Abstract
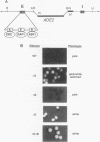
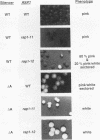
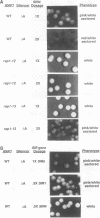
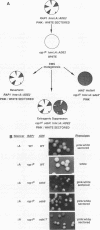
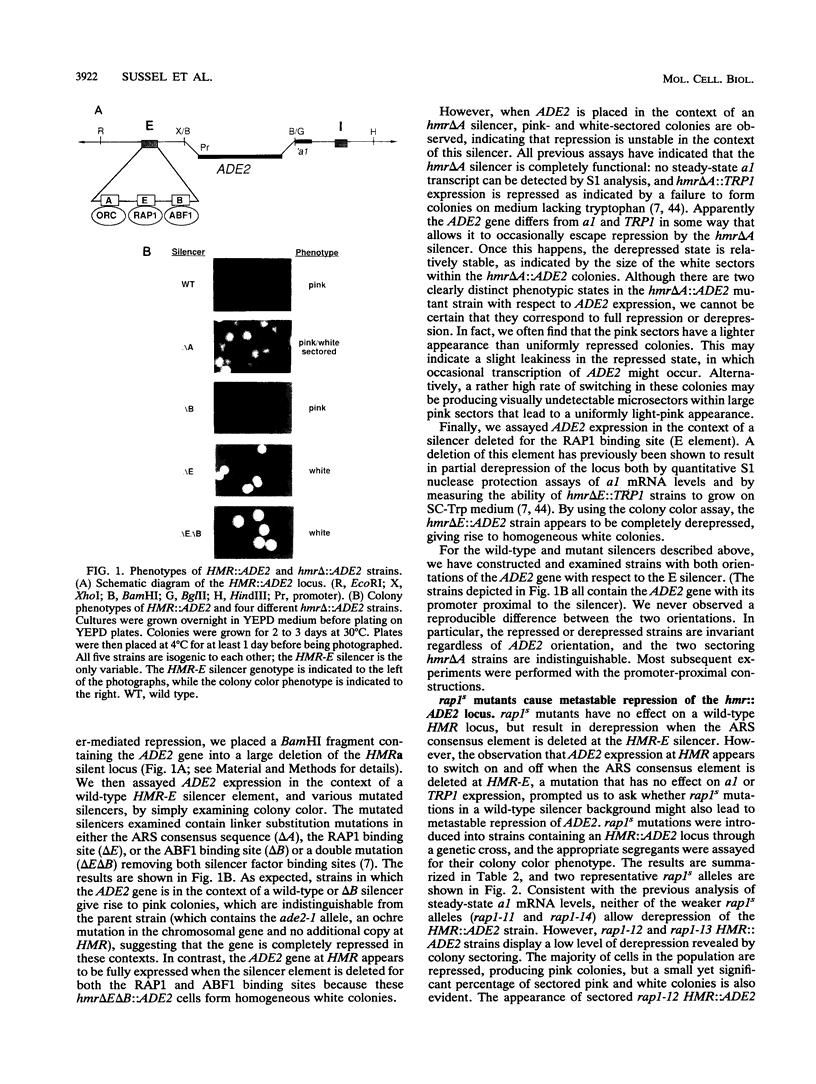
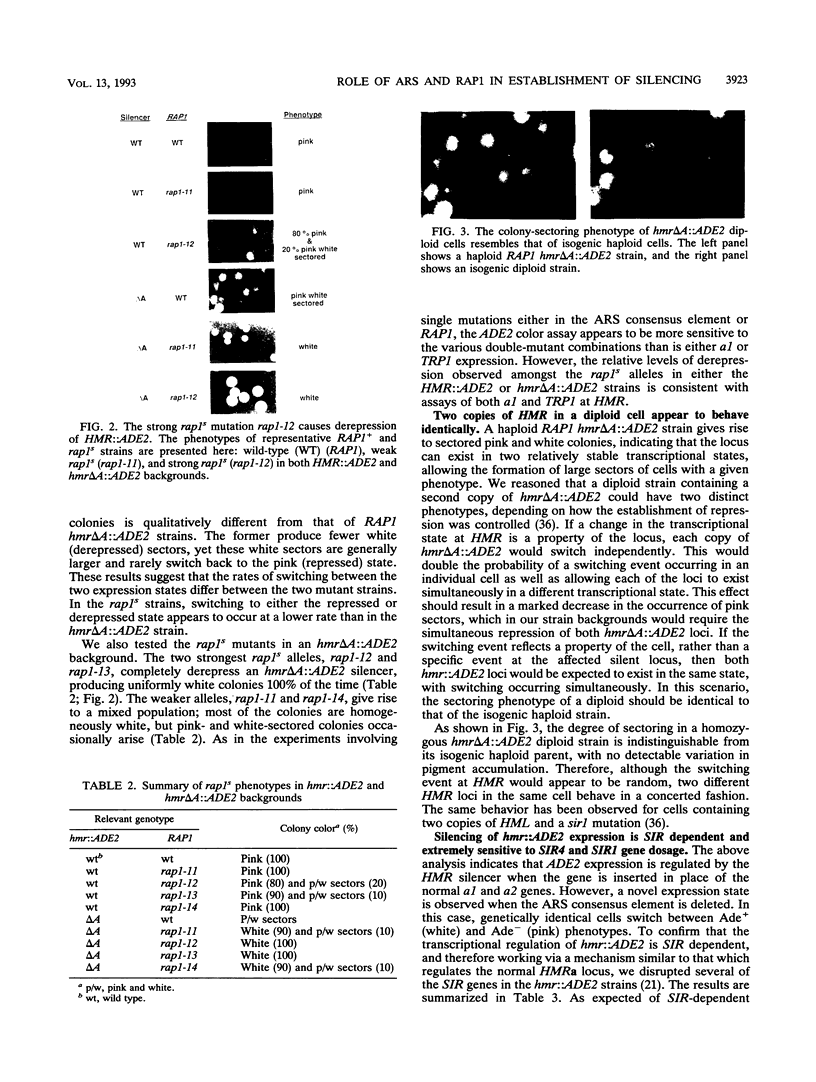
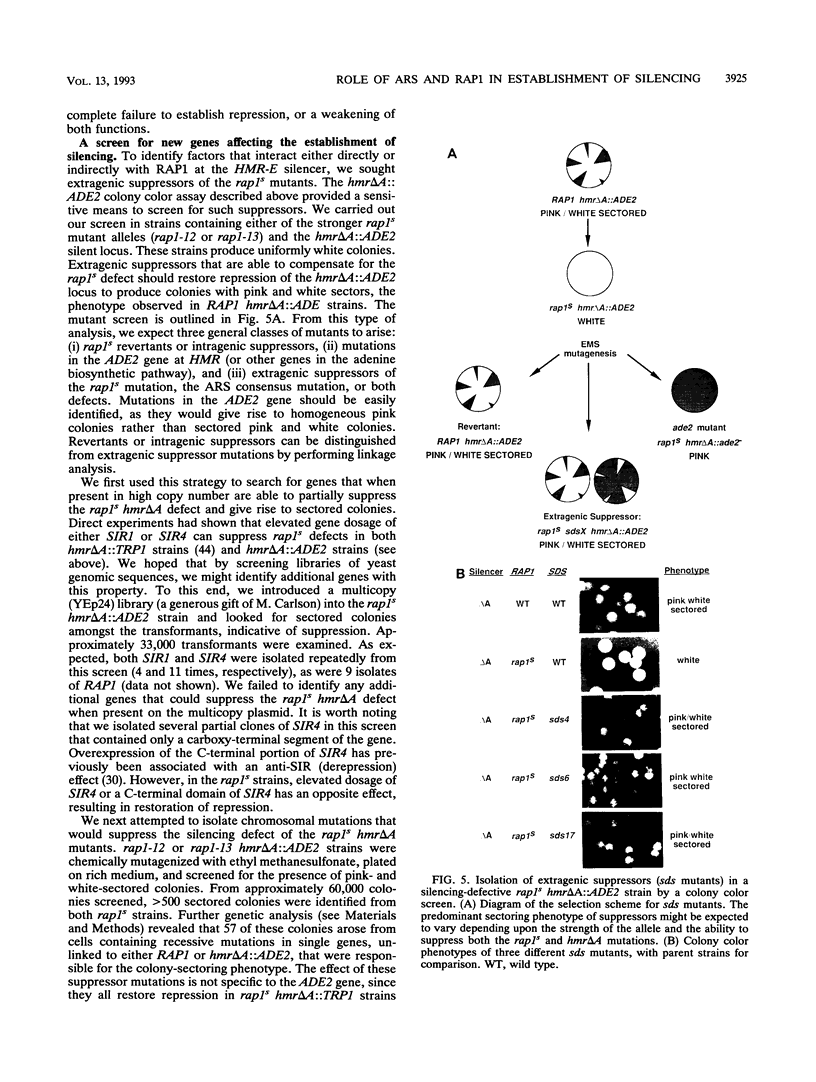
In this study, we used the ADE2 gene in a colony color assay to monitor transcription from the normally silent HMR mating-type locus in Saccharomyces cerevisiae. This sensitive assay reveals that some previously identified cis- and trans-acting mutations destabilize silencing, causing genetically identical cells to switch between repressed and derepressed transcriptional states. Deletion of the autonomously replicating sequence (ARS) consensus element at the HMR-E silencer or mutation of the silencer binding protein RAP1 (rap1s) results in the presence of large sectors within individual colonies of both repressed (Ade-, pink) and derepressed (Ade+, white) cells. These results suggest that both the ARS consensus element and the RAP1 protein play a role in the establishment of repression at HMR. In diploid cells, the two copies of HMR appear to behave identically, suggesting that the switching event, though apparently stochastic, reflects some property of the cell rather than a specific event at each HMR locus. In the ADE2 assay system, silencing depends completely upon the function of the SIR genes, known trans-acting regulators of the silent loci, and is sensitive to the gene dosage of two SIR genes, SIR1 and SIR4. Using the ADE2 colony color assay in a genetic screen for suppressors of rap1s, silencer ARS element deletion double mutants, we have identified a large number of genes that may affect the establishment of repression at the HMR silent mating-type locus.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J., Nasmyth K. A., Strathern J. N., Klar A. J., Hicks J. B. Regulation of mating-type information in yeast. Negative control requiring sequences both 5' and 3' to the regulated region. J Mol Biol. 1984 Jul 5;176(3):307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- Aparicio O. M., Billington B. L., Gottschling D. E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991 Sep 20;66(6):1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Axelrod A., Rine J. A role for CDC7 in repression of transcription at the silent mating-type locus HMR in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):1080–1091. doi: 10.1128/mcb.11.2.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Breeden L., Abraham J., Sternglanz R., Nasmyth K. Characterization of a "silencer" in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985 May;41(1):41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A., Tsang J. S., Stanway C., Kingsman A. J., Kingsman S. M. Transcriptional control of the Saccharomyces cerevisiae PGK gene by RAP1. Mol Cell Biol. 1989 Dec;9(12):5516–5524. doi: 10.1128/mcb.9.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams C. D., Norris D., Osley M. A., Fassler J. S., Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988 Feb;2(2):150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- Dubey D. D., Davis L. R., