Abstract
Background
The central function of chloroplasts is to carry out photosynthesis, and its gene content and structure are highly conserved across land plants. Parasitic plants, which have reduced photosynthetic ability, suffer gene losses from the chloroplast (cp) genome accompanied by the relaxation of selective constraints. Compared with the rapid rise in the number of cp genome sequences of photosynthetic organisms, there are limited data sets from parasitic plants.
Principal Findings/Significance
Here we report the complete sequence of the cp genome of Cistanche deserticola, a holoparasitic desert species belonging to the family Orobanchaceae. The cp genome of C. deserticola is greatly reduced both in size (102,657 bp) and in gene content, indicating that all genes required for photosynthesis suffer from gene loss and pseudogenization, except for psbM. The striking difference from other holoparasitic plants is that it retains almost a full set of tRNA genes, and it has lower dN/dS for most genes than another close holoparasitic plant, E. virginiana, suggesting that Cistanche deserticola has undergone fewer losses, either due to a reduced level of holoparasitism, or to a recent switch to this life history. We also found that the rpoC2 gene was present in two copies within C. deserticola. Its own copy has much shortened and turned out to be a pseudogene. Another copy, which was not located in its cp genome, was a homolog of the host plant, Haloxylon ammodendron (Chenopodiaceae), suggesting that it was acquired from its host via a horizontal gene transfer.
Introduction
The chloroplast is an important organelle in the plant cell, and its central function is to carry out photosynthesis and carbon fixation. In general, the chloroplast (cp) genome is highly conserved among seed plants with two copies of a large inverted repeat (IR) separated by small single copy (SSC) and large single copy (LSC) regions [1]. It usually contains 110–130 unique genes, which can be roughly divided into three large groups according to their functions: genetic system genes, photosynthesis genes and conserved open reading frames with miscellaneous functions [2].
However, a small group of angiosperm plants appear to have escaped from this dominant pattern by evolving the capacity to gain the water, carbon and nutrients via the vascular tissue of the parasitized host’s roots or shoots. This means that these parasitic plants have reduced (or no) photosynthetic ability, and no longer need genes that encode photosynthetic proteins. With the selective constraints on their cp coding genes relaxed, gene losses occur in these parasitic plants [3]. It is estimated that approximately 1% of all angiosperm species have resorted to a parasitic lifestyle, which has independently evolved 12 or 13 times [4]. Compared with a rapid rise in the number of cp genomes of photosynthetic organisms available on NCBI (254 in Viridiplantae, as of December 4, 2012), there are limited data sets from parasitic plants, especially from the completely non-photosynthetic species. In higher plants, the cp genome of holoparasite Epifagus virginiana in the family Orobanchaceae was sequenced first [5], followed by four species from the holoparasitic genus Cuscuta [6], [7], and three mycoheterotrophic plants, including Aneura mirabilis [8], Rhizanthella gardneri [9], and Neottia nidus-avis [10]. However, only one species of the completely non-photosynthetic plants, E. virginiana, which exploit other plants via direct connections rather than by mycorrhizal fungi, has been comprehensively analyzed in its cp genome structure and composition.
Orobanchaceae, as taxonomically redefined by a series of recent molecular studies, comprise around 89 genera and more than 2,000 species, making it the largest predominantly parasitic angiosperm family, the majority of which are facultative or obligate root parasites [11]–[16]. It contains all levels of parasitic ability ranging from nonparasitic to hemiparasitic and holoparasitic [12], [17]. Therefore, analyses of cp genomes of other holoparasitic species within the family Orobanchaceae could confirm the common attributes of non-photosynthetic evolution and provide point for genetic analysis of cp genome evolution.
In Orobanchaceae, Cistanche is a worldwide genus of holoparasitic desert plants. Specifically, C. deserticola, commonly known as desert-broomrape and traditionally used as an important tonic in China and Japan, is distributed in Northwest China and the Mongolian People’s Republic, and is also considered to be an endangered wild species in recent years due to increased consumption by humans [16]. C. deserticola is parasitized on the roots of psammophyte Haloxylon ammodendron (Chenopodiaceae), which mainly inhabit deserts and semi-deserts due to its high tolerance to drought and salinity. Similar to E. virginiana, C. deserticola is a completely non-photosynthetic species and usually grows underground. A number of studies about the chemical components or pharmacological effects of this species have been reported [1], [18], [19]. Further analysis of its cp genome structure and composition could provide new insights on the evolution of the parasitic cp genome.
Attributed to the direct connections between parasitic plants and their hosts, which allows the channelling of metabolites, such as sugars, amino acids and perhaps nucleic acids in the form of mRNA, direct haustorial contact between them usually facilitates a horizontal gene transfer (HGT) from a donor to a recipient plant [20]. HGT, known as exchange of genes across mating barriers, has played a major role in bacterial evolution. In recent years, increasing studies have reported HGT being recognized as a significant force in the evolution of eukaryotic genomes [21], [22]. In plants, the evolutionarily earliest examples of HGT might be the endosymbioses that gave rise to mitochondria and chloroplasts [23], [24]. Since the emergence of HGT events, usually detected as incongruences in molecular phylogenetic trees, a considerable number of studies have suggested gene exchanges between hosts and parasites [4], [25]–[30].
Although the HGT involving parasitic plants appears to have occurred in many parasitic lineages, the majority of reported cases of HGT have been limited to exchanges between mitochondrial genes among related species [4], [25]. Cases of HGT involving cp genomes are rare [29]. The disparity in frequency of plant-to-plant HGT between the mitochondrial and the cp genomes is considered due to an active homologous recombination system [31], [32]. It is reported that a chloroplast region including rps2, trnL-F, and rbcL among a group of nonphotosynthetic flowering plants, Phelipanche and Orobanche species, both from the family Orobanchaceae, were detected according to the phylogenetic trees based on available data [29].
In order to examine the effect of its non-photosynthetic life history on cp genome content, we sequenced the entire cp genome of C. deserticola. As a completely non-photosynthetic species from Orobanchaceae, it shows the same pattern in the process of gene loss as in chloroplasts of E. virginiana and other parasitic plants. We also found that C. deserticola has two copies of a cp gene rpoC2, one becoming a pseudogene, the other being horizontally acquired from the host H. ammodendron, according to a homology search and phylogenetic analysis.
Materials and Methods
Genome Sequencing and Assembly
The spikes of C. deserticola were collected from a plant base in Bayannur City of Inner-Mongolia area which was introduced from natural populations located in desert area of Inner Mongolia in northeastern China. The collecting permit was obtained from the owner (Jun Wei) of the plant base. The voucher specimen was deposited in the MOE Key Laboratory for Biodiversity Science and Ecological Engineering at Fudan University. For cp genome sequencing, total genomic DNA extraction was performed using the Plant Genomic DNA Kit (Tiangen Biotech Co., China), following the manufacturer’s instructions. The fragments of cp DNA were amplified by the polymerase chain reaction (PCR). In brief, due to loss and pseudogenizations in the cp genome of C. deserticola, PCR primers were designed using the reported PCR primers from several sources. The primers of the LSC region were designed using the reported conserved cp DNA primer pairs, which including 38 primer pairs as well as eight primer pairs flanking cpDNA microsatellites tested on 20 plant species from 13 families [33]. Only 14 of the 38 primer pairs are useable in C. deserticola. Then the primers for other regions were designed according to the primers of the cp genome available in the cp genome database [34]. Some primers were also developed from the cp genome sequences of related species (Olea europaea and E. virginiana) for specific regions. In order to amplify longer fragments, some of these primers were used combined, and some of them were designed based on the newly determined sequences of adjacent regions. By using all above primers, we covered the entire cp genome of C. deserticola with PCR fragments ranging in size from 500 bp to 3 kb. The overlapping regions of each pair of adjacent PCR fragments exceeded 150 bp. The amplified product was purified, and ligated into TaKaRa pMD19-T plasmids (TaKaRa BioInc, Shiga, Japan), which were then cloned into Escherichia coli strain DH5a. Multiple (≥6) clones were randomly selected and followed by automated sequencing using ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA). All fragments were sequenced 2–10 times (6-fold coverage of the C. deserticola cp genome on average). All these individual sequences were excluded vector, primer and low-quality reads, and then assembled using Sequencher 3.0 software (Gene Codes Corporation, USA). The inverted repeat regions (IRs) of the cpDNA were not amplified separately, but primers were designed to amplify the regions spanning the junctions of LSC/IRA, LSC/IRB, SSC/IRA and SSC/IRB. Considering two IRs cannot be distinguished by automated assembly software, we input the reads as two groups and obtained two large contigs, with each contig including one IR and its adjacent partial LSC and SSC regions. Then, the two large contigs were manually assembled into the complete circular genome sequence.
Genome Annotation and Molecular Evolutionary Analyses
Initial gene annotations were performed using the chloroplast annotation package DOGMA (http://phylocluster.biosci.utexas.edu/dogma/) [35]. Genes that were undetected by DOGMA, such as psbB, psbK, trnG-GCC, rpoC2, atpB, accD, and ycf1, were identified by Blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The correctness of the annotation for all genes was additionally verified by a similarity search against the available plant cp genome sequences. The regions with similarity to known protein coding genes but lacking intact open reading frames (ORF) were identified as pseudogenes. tRNA genes were annotated using DOGMA and ARAGORN v1.2 (http://130.235.46.10/ARAGORN/) [36], and then confirmed by ERPIN (http://tagc.univ-mrs.fr/erpin/) [37]. The circular gene map of the C. deserticola cp genome was drawn by GenomeVx [38] followed by manual modification. An assembled and corrected sequence of C. deserticola cp genome was deposited in GenBank.
To estimate the selection constraint on the genes remaining in C. deserticola cp genome, the protein-coding genes that shared between C. deserticola, E. virginiana, and related photosynthetic species O. europaea were chosen to calculate the ratio of the rates of nonsynonymous and synonymous changes (dN/dS). Nicotiana tabacum was also included in the analyses to calculate dN/dS for photosynthetic plants. Alignment was performed using ClustalW [39]. The pairwise dS, dN and dN/dS ratios were calculated using DnaSP ver. 5 [40].
Isolation and Sequencing of Potential HGT Gene (rpoC2)
In this study, we found that C. deserticola harbours another copy of rpoC2 outwith its own, which corresponds to the phylogenetic position of the host H. ammodendron. We propose that this copy arose via horizontal gene transfer. In order to confirm if HGT occurred across the range of C. deserticola, we sampled 50 accessions from the same plant base in Bayannur City which were introduced from five natural populations located in Alxa Left Banner, Alxa Right banner (two populations), and Urad Rear Banner in the Inner-Mongolia area, as well as Hetian in the Xinjiang area. In order to confirm if the HGT occurred across the range of C. deserticola, total DNA extractions from these materials and cp rpoC2 genes amplified by standard PCR were performed in two different labs, thus, eliminating laboratory contamination. The rpoC2 gene was amplified using primers f4 (5′-GATAGACATCGGTACTCCAGTGC-3′) and r6 (3′-TCATTATGGGAATGTACACGCG-5′) with the following conditions: 94°C for 2.5 min; 35 cycles each at 94°C for 1 min,55°C for 30 s, 72°C for 1 min. In H. ammodendron, the five clones of rpoC2 gene were also checked.
Phylogenetic Analyses of the Potential HGT Gene, rpoC2
All the copies of rpoC2 sequences detected in C. deserticola and H. ammodendron were used as queries for BLASTN searches against the NCBI database (E-value <10−3) [41] to identify and retrieve their homologs. On the basis of Angiosperm Phylogeny Group III [42], sampling for the present study focused on members of the clade Lamiales and Caryophyllales that includes two families Orobanchaceae and Chenopodiaceae. Finally, 29 sequences were sampled for the rpoC2 phylogenetic analyses, using Oryza nivara (NC_005973) as outgroups. Sequences were unambiguously aligned manually in BioEdit 7.0.4.1 [43].
Phylogenetic analyses were performed using maximum likelihood in PAUP v. 4.0b10 [44] and Bayesian inference in MrBayes v. 3.1 [45]. The appropriate ML model of nucleotide substitution (GTR+I+G) was determined by Modeltest 3.7 [46] according to the Akaike information criteria (AIC) [47]. Relative clade support was estimated by ML bootstrap analysis of 100 replicates of heuristic searches with settings as above. Bayesian analysis was performed with MrBayes 3.1 using same model (GTR+I+G) suggested by MrModeltest v2.2 [48]. The settings for the Metropolis-coupled Markov chain Monte Carlo process were: three runs with four chains each were run simultaneously for 1*107 generations, which were logged every 1000 generations. Convergence was considered to have been reached when the variance of split frequencies was <0.01. The first 2500 generations were discarded as the transient burn-in period. The 50%-majority-rule consensus of trees sampled in the Bayesian phylogenetic analysis was used to construct a phylogram.
