrid and farmed origin, in the control treatment and the stress treatment, replicated tanks are pooled. A significant positive relationship was detected in the wild salmon, in both treatments. The regression line is shown with a 95% confidence interval.
Discussion
Overall, no significant differences in mRNA expression of the seven genes investigated here were detected in salmon reared under standard hatchery conditions and salmon exposed to environmentally induced stress. One exception was detected in the insulin-like growth factor-1 (IGF-1), which was significantly different between treatments. As the mRNA expression level of the IGF-1 gene was downregulated in the stress treatment relative to the control treatment, as well as growth being lower, this indicates that nutritional stress [31,32,66-69], e.g., impaired feed intake, was induced in this study. The corresponding receptor IGF-1R was however, similarly expressed in both treatments. Expression of the four antioxidant genes, glutathione reductase (GR), Cu/Zn superoxide dismutase (Cu/Zn SOD), Mn superoxide dismutase (Mn SOD), glutathione peroxidase (GP), as well as the heat-shock protein 70 (HSP70) were similar among treatments, thus oxidative stress was not detected [28,29]. In general, the crowding stressor used to induce environmental stress upon salmon in this study had no clear effect upon mRNA expression levels of the genes studied here, and we were therefore not able to verify our hypothesis by evaluating the selected genes. However, despite little to no significant differences in mRNA expression levels between treatments there could still be regulations at the protein level. Further, as Atlantic salmon is partially tetraploid [70], differences in mRNA levels between treatments could potentially be masked if gene copies are regulated differently.
Significant differences in mRNA levels between farmed, hybrid and wild salmon were detected in the antioxidant genes GR, Cu/Zn SOD, Mn SOD, and GP, as well as in IGF-1. Thus, genetic origin of the salmon used in this study had an impact on the mRNA expression levels of five of the seven genes investigated. Here we discuss the genes where mRNA levels were affected by genetic origin, with a primary focus on the IGF-1 gene. IGF-1, in addition to being expressed significantly different between treatments and origins, also displayed a positive relationship to fish size in the wild salmon, in both treatments, while no such relationship were detected in the farmed and hybrid salmon where IGF-1 levels were significantly elevated.
Insulin- like growth factor −1
In salmonids, as well as in other fish species, positive correlations between feed ration and IGF-1 plasma levels [32,66-69,71], as well as IGF-1 mRNA levels in liver [33] and muscle [72,73], has been documented. As salmonids have been documented to reduce their feeding activity as a response to stress [14,19,74], the IGF-1 mRNA levels were expected to be downregulated in the stress treatment. In accordance to this expectation, the IGF-1 mRNA levels were downregulated in the stress contra the control treatment in this study. This indicates that feeding activity was suppressed in the salmon exposed to environmentally induced stress.
Head kidney IGF-1 mRNA levels were significantly elevated in the farmed and hybrid salmon relative to the wild salmon studied here, in both treatments, which was expected as there is documented a positive relationship between IGF-1 mRNA levels, as well as plasma levels, and growth rate in salmonids and other teleosts [54,66,69,71,75-77]. This indicates that growth-selection for approximately ten generations has not only resulted in increased growth rates in farmed salmon, but also elevated IGF-1 mRNA levels. Consistent with our results, elevated IGF-1 mRNA levels have been observed in domesticated relative to wild coho salmon Oncorhynchus kisutch, in both liver and muscle tissue [78,79]. Plasma IGF-1 levels were also elevated in the domesticated relative to the wild coho salmon [79], which also has been documented in other salmonids, i.e., rainbow trout Oncorhynchus mykiss[80]. In contrast to these studies, no differences in plasma IGF-1 levels [81,82], nor IGF-1 mRNA levels in liver, muscle or gill [81] of farmed and wild Atlantic salmon has been detected. The difference in the results between the study conducted by Neregard and colleagues [81] and the present study could be caused by tissue specific differences in regulation of IGF-1 mRNA levels [83,84]. However, in their study [81], mRNA levels, as well as plasma IGF-1 levels, were measured in Atlantic salmon sampled at cold temperatures < 5°C. As plasma IGF-1 levels have been documented to decline with decreasing temperatures [67,75,85], the relatively low plasma IGF-1 levels detected in their study could make variations among the strains harder to detect. This may also explain the differences between their study and other studies documenting differing IGF-1 mRNA levels in domesticated and wild salmonids.
Plasma IGF-1 levels have been documented to be correlated with body size [80]. However, the relationship between IGF-1 and body size seems to be weaker than the relationship between IGF-1 and growth rate [69,75,85,86]. Thus, IGF-1 is an indicator of growth performance at the time of measuring. A clear and positive relationship between IGF-1 mRNA expression level and body size was detected in the wild salmon in both environments in this study. In contrast, no such relationship was observed for either the farmed or the hybrid salmon. Theoretically, this striking contrast could have been caused by sudden differences in growth rates between small and large wild fish at the time of sampling. Alternatively, this may reflect genetic differences between wild and farmed salmon in the way in which IGF-1 influences growth rate. First, the potential for genetic differences influencing this trend are discussed.
As farmed salmon display higher mRNA levels of IGF-1 than wild fish, it appears that selection for growth has increased the growth hormone GH:IGF-1 pathway activity which is the main endocrine regulator of growth in salmonids [30]. In turn, this could explain the lack of relationship between fish size and IGF-1 mRNA levels in the farmed salmon studied here as these elevated levels may not be limiting growth rate. In support of this suggestion is the fact that domesticated Atlantic salmon display a smaller growth-response to GH treatment than wild salmon [81]. This theory is further supported by the study of Devlin and colleagues [79], where mRNA levels of IGF-1, as well as other genes involved in growth regulation, was regulated alike in farmed and GH transgenic coho salmon.
In addition to potential genetic differences causing the clear difference in relationship between fish size and IGF-1 mRNA levels between farmed and wild salmon, it is possible that this difference may have been caused by specific conditions in the present experiment. Farmed salmon outgrew wild salmon by 2.56:1 in the control treatment, and 2.49:1 in the stress treatment, while hybrids were outgrown by 1.66:1 and 1.58:1, respectively. Two weeks earlier the corresponding numbers were 2.93:1 and 3.42:1 for the wild salmon, and 1.54:1 and 1.61:1 for the hybrid salmon (based upon more than 2000 individuals sampled for a comprehensive growth reaction norm study, [27]. Thus, the difference in weight between wild and farmed salmon decreased after the first samples were taken, while the weight difference between hybrid and farmed salmon were stable. Also, similar growth reaction norm slopes were detected in salmon of all origin in this study, in contrast to the salmon sampled two weeks earlier, where the wild salmon displayed a significantly steeper slope than the farmed salmon (Figure 1). This could indicate that wild salmon in addition to displaying a positive relationship between IGF-1 levels and body size, displayed an increased growth rate at the time of sampling. This could be due to biased sampling, if the smallest individuals were unintentionally left in the tank at the time of sampling and therefore not used in the study. Furthermore, a sudden increase in growth rate could be caused by compensatory growth, where accelerated growth rates are experienced after a period of growth depressions [87,88]. In this study, salmon of all origin were communally reared in order to avoid strain-specific tank effects. As farmed salmon are more competitive and aggressive than wild salmon [14,18,19], growth depression could unintentionally have been induced in the wild salmon in this study, due to high inter-strain competition for feed. As the within-tank biomass was significantly reduced two weeks prior to our terminal sampling, this might have caused a reduction in the competition level, causing a sudden increase in feeding activity in the wild salmon. Increased IGF-1 mRNA levels in muscle of starved salmonids have previously been documented as a response to re-feeding [72,73]. Thus, compensatory growth could also, in part, explain the positive relationship between IGF-1 mRNA levels and body size detected in the wild salmon.
Antioxidant genes
In general, oxidative stress was not detected in the salmon exposed to the crowding stressor in the present study. Although mRNA expression levels were not upregulated or downregulated as a response to treatment, the four antioxidant genes, GR, Cu/Zn SOD, Mn SOD and GP, were expressed significantly different between the origins. In fish, oxidative stress can be induced by abiotic factors like toxins in the water [26], dissolved oxygen [53,89,90], temperature [38] and diet type [52], as well as biotic factors like age and feeding behaviour [91]. In the present study, water circulation was maintained during stressing, thus avoiding alterations in dissolved oxygen levels. However, oxidative stress generated by starvation or food deprivation has been documented in fish [92-94]. Thus, if food deprivation/compensatory growth were unintentionally induced in this study, this could have had an impact of the mRNA expression levels of the antioxidant genes that were expressed significantly different between the origins. However, differing antioxidant defense responses to starvation has been documented in salmonids when studied with respect to enzymatic activity of GR, SOD and GP [92,93,95]. For instance, a decrease in liver GR, SOD and GP enzymatic activity were detected in starved rainbow trout [92,95], while in contrast enzymatic activity in liver of brown trout increased during starvation [93], thus making it hard to generalize on the effect of oxidative stress, induced by food deprivations, in salmonids.
In this study, mRNA expression level of the antioxidant stress gene Mn SOD was expressed similar in the farmed and wild farmed salmon, while GR, Cu/Zn SOD and GP, were significantly elevated in the farmed relative to the wild salmon. This result is in contrast to a common-garden study documenting GR mRNA expression levels in Atlantic salmon originating from a domesticated Canadian strain, a wild Canadian strain and their first generation hybrids, reared under standard hatchery conditions [26]. In the study by Debes and colleagues [26] wild salmon displayed elevated mRNA levels compared to the farmed and hybrid salmon. Although, consistent with our study, hybrid and domesticated salmon displayed similar GR mRNA levels [26].
Hybrids
Hybrid salmon displayed body weights at an intermediate level of the wild and farmed salmon, however the mRNA levels expressed in head kidney tissue were only displayed at an intermediate level in one of the five genes regulated in this study. In three of the genes, mRNA expression levels were similar to the levels observed in the farmed salmon, while in one of the genes, expression levels were elevated compared to both the farmed and the wild salmon.
Non additive gene expression profiles in hybrids has been documented in hybrids created from wild and farmed Atlantic salmon strains of Norwegian [24] and Canadian [96] origin. Based on these studies [24,96] and on studies on other organisms, e.g., Drosophila[97], maize [98], Pacific oysters Crassostrea gigas[99], it has been suggested that most gene expression profiles appears to be regulated as non additive traits, while most phenotypic traits, e.g., growth, display additive genetic variation. However, in contrast, other studies have documented larger portions of additive relative to non additive pattern of inheritance of gene expression profiles in both Atlantic salmon [26] and maize [100], as well as in mice [101].
When quantified by microarrays in liver tissue [96] and whole fry [24], more than 80% of genes regulated in farmed, relative to wild Atlantic salmon displayed gene expressions in hybrids that departed from additive inheritance. However, in a microarray performed on gill tissue of Atlantic salmon, only one third of the expressions regulated in gill of farmed relative to wild salmon displayed a non additive pattern of inheritance in the hybrids [26]. In the study by Normandeau and colleagues [96], as well as in the present study, non additive expression levels in hybrids were similar to the expressions of farmed salmon, while in the study by Debes and colleagues [26], the majority of the non additive expression levels were displayed at levels closer to the wild than the farmed salmon. The presence of both additive and non additive gene regulations in hybrids, as well as non additive expressions being displayed similar to both farmed and wild origin, suggest that the pattern of inheritance in gene expression profiles in Atlantic salmon is both gene and tissue-specific [26].
Conclusions
In general, mRNA expression levels of the seven selected genes investigated in this study were not differentially regulated between treatments. One exception was detected in the IGF-1 gene, which was downregulated in the stress treatment where growth was lower. Although the effect of treatment was weak, genetic origin had an effect upon mRNA expression levels of the four antioxidant genes GR, Cu/Zn SOD, Mn SOD, and GP, as well as IGF-1. The farmed Mowi strain displayed elevated mRNA levels for GR, Cu/Zn SOD, GP, and IGF-1, compared to the wild Etne strain, while Mn SOD was expressed at a similar level. Hybrids displayed both additive and non additive gene regulations.
In the wild salmon, a clear positive relationship between IGF-1 mRNA expression levels and body size was observed in both replicates in both treatments. This is in contrast to the farmed and hybrid salmon where no such relationship was detected. It is not possible to exclude the possibility that this was caused by large wild salmon displaying increased growth rates at the time of sampling. However, it is suggested that the most plausible explanation for this clear difference is that as farmed salmon display higher levels of IGF-1 than the wild fish, these elevated levels may not be limiting growth rate. This deserves further scientific attention.
Availability of supporting data
The data set supporting the results of this article are available as an additional file (Additional file 7).
Abbreviations
mRNA: Messenger ribonucleic acid;GR: Glutathione reductase;Cu/Zn SOD: Cu/Zn superoxide dismutase;Mn SOD: Mn superoxide dismutase;GP: Glutathione peroxidase;HSP70: Heat-shock protein 70;IGF-1: Insulin-like growth factor-I;IGF-1R: Insulin-like growth factor 1 receptor;EF1AA: Elongation factor 1 alpha A;NARA: Norwegian Animal Research Authority;DNA: Deoxyribonucleic acid;RIN: RNA integrity number;dH20: Distilled water;nRT: Negative reverse transcriptase;PK: Positive control;NTC: No template control;qPCR: Quantitative real time polymerase chain reaction;MIQE: Minimum Information for Publication of Quantitative Real-Time PCR Experiments;Cq: Quantification Cycle;IQR: Interquartile range;SD: Standard deviation;LME: Linear mixed effect models;AIC: Akaike information criterion;RMLE: Restricted maximum likelihood;GH: Growth hormone;SOD: Superoxide dismutase;GP: Glutathione peroxidase;DDCq: ΔΔCq
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MFS, BOK, FN and KAG participated in the design and sampling of the study. MFS and BOK carried out the molecular studies and performed the statistical analysis. MFS, BOK, FN and KAG drafted the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
AIC comparisons of the LME models.
Summary of linear mixed effect models testing for differences in log-weight and expression of the seven target genes in farmed versus hybrid salmon, hybrid versus wild salmon and wild versus farmed salmon.
Cq values of the reference gene EF1AA and ΔΔCq values of the seven target genes in salmon of farmed, hybrid and wild origin, in both treatments.
Linear regression between ΔΔDCq values on the y-axis and fish size (weight in grams) on the x-axis, for the seven selected genes, performed with a 95% confidence interval.
Linear regression between ΔΔDCq values on the y-axis and sampling period (1–3) on the x-axis, for the seven selected genes, performed with a 95% confidence interval.
Linear regression between ΔΔDCq values on the y-axis and time in anaesthesia (minutes) on the x-axis, for the seven selected genes, performed with a 95% confidence interval.
Full data set supporting the results of this article, with index.
Contributor Information
Monica F Solberg, Email: monica.solberg@imr.no.
Bjørn Olav Kvamme, Email: bjornok@imr.no.
Frank Nilsen, Email: frank.nilsen@bio.uib.no.
Kevin A Glover, Email: kevin.glover@imr.no.
Acknowledgements
We greatly acknowledge Dr. Øystein Skaala for his contributions towards the design of the salmon crosses and the production of the 29 families used in this study. We thank Ivar Helge Matre and Lise Dyrhovden for their excellent assistant in conducting the experiment at Matre research station and Heidi Kongshaug for her indispensable support in the laboratory. This project was conducted within the project INTERACT, funded by the Research Council of Norway.
References
- Gjedrem T, Gjoen HM, Gjerde B. Genetic-origin of Norwegian farmed Atlantic salmon. Aquaculture. 1991;98(1–3):41–50. [Google Scholar]
- The Norwegian Directorate of Fisheries. Oppdaterte rømmingstall (in Norwegian) http://www.fiskeridir.no/statistikk/akvakultur/oppdaterte-roemmingstall.