Results
The cp Genome Structure of C. deserticola
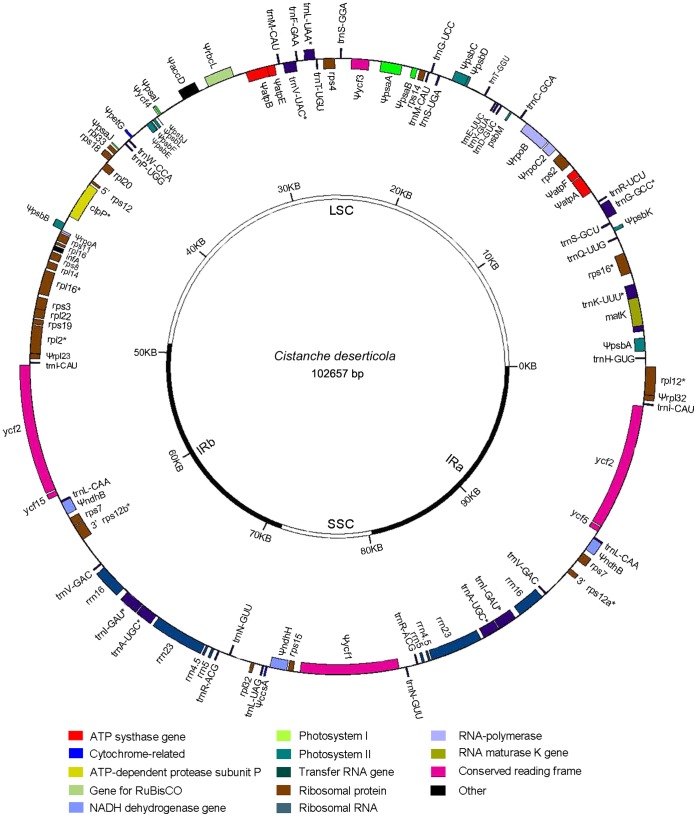
As expected, the cp genome of C. deserticola [GenBank number: KC128846] is greatly reduced in size (102,657 bp) and in gene content. It is a quadripartite structure typical of the majority of land plant chloroplast chromosomes with a large single copy (LSC) region of 49,130 bp separated from 8,819 bp small single copy (SSC) region by two inverted repeats (IRs), each of 22,354 bp (Fig. 1). In angiosperms, it is the fourth completely nonphotosynthetic species and the eighth parasitic species of which complete sequences of the cp genome are now available. Among these species, the cp genome of C. deserticola is larger than those of other five holoparasites species (E. virginiana, R. gardneri, N. nidus-avis, Cuscuta obtusiflora and C. gronovii), but is smaller than those of other hemiparasitic Cuscuta species, which has more or less green color distributed throughout the stems and inflorescences (Tables 1, 2).
Figure 1. Gene Maps of the plastid chromosomes of C. deserticola.
Genes shown inside the circle are transcribed clockwise, those outside the circle are transcribed counterclockwise. The large single copy region (LSC) and the small single copy region (SSC) are separated by two inverted repeats (IRa and IRb). Asterisks indicate intron containing genes. Pseudogenes are marked by Ψ.
Table 1. Gene contents of plastidome of Cistanche deserticola compared to Nicotiana tabacum and other parasitic plants (based on this research and previous reports [6], [9], [67]).
| Photosynthesis and energyproduction genes | RibosomalRNA genes | TransferRNA genes | |||||||||||||||||||||||||||
| Nt | Cr | Co | Cg | Ce | Cd | Ev | Rg | Nn | Nt | Cr | Co | Cg | Ce | Cd | Ev | Rg | Nn | Nt | Cr | Co | Cg | Ce | Cd | Ev | Rg | Nn | |||
| atpA | • | • | • | • | • | Ψ | Ψ | • | Ψ | rrn16 | • | • | • | • | • | • | • | • | • | trnA-ugc | • | • | ○ | Ψ | • | • | Ψ | ○ | • |
| atpB | • | • | • | • | • | Ψ | Ψ | ○ | Ψ | rrn23 | • | • | • | • | • | • | • | • | • | trnC-gca | • | • | • | • | • | • | Ψ | • | • |
| atpE | • | • | • | • | • | Ψ | ○ | ○ | Ψ | rrn4.5 | • | • | • | • | • | • | • | • | • | trnD-guc | • | • | • | • | • | • | • | • | • |
| atpF | • | • | • | • | • | Ψ | ○ | ○ | ○ | rrn5 | • | • | • | • | • | • | • | • | • | trnE-uuc | • | • | • | • | • | • | • | • | • |
| atpH | • | • | • | • | • | ○ | ○ | ○ | ○ | trnF-gaa | • | • | • | • | • | • | • | • | • | ||||||||||
| atpI | • | • | • | • | • | ○ | ○ | ○ | Ψ | trnfM-cau | • | • | • | • | • | • | • | • | • | ||||||||||
| ndhA | • | ○ | ○ | ○ | ○ | ○ | ○ | ○ | Ψ | trnG-gcc | • | • | • | • | • | • | ○ | ○ | • | ||||||||||
| ndhB | • | Ψ | ○ | ○ | Ψ | Ψ | Ψ | ○ | Ψ | RNA polymerase and intron maturase genes | trnG-ucc | • | • | ○ | Ψ | • | • | ○ | ○ | • | |||||||||
| ndhC | • | ○ | ○ | ○ | ○ | ○ | ○ | ○ | Ψ | trnH-gug | • | • | • | • | • | • | • | ○ | • | ||||||||||
| ndhD | • | ○ | ○ | ○ | Ψ | ○ | ○ | ○ | ○ | Nt | Cr | Co | Cg | Ce | Cd | Ev | Rg | Nn | trnI-cau | • | • | • | • | • | • | • | • | • | |
| ndhE | • | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | trnI-gau | • | • | ○ | Ψ | • | • | Ψ | ○ | ○ | ||||||||||
| ndhF | • | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | matK | • | • | ○ | ○ | • | • | • | ○ | Ψ | trnK-uuu | • | ○ | ○ | ○ | ○ | • | ○ | ○ | • |
| ndhG | • | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | rpoA | • | • | Ψ | ○ | • | Ψ | Ψ | ○ | ○ | trnL-caa | • | • | • | • | • | • | • | ○ | • |
| ndhH | • | ○ | ○ | ○ | ○ | Ψ | ○ | ○ | Ψ | rpoB | • | • | ○ | ○ | • | Ψ | ○ | ○ | Ψ | trnL-uaa | • | • | • | • | • | • | ○ | Ψ | • |
| ndhI | • | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | rpoC1 | • | • | ○ | ○ | • | ○ | ○ | ○ | ○ | trnL-uag | • | • | • | • | • | • | • | ○ | • |
| ndhJ | • | ○ | ○ | ○ | ○ | ○ | ○ | ○ | Ψ | rpoC2 | • | • | ○ | ○ | • | Ψ | ○ | ○ | Ψ | trnM-cau | • | • | • | • | • | • | • | ○ | • |
| ndhK | • | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | trnN-guu | • | • | • | • | • | • | • | ○ | • | ||||||||||
| petA | • | • | • | • | • | ○ | ○ | ○ | Ψ | trnP-ugg | • | • | • | • | • | • | • | ○ | Ψ | ||||||||||
| petB | • | • | • | • | • | ○ | ○ | ○ | ○ | Ribosomal protein and initiation factor genes | trnQ-uug | • | • | • | • | • | • | • | • | • | |||||||||
| petD | • | • | • | • | • | ○ | ○ | ○ | ○ | trnR-acg | • | • | ○ | Ψ | • | • | • | ○ | • | ||||||||||
| petG | • | • | • | • | • | Ψ | ○ | ○ | Ψ | Nt | Cr | Co | Cg | Ce | Cd | Ev | Rg | Nn | trnR-ucu | • | • | • | • | • | • | Ψ | ○ | • | |
| petL | • | • | • | • | • | ○ | ○ | ○ | Ψ | infA | Ψ | ○ | ○ | ○ | Ψ | • | • | • | • | trnS-gcu | • | • | • | • | • | • | • | ○ | • |
| petN | • | • | • | • | • | ○ | ○ | ○ | ○ | rpl14 | • | • | • | • | • | • | Ψ | • | • | trnS-gga | • | • | • | • | • | • | Ψ | ○ | • |
| psaA | • | • | • | • | • | Ψ | ○ | ○ | Ψ | rpl16 | • | • | • | • | • | • | • | • | • | trnS-uga | • | • | • | • | • | • | • | ○ | • |
| psaB | • | • | • | • | • | Ψ | ○ | Ψ | Ψ | rpl2 | • | • | • | • | • | • | • | • | • | trnT-ggu | • | • | • | • | • | • | ○ | ○ | • |
| psaC | • | • | • | • | • | ○ | ○ | ○ | ○ | rpl20 | • | • | • | • | • | • | • | • | • | trnT-ugu | • | • | • | • | • | • | ○ | ○ | • |
| psaI | • | • | ○ | ○ | • | ○ | ○ | ○ | ○ | rpl22 | • | • | • | • | • | • | ○ | ○ | • | trnV-gac | • | • | • | • | • | • | ○ | ○ | • |
| psaJ | • | • | • | • | • | ○ | ○ | ○ | Ψ | rpl23 | • | Ψ | ○ | ○ | Ψ | Ψ | Ψ | • | • | trnV-uac | • | • | ○ | ○ | • | • | ○ | ○ | • |
| psbA | • | • | • | • | • | Ψ | Ψ | ○ | ○ | rpl32 | • | • | ○ | ○ | • | • | ○ | ○ | • | trnW-cca | • | • | • | • | • | • | • | • | • |
| psbB | • | • | • | • | • | Ψ | Ψ | ○ | ○ | rpl33 | • | • | • | • | • | • | • | Ψ | • | trnY-gua | • | • | • | • | • | • | • | • | • |
| psbC | • | • | • | • | • | Ψ | ○ | ○ | Ψ | rpl36 | • | • | • | • | • | • | • | • | • | ||||||||||
| psbD | • | • | • | • | • | Ψ | ○ | ○ | Ψ | rps11 | • | • | • | • | • | • | • | • | • | Other essential genes | |||||||||
| psbE | • | • | • | • | • | ○ | ○ | ○ | ○ | rps12 | • | • | • | • | • | • | • | Ψ | • | ||||||||||
| psbF | • | • | • | • | • | Ψ | ○ | ○ | ○ | rps14 | • | • | • | • | • | • | • | • | • | Nt | Cr | Co | Cg | Ce | Cd | Ev | Rg | Nn | |
| psbH | • | • | • | • | • | ○ | ○ | ○ | ○ | rps15 | • | • | • | • | • | • | ○ | ○ | • | ClpP | • | • | • | • | • | • | • | • | • |
| psbI | • | • | • | • | • | ○ | ○ | ○ | ○ | rps16 | • | Ψ | ○ | ○ | Ψ | • | ○ | ○ | Ψ | accD | • | • | • | • | • | Ψ | • | • | • |
| psbJ | • | • | • | • | • | Ψ | ○ | ○ | ○ | rps18 | • | • | • | • | • | • | • | • | Ψ | cemA | • | • | • | • | • | ○ | ○ | ○ | ○ |
| psbK | • | • | • | • | • | Ψ | ○ | ○ | Ψ | rps19 | • | • | • | • | • | • | • | • | • | ccsA | ○ | • | • | • | • | Ψ | ○ | ○ | ○ |
| psbL | • | • | • | • | • | Ψ | ○ | ○ | ○ | rps2 | • | • | • | • | • | • | • | • | • | ycf1 | • | • | • | • | • | Ψ | • | • | • |
| psbM | • | • | • | • | • | • | ○ | ○ | Ψ | rps3 | • | • | • | • | • | • | • | • | • | ycf2 | • | • | • | • | • | • | • | • | • |
| psbN | • | • | • | • | • | ○ | ○ | ○ | ○ | rps4 | • | • | • | • | • | • | • | • | • | ycf3 | • | • | • | • | • | Ψ | ○ | ○ | Ψ |
| psbT | • | • | • | • | • | ○ | ○ | ○ | ○ | rps7 | • | • | • | • | • | • | • | • | • | ycf4 | • | • | • | • | • | Ψ | ○ | ○ | ○ |
| rbcL | • | • | • | • | • | Ψ | Ψ | ○ | Ψ | rps8 | • | • | • | • | • | • | • | • | • | ycf15 | • | Ψ | ○ | Ψ | Ψ | • | Ψ | ○ | ○ |
Nt, Nicotiana tabacum; Cr, Cuscuta reflexa; Co, Cuscuta obtusiflora; Cg, Cuscuta gronovii; Ce, Cuscuta exaltata; Cd, Cistanche deserticola; Ev, Epifagus virginiana; Rg, Rhizanthella gardneri; Nn, Neottia nidus-avis; Ψ, pseudogene; •, present; ○, missing.
Table 2. Global features of Cistanche deserticola plastidome compared to Nicotiana tabacum and other parasitic plants.
| Species | Accession no | Size (nt) | Numberof genesa | Number of pseudogenes | Number of tRNAsb | number of genes with introns | Reference |
| Rhizanthella gardneri | GQ413967 | 59,190 | 37 | 6 | 9 | 4 | [9] |
| Epifagus virginiana | NC_001568 | 70,028 | 53 | 18 | 17 | 4 | [5] |
| Cuscuta obtusiflora | NC_009949 | 85,280 | 98 | 1 | 24 | 5 | [7] |
| Cuscuta gronovii | AM711639 | 86,744 | 99 | 5 | 24 | 5 | [6] |
| Neottia nidus-avis | JF325876 | 92,060 | 74 | 28 | 28 | 8 | [10] |
| Cistanche deserticola | KC128846 | 102,657 | 77 | 32 | 30 | 14 | this study |
| Cuscuta reflexa | AM711640 | 121,521 | 115 | 4 | 29 | 16 | [6] |
| Cuscuta exaltata | NC_009963 | 125,373 | 112 | 8 | 29 | 16 | [7] |
| Nicotiana tabacum | NC_001879 | 155,939 | 151 | 1 | 30 | 23 | [68] |
Data in this table came from analyses using sequences on GenBank or plastid chromosomes gene maps in their original report.
Number of genes excluding the pseudogenes. Duplicated genes in the IR regions were counted twice.
Duplicated tRNAs were counted both.
When the IR is considered only once, the cp genome of C. deserticola contains 60 genes, encoding 27 proteins, 4 ribosomal RNAs (rRNA) and 30 transfer RNAs (tRNA). The positions of 61 genes, including 29 unique and 16 duplicated ones in the IRs regions, were localized on the map (Fig. 1). The cp genome of C. deserticola has an overall GC content of 36.8%, which is similar to E. virginiana (36%) but slightly lower than the photosynthetic species Nicotiana tabacum (37.8%). Like other land plants [49], [50], GC content is unevenly distributed across the C. deserticola cp genome. The highest GC content is in the IRs (43%), reflecting the high GC content of rRNA genes, and the lowest is in the SSC (27.5%) region.