- Saegrov H, Hindar K, Kalas S, Lura H. Escaped farmed Atlantic salmon replace the original salmon stock in the River Vosso, western Norway. ICES J Mar Sci. 1997;54(6):1166–1172. [Google Scholar]
- Crozier WW. Escaped farmed salmon, Salmo salar L., in the Glenarm River, Northern Ireland: genetic status of the wild population 7 years on. Fish Manag Ecol. 2000;7(5):437–446. doi: 10.1046/j.1365-2400.2000.00219.x. [DOI] [Google Scholar]
- Crozier WW. Evidence of genetic interaction between escaped farmed salmon and wild Atlantic salmon (Salmo salar L.) in a northern Irish river. Aquaculture. 1993;113(1–2):19–29. [Google Scholar]
- Clifford SL, McGinnity P, Ferguson A. Genetic changes in an Atlantic salmon population resulting from escaped juvenile farm salmon. J Fish Biol. 1998;52(1):118–127. doi: 10.1111/j.1095-8649.1998.tb01557.x. [DOI] [Google Scholar]
- Clifford SL, McGinnity P, Ferguson A. Genetic changes in Atlantic salmon (Salmo salar) populations of northwest Irish rivers resulting from escapes of adult farm salmon. Can J Fish Aquat Sci. 1998;55(2):358–363. doi: 10.1139/f97-229. [DOI] [Google Scholar]
- Skaala O, Wennevik V, Glover KA. Evidence of temporal genetic change in wild Atlantic salmon, Salmo salar L., populations affected by farm escapees. ICES J Mar Sci. 2006;63(7):1224–1233. doi: 10.1016/j.icesjms.2006.04.005. [DOI] [Google Scholar]
- Bourret V, O'Reilly PT, Carr JW, Berg PR, Bernatchez L. Temporal change in genetic integrity suggests loss of local adaptation in a wild Atlantic salmon (Salmo salar) population following introgression by farmed escapees. Heredity. 2011;106(3):500–510. doi: 10.1038/hdy.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover KA, Quintela M, Wennevik V, Besnier F, Sørvik AGE, Skaala O. Three decades of farmed escapees in the wild: a spatio-temporal analysis of Atlantic salmon population genetic structure throughout Norway. PLoS One. 2012;7(8):e43129. doi: 10.1371/journal.pone.0043129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover KA, Ottera H, Olsen RE, Slinde E, Taranger GL, Skaala O. A comparison of farmed, wild and hybrid Atlantic salmon (Salmo salar L.) reared under farming conditions. Aquaculture. 2009;286(3–4):203–210. [Google Scholar]
- McGinnity P, Stone C, Taggart JB, Cooke D, Cotter D, Hynes R, McCamley C, Cross T, Ferguson A. Genetic impact of escaped farmed Atlantic salmon (Salmo salar L.) on native populations: use of DNA profiling to assess freshwater performance of wild, farmed, and hybrid progeny in a natural river environment. ICES J Mar Sci. 1997;54(6):998–1008. [Google Scholar]
- Thodesen J, Grisdale-Helland B, Helland SJ, Gjerde B. Feed intake, growth and feed utilization of offspring from wild and selected Atlantic salmon (Salmo salar) Aquaculture. 1999;180(3–4):237–246. [Google Scholar]
- Fleming IA, Einum S. Experimental tests of genetic divergence of farmed from wild Atlantic salmon due to domestication. ICES J Mar Sci. 1997;54(6):1051–1063. [Google Scholar]
- Gross MR. One species with two biologies: Atlantic salmon (Salmo salar) in the wild and in aquaculture. Can J Fish Aquat Sci. 1998;55:131–144. doi: 10.1139/d98-024. [DOI] [Google Scholar]
- Gjedrem T. Genetic improvement of cold-water fish species. Aquacult Res. 2000;31(1):25–33. doi: 10.1046/j.1365-2109.2000.00389.x. [DOI] [Google Scholar]
- McGinnity P, Prodohl P, Ferguson K, Hynes R, O'Maoileidigh N, Baker N, Cotter D, O'Hea B, Cooke D, Rogan G. Fitness reduction and potential extinction of wild populations of Atlantic salmon, Salmo salar, as a result of interactions with escaped farm salmon. Proc R Soc Lond B Biol Sci. 2003;270(1532):2443–2450. doi: 10.1098/rspb.2003.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde ALS, Fraser DJ, Hutchings JA. Fitness-related consequences of competitive interactions between farmed and wild Atlantic salmon at different proportional representations of wild-farmed hybrids. ICES J Mar Sci. 2009;67(4):657–667. [Google Scholar]
- Einum S, Fleming IA. Genetic divergence and interactions in the wild among native, farmed and hybrid Atlantic salmon. J Fish Biol. 1997;50(3):634–651. doi: 10.1111/j.1095-8649.1997.tb01955.x. [DOI] [Google Scholar]
- Houde ALS, Fraser DJ, Hutchings JA. Reduced anti-predator responses in multi-generational hybrids of farmed and wild Atlantic salmon (Salmo salar L.) Conserv Genet. 2010;11(3):785–794. doi: 10.1007/s10592-009-9892-2. [DOI] [Google Scholar]
- Skaala O, Taggart JB, Gunnes K. Genetic differences between five major domesticated strains of Atlantic salmon and wild salmon. J Fish Biol. 2005;67:118–128. doi: 10.1111/j.0022-1112.2005.00843.x. [DOI] [Google Scholar]
- Skaala O, Hoyheim B, Glover KA, Dahle G. Microsatellite analysis in domesticated and wild Atlantic salmon (Salmo salar L.): allelic diversity and identification of individuals. Aquaculture. 2004;240(1–4):131–143. [Google Scholar]
- Karlsson S, Moen T, Lien S, Glover KA, Hindar K. Generic genetic differences between farmed and wild Atlantic salmon identified from a 7 K SNP-chip. Mol Ecol Resour. 2011;11:247–253. doi: 10.1111/j.1755-0998.2010.02959.x. [DOI] [PubMed] [Google Scholar]