Although C. deserticola has a relatively larger cp genome sequence, it also exhibited severe physiological reductions: all genes required for photosynthesis (encoding photosystem I and II components, cytochrome b6f complex, NAD(P)H dehydrogenase, photosystem assembly factors (ycf3, ycf4) and ATP synthase) suffered gene losses and pseudogenizations except for psbM. Additional pseudogenization is also seen in genes encoding cp-encoded RNA polymerase (rpo), Cytochrome c biogenesis protein (ccsA), and Acetyl-CoA carboxylase (accD). C. deserticola retains many genes of the translation machinery, including 8 rpl genes, 11 rps genes, and an initiation factor, infA. Only rpl23 is apparently a pseudogene with nonfunctional reading frames (Table 1).
In E. virginiana, a total of five tRNAs were pseudogenized and eight tRNAs were lost [51]. In contrast, C. deserticola retains almost all the rRNA and tRNA genes: two identical copies of rRNA gene clusters (16S-23S-4.5S-5S) were found in the IR regions; 30 different tRNAs, which can recognize all 61 codons present in the cp genes, were identified (Table 1, Fig. 1). Intron content of genes retained in the C. deserticola cp genome is conserved with other seed plants: it has 11 genes with introns, six in tRNAs and 5 in protein coding genes. Two of the 12 intron-containing genes have a single intron and two genes, clpP and rps12, have two introns. All of these belong to the group II intron, whereas trnL-UAA is the only group I intron. Among the tRNA genes, trnK-UUU has a special role, since the only RNA maturase gene (matK) found on the cp genome was located in its intron [52]. Unlike other parasitic plants, C. deserticola harbours the complete trnK-UUU gene, including its intron matK gene (Table 1, Fig. 1).
The examination of pairwise dN/dS ratio for the alignable genes shared between C. deserticola, E. virginiana and their autotrophic relatives demonstrates that most of genes are under greater constraint in fully nonphotosynthetic E. virginiana and C. deserticola than photosynthetic species O. europaea and N. tabacum (Fig. 2). In addition, of 15 protein-coding genes shared between E.virginiana and C. deserticola, 13 genes have a higher dN/dS in E.virginiana than in C. deserticola (Fig. 2).
Figure 2. Pairwise dN/dS value of C. deserticola (Cd), E. virginiana (Ev) and N. tabacum (Nt) vs. O. europaea (Oe) for all shared protein-coding genes.
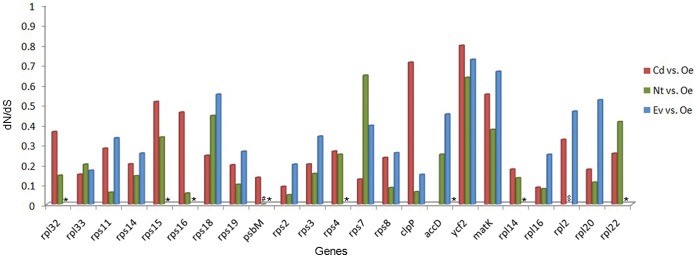
*indicates gene lost, #indicates the absence of non synonymous substitutions, $indicates the absence of synonymous substitutions.
Horizontal Transfer of rpoC2 from Host to Parasitic Plants
We used the general primers of rpoC2 to amplify the total DNA of C. deserticola and subsequent cloning. Contrary to our expectation, the sequences obtained resemble the genes of H. ammodendron but not C. deserticola, based on sequence similarity through BLAST and phylogenetic trees (GenBank number: KC543998, Fig. 3). This raised the possibility that HGT may have occurred between the parasite and its host. In order to confirm this result, we ruled out that the results were due to contamination or mixing-up of templates by repeating the experiment in a different laboratory. The results of the amplification are congruent with the previous results. Then, we confirmed the presence of the H. ammodendron type copy in 46 accessions out of 50 samples from five C. deserticola populations by using the same specific primers. Four of the accessions’ lack of amplification was probably due to poor DNA quality. The transferred rpoC2 copy (H. ammodendron type) amplified from C. deserticola is about 1050 bp, and covers amino acid positions 98–443 (nucleotide positions 294–1329) of the rpoC2 gene of O. europea. To clarify the evolutionary characteristics of the rpoC2 fragment transferred from H. ammodendron to C. deserticola, we aligned the nucleotide sequences of H. ammodendron type rpoC2 amplified from both of two plants with intact open reading frames of other related species. The results indicated that the transferred rpoC2 fragment differed from functional copies in a few point mutations and one key nucleotide insertion (C in 927 bp), which resulted in several subsequent premature termination codons and frame shifts mutations (Fig. 4). Because the H. ammodendron type rpoC2 was not found in the complete cp genome of C. deserticola, we speculated it should be transferred into the nuclear or mitochondrial genome.
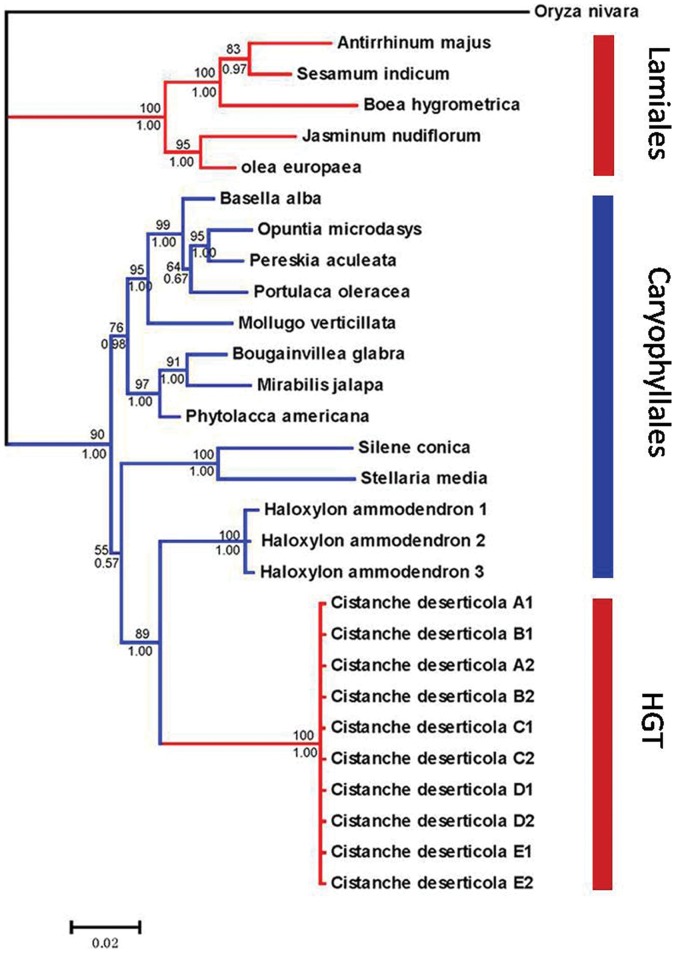
Figure 3. Phylogenetic evidence for horizontal gene transfer of the plastid rpoC2 from Haloxylon ammodendron to Cistanche deserticola.
Lamiales are coloured in red, and Caryophyllales are coloured in blue. While the ten C. deserticola sequences involved in horizontal gene transfer are coloured in red. Numbers at nodes are posterior probabilities >0.60 and maximum likelihood bootstrap values >60. The Genebank number: Oryza nivara, NC_005973; Antirrhinum indicum, GQ997028; Sesamum indicum, NC_016433; Boea hygrometrica, NC_016468; Jasminum nudiflorum, NC_008407; Olea europaea, NC_013707; Basella alba, HQ843359; Opuntia microdasys, HQ843375; Pereskia aculeata, HQ843376; Portulaca oleracea, HQ843380; Mollugo verticillata, HQ843373; Bougainvillea glabra, HQ843360; Mirabilis jalapa, HQ843372; Phytolacca americana, HQ843378; Celosia cristata, HQ843361; Spinacia oleracea, NC_002202; Silene conica, NC_016729; Stellaria media, HQ843386; Cistanche deserticola (HGT), KC543998.
Figure 4. Position of inserted cytosine within the transferred rpoC2 gene.
(A) Alignment of the nucleotide sequences of transferred rpoC2 gene amplified from parasite and host with intact open reading frames of other related species. The inserted cytosine was labeled with colored vertical lines. (B) Inserted cytosine resulted in followed premature termination codon in the transferred rpoC2 in Cistanche deserticola.
However, C. deserticola’s own rpoC2 copies were not detected by PCR amplification using specific primers, which make us consider that this gene was lost or turned out to be a pseudogene. Thus, we searched the finished cp genome of C. deserticola with rpoC2 homologues by the BLAST method. The results shown that C. deserticola also retains its own significant shortened rpoC2, which has turned out to be a pseudogene of only 439 bp.
Phylogenetic Analysis of Transferred rpoC2 Gene
The HGT result was further supported by our phylogenetic analysis. Maximum likelihood and Bayesian trees constructed using the two methods described earlier gave congruent results (Fig. 3). The two orders Lamiales and Caryophyllales confirmed as well as supported distinct clades in the phylogenetic tree. The transferred rpoC2 is located in the clade Caryophyllales (host clade) but does not cluster inside Lamiales (parasitic clade), which forms a clade with a relatively strong bootstrap support. The retained rpoC2 (C. deserticola type copy) was not used in this analysis because its sequence was severely fragmented when align with other homologs. The sequence alignment and the phylogenetic distribution of the rpoC2 in Chenopodiaceae suggest that the horizontal gene transfer happened between the host H. ammodendron and parasitic plant C. deserticola.
Discussion
Gene Losse in the cp Genome of C. deserticola
Compared to more than 250 completely sequenced cp genomes of photosynthetic plants, the number of fully sequenced cp genomes of non-photosynthetic plants is very small. To date, only eight heterotrophic species, exhibiting parasitic lifestyles and having strongly reduced cp genomes, have been thoroughly investigated with respect to their cp genome sequences [5]. In this study, we have sequenced the cp genome of C. deserticola, a holoparasitic species from Orobanchaceae with the expectation that comparison of cp genomic features between these two relatives will provide further insights on parasitic cp genome evolution.
The overlapping PCR products have indicated the reduced circular form of the cp chromosomes in C. deserticola. Similar to E. virginiana, almost all of its photosynthetic genes have been lost or have become pseudogenes after the loss of a major metabolic function. It is different from other heterotrophic plants in many ways: it retains almost all the tRNA genes; the photosynthetic gene psbM remains as residues and others suffered gene pseudogenizations rather than losses as E. virginiana; C. deserticola harbours complete trnK-UUU gene but not its intron matK gene, and so on.
Some parasitic species exhibit extensive losses of tRNA genes (Table 1). In E. virginiana, a total of 13 tRNAs were pseudogenized or lost. As in photosynthetic plants, C. deserticola encompasses around 30 tRNA genes in cp genomes, and it is the only one of parasitic plants which possessed a full cp tRNA set as nonparasitic plants. This suggests that the loss of the transfer RNA genes from the cp genome occurred later than those of photosynthesic genes.
Most of the splicing factors are nuclear-encoded, but one maturase protein is encoded by a cp gene, matK, which was located within an intron of trnK-UUU [52], [53]. The trnK gene is lost in all parasitic angiosperm cp genomes except for C. deserticola and Neottia nidus-avis (Table 1). In the Neottia cp genome, the intron matK is a pseudogene with strong divergence of its 5′end compared to other photosynthetic orchids [10]. In contrast, in C. reflexa, C. exaltata, and E. virginiana, matK has been retained as a free-standing gene [52], [54]. Unlike other parasitic angiosperm species, neither the trnK-UUU gene nor its intron matK gene was missing in C. deserticola. It has been reported that matK is also needed for splicing other chloroplast group II introns in the cp genome [55]. Thus the retaining of matK in C. deserticola is not surprising because its cp genome has retained 9 group IIa introns (including rpl2, rpl16, rps12, clpP, trnA-UGC, trnI-GAU, trnK-UUU, trnG-UCC, trnV-UAC). While the trnK gene exists in the C. deserticola cp genome, which was similar to photosynthetic plants, this may suggest the plant has undergone fewer losses, either due to a function of reduced level of holoparasitism, or a recent switch to this life history [56].
The entire set of chloroplast NAD (P) H dehydrogenase consisting of 11 genes has been lost or turned into pseudogenes without exception in C. deserticola. What is interesting is that a loss of ndh genes was also present in all sequenced cp genomes of parasitic plants investigated to date, regardless of the degree of evolutionary degradation of photosynthetic capacity (Table 1). It was confirmed that cp-encoded ndh genes were first lost in the transition to heterotrophy [7], [57]. It has been speculated that the condensation of the genome by loss of many non-coding regions and unimportant parts of the cp genome is an early reaction of the cp genome to the parasitic lifestyle [6].
After calculating dN/dS for shared cp genes between E. virginiana, C. deserticola and two photosynthetic species, an obvious trend of relaxed selection was revealed in both fully nonphotosynthetic species with higher dN/dS. It may indicate that these genes were suffering an initial stage of pseudogenization. However, C. deserticola has lower dN/dS for more genes than E. virginiana, which suffered a high degree of gene loss and pseudogenization, further indicating C. deserticola may undergo reduced level of holoparasitism or a recent switch to this life history. The gene psbM, which was the only one photosynthetic gene retained in C. deserticola, showed a higher dN/dS than in photosynthetic species (dN/dS = 0), suggesting advent of relaxed selection and initial stage of pseudogenization in this gene in C. deserticola. However, some unexpected high dN/dS were also found in rpl33, rps7 and rpl22 in photosynthetic species. The short length of sequences may reduce the reliability of dN/dS estimation in these genes [58].