- Roberge C, Normandeau E, Einum S, Guderley H, Bernatchez L. Genetic consequences of interbreeding between farmed and wild Atlantic salmon: insights from the transcriptome. Mol Ecol. 2008;17(1):314–324. doi: 10.1111/j.1365-294X.2007.03438.x. [DOI] [PubMed] [Google Scholar]
- Roberge C, Einum S, Guderley H, Bernatchez L. Rapid parallel evolutionary changes of gene transcription profiles in farmed Atlantic salmon. Mol Ecol. 2006;15(1):9–20. doi: 10.1111/j.1365-294X.2005.02807.x. [DOI] [PubMed] [Google Scholar]
- Debes PV, Normandeau E, Fraser DJ, Bernatchez L, Hutchings JA. Differences in transcription levels among wild, domesticated, and hybrid Atlantic salmon (Salmo salar) from two environments. Mol Ecol. 2012;21:2574–2587. doi: 10.1111/j.1365-294X.2012.05567.x. [DOI] [PubMed] [Google Scholar]
- Solberg MF, Skaala Ø, Nilsen F, Glover KA. Does domestication cause changes in growth reaction norms? A study of farmed, wild and hybrid atlantic Salmon families exposed to environmental stress. PLoS ONE. 2013;8(1):e54469. doi: 10.1371/journal.pone.0054469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Iwama GK, Afonso LOB, Todgham A, Ackerman P, Nakano K. Are hsps suitable for indicating stressed states in fish? J Exp Biol. 2004;207(1):15–19. doi: 10.1242/jeb.00707. [DOI] [PubMed] [Google Scholar]
- Bjornsson BT. The biology of salmon growth hormone: from daylight to dominance. Fish Physiol Biochem. 1997;17(1–6):9–24. [Google Scholar]
- Reinecke M, Bjornsson BT, Dickhoff WW, McCormick SD, Navarro I, Power DM, Gutierrez J. Growth hormone and insulin-like growth factors in fish: Where we are and where to go. Gen Comp Endocrinol. 2005;142(1–2):20–24. doi: 10.1016/j.ygcen.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Moriyama S, Swanson P, Nishii M, Takahashi A, Kawauchi H, Dickhoff WW, Plisetskaya EM. Development of a homologous radioimmunoassay for coho salmon insulin-like growth-factor-I. Gen Comp Endocrinol. 1994;96(1):149–161. doi: 10.1006/gcen.1994.1167. [DOI] [PubMed] [Google Scholar]
- Duan C, Plisetskaya EM. Nutritional regulation of insulin-like growth factor-I messenger-RNA expression in salmon tissues. J Endocrinol. 1993;139(2):243–252. doi: 10.1677/joe.0.1390243. [DOI] [PubMed] [Google Scholar]
- Olsvik PA, Lie KK, Jordal AEO, Nilsen TO, Hordvik I. Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon. BMC Mol Biol. 2005;6:21. doi: 10.1186/1471-2199-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LJ, Somamoto T, Lie KK, Dijkstra JM, Hordvik I. Characterisation of salmon and trout CD8 alpha and CD8 beta. Mol Immunol. 2005;42(10):1225–1234. doi: 10.1016/j.molimm.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 2010;10:4. doi: 10.1186/1471-2229-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost P, Nilsen F. Validation of reference genes for transcription profiling in the salmon louse, Lepeophtheirus salmonis, by quantitative real-time PCR. Vet Parasitol. 2003;118(1–2):169–174. doi: 10.1016/j.vetpar.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Aursnes IA, Rishovd AL, Karlsen HE, Gjøen T. Validation of reference genes for quantitative RT-qPCR studies of gene expression in Atlantic cod (Gadus morhua l.) during temperature stress. BMC Research Notes. 2011;4:104. doi: 10.1186/1756-0500-4-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamalainen HK, Tubman JC, Vikman S, Kyrola T, Ylikoski E, Warrington JA, Lahesmaa R. Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal Biochem. 2001;299(1):63–70. doi: 10.1006/abio.2001.5369. [DOI] [PubMed] [Google Scholar]
- Zahl IH, Kiessling A, Samuelsen OB, Olsen RE. Anesthesia induces stress in Atlantic salmon (Salmo salar), Atlantic cod (Gadus morhua) and Atlantic halibut (Hippoglossus hippoglossus) Fish Physiol Biochem. 2010;36(3):719–730. doi: 10.1007/s10695-009-9346-2. [DOI] [PubMed] [Google Scholar]
- Sanchez JA, Clabby C, Ramos D, Blanco G, Flavin F, Vazquez E, Powell R. Protein and microsatellite single locus variability in Salmo salar L. (Atlantic salmon) Heredity. 1996;77:423–432. doi: 10.1038/hdy.1996.162. [DOI] [PubMed] [Google Scholar]
- O'Reilly PT, Hamilton LC, McConnell SK, Wright JM. Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can J Fish Aquat Sci. 1996;53(10):2292–2298. [Google Scholar]
- Grimholt U, Drablos F, Jorgensen SM, Hoyheim B, Stet RJM. The major histocompatibility class I locus in Atlantic salmon (Salmo salar L.): polymorphism, linkage analysis and protein modelling. Immunogenetics. 2002;54(8):570–581. doi: 10.1007/s00251-002-0499-8. [DOI] [PubMed] [Google Scholar]
- Stet RJM, de Vries B, Mudde K, Hermsen T, van Heerwaarden J, Shum BP, Grimholt U. Unique haplotypes of co-segregating major histocompatibility class II A and class II B alleles in Atlantic salmon (Salmo salar) give rise to diverse class II genotypes. Immunogenetics. 2002;54(5):320–331. doi: 10.1007/s00251-002-0477-1. [DOI] [PubMed] [Google Scholar]
- Slettan A, Olsaker I, Lie O. Atlantic salmon, Salmo salar, microsattelites at the SsOSL25, SsOSL85, SsOSL311, SsOSL417 loci. Anim Genet. 1995;26(4):281–282. doi: 10.1111/j.1365-2052.1995.tb03262.x. [DOI] [PubMed] [Google Scholar]
- Taggart JB. FAP: an exclusion-based parental assignment program with enhanced predictive functions. Molecular Ecol Notes. 2007;7(3):412–415. [Google Scholar]
- Glover KA, Taggart JB, Skaala O, Teale AJ. Comparative performance of juvenile sea trout families in high and low feeding environments. J Fish Biol. 2001;59(1):105–115. doi: 10.1111/j.1095-8649.2001.tb02341.x. [DOI] [Google Scholar]
- Glover KA, Taggart JB, Skaala O, Teale AJ. A study of inadvertent domestication selection during start-feeding of brown trout families. J Fish Biol. 2004;64(5):1168–1178. doi: 10.1111/j.0022-1112.2004.00376.x. [DOI] [Google Scholar]
- Glover KA, Hansen MM, Lien S, Als TD, Hoyheim B, Skaala O. A comparison of SNP and STR loci for delineating population structure and performing individual genetic assignment. BMC Genet. 2010;11:2. doi: 10.1186/1471-2156-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover KA. Forensic identification of fish farm escapees: the Norwegian experience. Aquaculture Environ Interact. 2010;1(1):1–10. [Google Scholar]
- Glover KA, Skilbrei OT, Skaala O. Genetic assignment identifies farm of origin for Atlantic salmon Salmo salar escapees in a Norwegian fjord. ICES J Mar Sci. 2008;65(6):912–920. doi: 10.1093/icesjms/fsn056. [DOI] [Google Scholar]
- Olsvik PA, Torstensen BE, Berntssen MHG. Effects of complete replacement of fish oil with plant oil on gastrointestinal cell death, proliferation and transcription of eight genes' encoding proteins responding to cellular stress in Atlantic salmon Salmo salar L. J Fish Biol. 2007;71(2):550–568. doi: 10.1111/j.1095-8649.2007.01521.x. [DOI] [Google Scholar]
- Huang TS, Olsvik PA, Krovel A, Tung HS, Torstensen BE. Stress-induced expression of protein disulfide isomerase associated 3 (PDIA3) in Atlantic salmon (Salmo salar L.) Comp Biochem Physiol B Biochem Mol Biol. 2009;154(4):435–442. doi: 10.1016/j.cbpb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Nordgarden U, Fjelldal PG, Hansen T, Bjornsson BT, Wargelius A. Growth hormone and insulin-like growth factor-I act together and independently when regulating growth in vertebral and muscle tissue of atlantic salmon postsmolts. Gen Comp Endocrinol. 2006;149(3):253–260. doi: 10.1016/j.ygcen.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Olsvik PA, Kristensen T, Waagbo R, Rosseland BO, Tollefsen KE, Baeverfjord G, Berntssen MHG. mRNA expression of antioxidant enzymes (SOD, CAT and GSH-Px) and lipid peroxidative stress in liver of Atlantic salmon (Salmo salar) exposed to hyperoxic water during smoltification. Comp Biochem Physiol C Toxicol Pharmacol. 2005;141(3):314–323. doi: 10.1016/j.cbpc.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FSB, Olsvik PA, Penning LC, Toegel S. MIQE precis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol. 2010;11:74. doi: 10.1186/1471-2199-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C-T method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Crawley MJ. The R Book. London: Wiley; 2007. [Google Scholar]
- Johnson JB, Omland KS. Model selection in ecology and evolution. Trends Ecol Evol. 2004;19(2):101–108. doi: 10.1016/j.tree.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Keene ON. The log transformation is special. Stat Med. 1995;14(8):811–819. doi: 10.1002/sim.4780140810. [DOI] [PubMed] [Google Scholar]
- Cole TJ. Sympercents: symmetric percentage differences on the 100 log(e) scale simplify the presentation of log transformed data. Stat Med. 2000;19(22):3109–3125. doi: 10.1002/1097-0258(20001130)19:22<3109::AID-SIM558>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Hairston NG, Holtmeier CL, Lampert W, Weider LJ, Post DM, Fischer JM, Caceres CE, Fox JA, Gaedke U. Natural selection for grazer resistance to toxic cyanobacteria: Evolution of phenotypic plasticity? Evolution. 2001;55(11):2203–2214. doi: 10.1111/j.0014-3820.2001.tb00736.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team: R. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- Bate D, Maechler M, Bolker B. lme4: Linear mixed-effects models using S4 classes. R package version 0.999999-0. 2012. http://cran.r-project.org/web/packages/lme4/
- Pierce AL, Beckman BR, Schearer KD, Larsen DA, Dickhoff WW. Effects of ration on somatotropic hormones and growth in coho salmon. Comp Biochem Physiol B Biochem Mol Biol. 2001;128(2):255–264. doi: 10.1016/S1096-4959(00)00324-9. [DOI] [PubMed] [Google Scholar]
- Larsen DA, Beckman BR, Dickhoff WW. The effect of low temperature and fasting during the winter on metabolic stores and endocrine physiology (Insulin, insulin-like growth factor-I and thyroxine) of coho salmon, Oncorhynchus kisutch. Gen Comp Endocrinol. 2001;123(3):308–323. doi: 10.1006/gcen.2001.7677. [DOI] [PubMed] [Google Scholar]