HGT from H. ammodendron to C. deserticola
HGT in parasitic systems has been detected by using phylogenetic trees when a DNA sequence obtained from a parasite is placed closer to its host rather than with its closest relatives. Unexpectedly we had a windfall in the process of amplifying the cp genome sequence of C. deserticola. One of these sequences, rpoC2 gene, was present in two copies within this parasite and one of them was a homolog of their host and led to conflicting phylogenies. The most reasonable explanation for our results is that cp rpoC2 gene in C. deserticola was acquired from its host, H. ammodendron via HGT. In order to confirm the results and provide special opportunities for studying the evolutionary dynamics of HGT at the population level, we also collected 50 samples from five populations and successfully amplified transferred the rpoC2 gene from 46 accessions. In addition, the events present in most individuals spanning Xinjiang and Inner Mongolia, may suggest that the HGT of rpoC2 probably occurred in a C. deserticola common ancestor of these populations, which expanded into its present wide distribution quickly.
So far, the incidence of HGT in the family Orobanchaceae is high, including one nuclear HGT event which occurred between parasitic Striga hermonthica (Orobanchaceae) and its host Sorghum bicolor (Poaceae), as well as a chloroplast region including rps2, trnL-F, and rbcL in a group of non-photosynthetic members (Orobanche and Phelipanche) of Orobanchaceae [29], [59]. Our study shows that cp rpoC2 has transferred from H. ammodendron to C. deserticola via HGT. However, it is impossible to presume the localization of the transferred rpoC2 based on the available data. We just could rule out its location in the cp genome according to our completed cp genome of C. deserticola. This agrees with the reports that events of foreign DNA transferred into the cp genome are rare [60], [61]. The possibility of disparity between plant mitochondrial and nuclear genomes vs. cp genomes in rates of HGT is that the mitochondrion and nuclear genomes contain much more non-coding DNA than compact cp genomes [62], [63].
As desert plants, H. ammodendron and C. deserticola have developed an extremely specialized set of morphological, biochemical and molecular traits to adapt scare nutrients and water in the soil, such as loss of leaves and the development of haustoria in C. deserticola. With this feeding organ, C. deserticola can extract water and nutrients from the parasitized host, including the nucleic acids in the form of mRNA. It is why HGT appears to be facilitated by the direct physical association between the parasite and its host in the parasitic systems.Moreover, C. deserticola is a typical root parasite, meaning they are usually in contact with its host through meristems. In plants, meristems are less protected than the germlines in most multicellular animals [22]. Therefore, the genes, which transferred to the root apical meristem, could have the opportunity to be integrated in the genome and transmitted to the next generation.
In our study, either the transferred rpoC2 or its native copy appear to be non-functional pseudogenes in C. deserticola. Previous work has reported plant mtDNA pseudogenes that are transcribed and edited, so this raises the possibility that some of these genes may actually be functional [64], [65]. The fact is that acquiring a new gene can lead to an obvious benefit to living in that particular environment. H. ammodendron, which is distributed across dry deserts and salt pans, has high tolerance to osmotic and salt stress [66]. We postulate that C. deserticola could not only obtain the carbohydrate, minerals and water, but also the straightforward source of useful genetic information from the neighbour already adapted to that environment. In the ‘genomic era’, future work is still needed to discover more HGT events in this pair of host and parasite by next generation sequencing, especially genes in mitochondrial and nuclear genomes.
Acknowledgments
The authors would like to thank Yiyao Hu and Jiaqi Wu for their helpful suggestions.
Funding Statement
This work is supported by the National Science Foundation of China [30925004, 91131901 to Zhong Y; 31070197 to Li JQ]; and China Postdoctoral Science Foundation [20100480550, 2011M500538 to Zhang TC and Qiao Q]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Raubeson LA, Jansen RK (2005) Chloroplast genomes of plants. In: Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Henry RJ (Ed.). CABI Publishing, London.
- 2.Krause K (2012) Plastid genomes of parasitic plants: a trail of reductions and losses. In: Organelle genetics. Bullerwell C (Ed.). Springer, Berlin.
- 3.Jansen RK, Ruhlman TA (2012) Plastid Genomes of Seed Plants. In: Genomics of Chloroplasts and Mitochondria, Advances in Photosynthesis and Respiration. Bock R, Knoop V (Eds.). Springer, Berlin.
- 4. Barkman TJ, McNeal JR, Lim SH, Coat G, Croom HB, et al. (2007) Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol 7: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wolfe KH, Morden CW, Palmer JD (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Nat Acad Sci USA 89: 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Funk HT, Berg S, Krupinska K, Maier UG, Krause K (2007) Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii . BMC Plant Biol 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McNeal JR, Kuehl JV, Boore JL, De Pamphilis CW (2007) Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta . BMC Plant Biol 7: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wickett NJ, Zhang Y, Hansen SK, Roper JM, Kuehl JV, et al. (2008) Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis . Mol Biol Evol 25: 393–401. [DOI] [PubMed] [Google Scholar]
- 9. Delannoy E, Fujii S, des Francs-Small CC, Brundrett M, Small I (2011) Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol 28: 2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Logacheva MD, Schelkunov MI, Penin AA (2011) Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis . Genome Biol Evol 3: 1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolfe AD, dePamphilis CW (1998) The effect of relaxed functional constraints on the photosynthetic gene rbcL in photosynthetic and nonphotosynthetic parasitic plants. Mol Biol Evol 15: 1243–1258. [DOI] [PubMed] [Google Scholar]
- 12.Oxelman B, Kornhall P, Olmstead RG, Bremer B (2005) Further disintegration of Scrophulariaceae. Taxon: 411–425.
- 13. Wolfe AD, Randle CP, Liu L, Steiner KE (2005) Phylogeny and biogeography of Orobanchaceae. Folia Geobot 40: 115–134. [Google Scholar]
- 14. Bennett JR, Mathews S (2006) Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am J Bot. 93: 1039–1051. [DOI] [PubMed] [Google Scholar]
- 15.Nickrent DL (2003) The parasitic plant connection website. Available: http://www.parasiticplants.siu.edu/. Accessed 2012 Dec 1.
- 16.Zhang Z, Tzvelev N (1990) Orobanchaceae. In: Flora reipublicae popularis sinicae. Vol 69, Science Press, Beijing.