- Gabillard JC, Kamangar BB, Montserrat N. Coordinated regulation of the GH/IGF system genes during refeeding in rainbow trout (Oncorhynchus mykiss) J Endocrinol. 2006;191(1):15–24. doi: 10.1677/joe.1.06869. [DOI] [PubMed] [Google Scholar]
- Dyer AR, Barlow CG, Bransden MP, Carter CG, Glencross BD, Richardson N, Thomas PM, Williams KC, Carragher JF. Correlation of plasma IGF-I concentrations and growth rate in aquacultured finfish: a tool for assessing the potential of new diets. Aquaculture. 2004;236(1–4):583–592. [Google Scholar]
- Bailey GS, Poulter RTM, Stockwell PA. Gene duplication in tetraploid fish - model for gene silencing at unlinked duplicated loci. Proc Natl Acad Sci USA. 1978;75(11):5575–5579. doi: 10.1073/pnas.75.11.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perezsanchez J, Martipalanca H, Kaushik SJ. Ration size and protein-intake affect circulating growth-hormone concentration, hepatic growth-hormone binding and plasma insulin-like growth-factor-I immunoreactivity in a marine teleost, the gilthead sea bream (Sparus aurata) J Nutr. 1995;125(3):546–552. doi: 10.1093/jn/125.3.546. [DOI] [PubMed] [Google Scholar]
- Chauvigne F, Gabillard JC, Weil C, Rescan PY. Effect of refeeding on IGFI, IGFII, IGF receptors, FGF2, FGF6, and myostatin mRNA expression in rainbow trout myotomal muscle. Gen Comp Endocrinol. 2003;132(2):209–215. doi: 10.1016/S0016-6480(03)00081-9. [DOI] [PubMed] [Google Scholar]
- Bower NI, Li XJ, Taylor R, Johnston IA. Switching to fast growth: the insulin-like growth factor (IGF) system in skeletal muscle of Atlantic salmon. J Exp Biol. 2008;211(24):3859–3870. doi: 10.1242/jeb.024117. [DOI] [PubMed] [Google Scholar]
- Woodward CC, Strange RJ. Physiological stress responses in wild and hatchery-reared rainbow-trout. Trans Am Fish Soc. 1987;116(4):574–579. doi: 10.1577/1548-8659(1987)116<574:PSRIWA>2.0.CO;2. [DOI] [Google Scholar]
- Beckman BR, Larsen DA, Moriyama S, Lee-Pawlak B, Dickhoff WW. Insulin-like growth factor-I and environmental modulation of growth during smoltification of spring chinook salmon (Oncorhynchus tshawytscha) Gen Comp Endocrinol. 1998;109(3):325–335. doi: 10.1006/gcen.1997.7036. [DOI] [PubMed] [Google Scholar]
- Wargelius A, Fjelldal PG, Benedet S, Hansen T, Bjornsson BT, Nordgarden U. A peak in gh-receptor expression is associated with growth activation in Atlantic salmon vertebrae, while upregulation of igf-I receptor expression is related to increased bone density. Gen Comp Endocrinol. 2005;142(1–2):163–168. doi: 10.1016/j.ygcen.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Nordgarden U, Hansen T, Hemre GI, Sundby A, Bjornsson BT. Endocrine growth regulation of adult Atlantic salmon in seawater: The effects of light regime on plasma growth hormone, insulin-like growth factor-I, and insulin levels. Aquaculture. 2005;250(3–4):862–871. [Google Scholar]
- Overturf K, Sakhrani D, Devlin RH. Expression profile for metabolic and growth-related genes in domesticated and transgenic coho salmon (Oncorhynchus kisutch) modified for increased growth hormone production. Aquaculture. 2010;307(1–2):111–122. [Google Scholar]
- Devlin RH, Sakhrani D, Tymchuk WE, Rise ML, Goh B. Domestication and growth hormone transgenesis cause similar changes in gene expression in coho salmon (Oncorhynchus kisutch) Proc Natl Acad Sci USA. 2009;106(9):3047–3052. doi: 10.1073/pnas.0809798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymchuk WE, Beckman B, Devlin RH. Altered Expression of Growth Hormone/Insulin-Like Growth Factor I Axis Hormones in Domesticated Fish. Endocrinology. 2009;150(4):1809–1816. doi: 10.1210/en.2008-0797. [DOI] [PubMed] [Google Scholar]
- Neregard L, Sundt-Hansen L, Hindar K, Einum S, Johnsson JI, Devlin RH, Fleming IA, Bjornsson BT. Wild Atlantic salmon Salmo salar L. strains have greater growth potential than a domesticated strain selected for fast growth. J Fish Biol. 2008;73(1):79–95. doi: 10.1111/j.1095-8649.2008.01907.x. [DOI] [Google Scholar]
- Fleming IA, Agustsson T, Finstad B, Johnsson JI, Bjornsson BT. Effects of domestication on growth physiology and endocrinology of Atlantic salmon (Salmo salar) Can J Fish Aquat Sci. 2002;59(8):1323–1330. doi: 10.1139/f02-082. [DOI] [Google Scholar]
- Pfaffl M, Schwarz F, Sauerwein H. Quantification of insulin-like growth factor-1 (IGF-1) mRNA: Modulation of growth intensity by feeding results in inter- and intra-tissue-specific differences of IGF-1 mRNA expression in steers. Exp Clin Endocrinol Diabetes. 1998;106(6):514–521. doi: 10.1055/s-0029-1212026. [DOI] [PubMed] [Google Scholar]
- Greene MW, Chen TT. Quantitation of IGF-I, IGF-II, and multiple insulin receptor family member messenger RNAs during embryonic development in rainbow trout. Mol Reprod Dev. 1999;54(4):348–361. doi: 10.1002/(SICI)1098-2795(199912)54:4<348::AID-MRD5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Beckman BR, Shimizu M, Gadberry BA, Parkins PJ, Cooper KA. The effect of temperature change on the relations among plasma IGF-1, 41-kDa IGFBP, and growth rate in postsmolt coho salmon. Aquaculture. 2004;241(1–4):601–619. [Google Scholar]