- 17. Olmstead RG, Wolfe AD, Young ND, Elisons WJ, Reeves PA (2001) Disintegration of the Scrophulariaceae. Am J Bot 88: 348–361. [PubMed] [Google Scholar]
- 18. Lin LW, Hsieh MT, Tsai FH, Wang WH, Wu CR (2002) Anti-nociceptive and anti-inflammatory activity caused by Cistanche deserticola in rodents. J Ethnopharmacol 83: 177–182. [DOI] [PubMed] [Google Scholar]
- 19. Wang X, Qi Y, Cai R, Li X, Yang M, et al. (2009) The effect of Cistanche deserticola polysaccharides (CDPS) on marcrophages activation. Chin Pharmacol Bull 25: 787–790. [Google Scholar]
- 20. Westwood JH, Yoder JI, Timko MP, dePamphilis CW (2010) The evolution of parasitism in plants. Trends Plant Sci 15: 227–235. [DOI] [PubMed] [Google Scholar]
- 21. Keeling PJ, Palmer JD (2008) Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9: 605–618. [DOI] [PubMed] [Google Scholar]
- 22. Bock R (2010) The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci 15: 11–22. [DOI] [PubMed] [Google Scholar]
- 23. Gray MW (1993) Origin and evolution of organelle genomes. Curr Opin Genet Dev 3: 884–890. [DOI] [PubMed] [Google Scholar]
- 24. Gray MW (2012) Mitochondrial evolution. Cold Spring Harb Perspect Biol 4(9): a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bergthorsson U, Adams KL, Thomason B, Palmer JD (2003) Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424(6945): 197–201. [DOI] [PubMed] [Google Scholar]
- 26. Olson LE, Yoder AD (2002) Using secondary structure to identify ribosomal numts: cautionary examples from the human genome. Mol Biol Evol 19: 93–100. [DOI] [PubMed] [Google Scholar]
- 27. Wolfe AD, Randle CP (2004) Recombination, heteroplasmy, haplotype polymorphism, and paralogy in plastid genes: implications for plant molecular systematics. Syst Bot 29: 1011–1020. [Google Scholar]
- 28. Woloszynska M, Bocer T, Mackiewicz P, Janska H (2004) A fragment of chloroplast DNA was transferred horizontally, probably from non-eudicots, to mitochondrial genome of Phaseolus . Plant Mol Biol 56: 811–820. [DOI] [PubMed] [Google Scholar]
- 29. Park JM, Manen JF, Schneeweiss GM (2007) Horizontal gene transfer of a plastid gene in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae). Mol Phylogenet Evol 43: 974–985. [DOI] [PubMed] [Google Scholar]
- 30. Mower JP, Stefanovi, cacute S, Young GJ, Palmer JD (2004) Plant genetics: gene transfer from parasitic to host plants. Nature 432: 165–166. [DOI] [PubMed] [Google Scholar]
- 31. Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA (2007) Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19: 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carlsson J, Leino M, Glimelius K (2007) Mitochondrial genotypes with variable parts of Arabidopsis thaliana DNA affect development in Brassica napus lines. Theor Appl Genet 115: 627–641. [DOI] [PubMed] [Google Scholar]
- 33. Grivet D, Heinze B, Vendramin G, Petit R (2005) Genome walking with consensus primers: application to the large single copy region of chloroplast DNA. Mol Ecol Notes 1: 345–349. [Google Scholar]
- 34. Heinze B (2007) A database of PCR primers for the chloroplast genomes of higher plants. Plant Methods 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255. [DOI] [PubMed] [Google Scholar]
- 36. Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32: 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gautheret D, Lambert A (2001) Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J Mol Biol 313: 1003–1004. [DOI] [PubMed] [Google Scholar]
- 38. Conant GC, Wolfe KH (2008) GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24: 861–862. [DOI] [PubMed] [Google Scholar]
- 39. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 41. Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bremer B, Bremer K, Chase M, Fay M, Reveal J, et al. (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161(2): 105–121. [Google Scholar]
- 43. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98. [Google Scholar]
- 44.Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony, version 4.0 b10.
- 45. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 46. Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 47. Akaike H (1974) A new look at the statistical model identification. IEEE Transactions on. Automatic Control 19(6): 716–723. [Google Scholar]
- 48.Nylander J (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- 49. Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, et al. (2006) Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol 6: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shimda H, Sugiuro M (1991) Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res 19: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wolfe KH, Mordent CW, Ems SC, Palmer JD (1992) Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J Mol Evol 35: 304–317. [DOI] [PubMed] [Google Scholar]
- 52. Vogel J, Hübschmann T, Börner T, Hess WR (1997) Splicing and intron-internal RNA editing of trnK-matK transcripts in barley plastids: support for matK as an essential splice factor. J Mol Biol 270: 179–187. [DOI] [PubMed] [Google Scholar]
- 53. Asakura Y, Barkan A (2006) Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol 142: 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hess WR, Müller A, Nagy F, Börner T (1994) Ribosome-deficient plastids affect transcription of light-induced nuclear genes: genetic evidence for a plastid-derived signal. Mol Gen Genet 242: 305–312. [DOI] [PubMed] [Google Scholar]
- 55. Zoschke R, Nakamura M, Liere K, Sugiura M, Börnera T, et al. (2010) An organellar maturase associates with multiple group II introns. Proc Nat Acad Sci USA 107: 3245–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krause K (2008) From chloroplasts to “cryptic” plastids : evolution of plastid genomes in parasitic plants. Curr Genet 54: 111–121. [DOI] [PubMed] [Google Scholar]
- 57. Martín M, Sabater B (2010) Plastid ndh genes in plant evolution. Plant Physiol Biochem 48: 636–645. [DOI] [PubMed] [Google Scholar]
- 58. Haddrill PR, Waldron FM, Charlesworth B (2008) Elevated levels of expression associated with regions of the Drosophila genome that lack crossing over. Biol Lett 4: 758–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Davis CC, Wurdack KJ (2004) Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science 305: 676–678. [DOI] [PubMed] [Google Scholar]
- 60. Haberle RC, Fourcade HM, Boore JL, Jansen RK (2008) Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol 66: 350–361. [DOI] [PubMed] [Google Scholar]
- 61. Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK (2010) Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol 70: 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koulintchenko M, Konstantinov Y, Dietrich A (2003) Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J 22: 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature Rev Genet 5: 123–135. [DOI] [PubMed] [Google Scholar]
- 64. Quiñones V, Zanlungo S, Moenne A, Gómez I, Holuigue L, et al. (1996) The rpl5-rps 14-cob gene arrangement in Solanum tuberosum: rps14 is a transcribed and unedited pseudogene. Plant Mol Biol 31: 937–943. [DOI] [PubMed] [Google Scholar]
- 65. Kim DH, Kim BD (2006) The organization of mitochondrial atp6 gene region in male fertile and CMS lines of pepper (Capsicum annuum L.). Curr Genet 49: 59–67. [DOI] [PubMed] [Google Scholar]
- 66. Tobe K, Li X, Omasa K (2000) Effects of sodium chloride on seed germination and growth of two Chinese desert shrubs, Haloxylon ammodendron and H. persicum (Chenopodiaceae). Aust J Bot 48: 455–460. [Google Scholar]
- 67. Krause K (2011) Piecing together the puzzle of parasitic plant plastome evolution. Planta 234: 647–656. [DOI] [PubMed] [Google Scholar]
- 68. Wakasugi T, Sugita M, Tsudzuki T, Sugiura M (1998) Updated gene map of tobacco chloroplast DNA. Plant Mol Biol Rep 16: 231–241. [Google Scholar]