- Sacobie CFD, Glebe BD, Barbeau MA, Lall SP, Benfey TJ. Effect of strain and ploidy on growth performance of Atlantic salmon, Salmo salar, following seawater transfer. Aquaculture. 2012;334:58–64. [Google Scholar]
- Ali M, Nicieza A, Wootton RJ. Compensatory growth in fishes: a response to growth depression. Fish Fish. 2003;4(2):147–190. doi: 10.1046/j.1467-2979.2003.00120.x. [DOI] [Google Scholar]
- Metcalfe NB, Monaghan P. Compensation for a bad start: grow now, pay later? Trends Ecol Evol. 2001;16(5):254–260. doi: 10.1016/S0169-5347(01)02124-3. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Oxygen toxicity: A radical explanation. J Exp Biol. 1998;201(8):1203–1209. doi: 10.1242/jeb.201.8.1203. [DOI] [PubMed] [Google Scholar]
- Waagbo R, Hosfeld CD, Fivelstad S, Olsvik PA, Breck O. The impact of different water gas levels on cataract formation, muscle and lens free amino acids, and lens antioxidant enzymes and heat shock protein mRNA abundance in smolting Atlantic salmon, Salmo salar L. Comp Biochem Physiol A Mol Integr Physiol. 2008;149(4):396–404. doi: 10.1016/j.cbpa.2008.01.034. [DOI] [PubMed] [Google Scholar]
- Martinez-Alvarez RM, Morales AE, Sanz A. Antioxidant defenses in fish: Biotic and abiotic factors. Rev Fish Biol Fisheries. 2005;15(1–2):75–88. [Google Scholar]
- Furne M, Garcia-Gallego M, Hidalgo MC, Morales AE, Domezain A, Domezain J, Sanz A. Oxidative stress parameters during starvation and refeeding periods in Adriatic sturgeon (Acipenser naccarii) and rainbow trout (Oncorhynchus mykiss) Aquacult Nutr. 2009;15(6):587–595. doi: 10.1111/j.1365-2095.2008.00626.x. [DOI] [Google Scholar]
- Bayir A, Sirkecioglu AN, Bayir M, Haliloglu HI, Kocaman EM, Aras NM. Metabolic responses to prolonged starvation, food restriction, and refeeding in the brown trout, Salmo trutta: Oxidative stress and antioxidant defenses. Comp Biochem Physiol B Biochem Mol Biol. 2011;159(4):191–196. doi: 10.1016/j.cbpb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Hidalgo MC, Exposito A, Palma JM, de la Higuera M. Oxidative stress generated by dietary Zn-deficiency: studies in rainbow trout (Oncorhynchus mykiss) Int J Biochem Cell Biol. 2002;34(2):183–193. doi: 10.1016/S1357-2725(01)00105-4. [DOI] [PubMed] [Google Scholar]
- Blom S, Andersson TB, Forlin L. Effects of food deprivation and handling stress on head kidney 17 alpha-hydroxyprogesterone 21-hydroxylase activity, plasma cortisol and the activities of liver detoxification enzymes in rainbow trout. Aquat Toxicol. 2000;48(2–3):265–274. doi: 10.1016/s0166-445x(99)00031-4. [DOI] [PubMed] [Google Scholar]
- Normandeau E, Hutchings JA, Fraser DJ, Bernatchez L. Population-specific gene expression responses to hybridization between farm and wild Atlantic salmon. Evol Appl. 2009;2(4):489–503. doi: 10.1111/j.1752-4571.2009.00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, Riley-Berger R, Harshman L, Kopp A, Vacha S, Nuzhdin S, Wayne M. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics. 2004;167(4):1791–1799. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger DL, Gray AD, Ream TS, Kato A, Coe EH, Birchler JA. Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics. 2005;169(1):389–397. doi: 10.1534/genetics.104.032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock D, Lin JZ, DeCola S, Haudenschild CD, Meyer E, Manahan DT, Bowen B. Transcriptomic analysis of growth heterosis in larval Pacific oysters (Crassostrea gigas) Proc Natl Acad Sci USA. 2007;104(7):2313–2318. doi: 10.1073/pnas.0610880104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. All possible modes of gene action are observed in a global comparison of gene expression in a maize F-1 hybrid and its inbred parents. Proc Natl Acad Sci USA. 2006;103(18):6805–6810. doi: 10.1073/pnas.0510430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XQ, Affourtit J, Shockley KR, Woo Y, Churchill GA. Inheritance patterns of transcript levels in F-1 hybrid mice. Genetics. 2006;174(2):627–637. doi: 10.1534/genetics.106.060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AIC comparisons of the LME models.
Summary of linear mixed effect models testing for differences in log-weight and expression of the seven target genes in farmed versus hybrid salmon, hybrid versus wild salmon and wild versus farmed salmon.
Cq values of the reference gene EF1AA and ΔΔCq values of the seven target genes in salmon of farmed, hybrid and wild origin, in both treatments.
Linear regression between ΔΔDCq values on the y-axis and fish size (weight in grams) on the x-axis, for the seven selected genes, performed with a 95% confidence interval.
Linear regression between ΔΔDCq values on the y-axis and sampling period (1–3) on the x-axis, for the seven selected genes, performed with a 95% confidence interval.
Linear regression between ΔΔDCq values on the y-axis and time in anaesthesia (minutes) on the x-axis, for the seven selected genes, performed with a 95% confidence interval.
Full data set supporting the results of this article, with index.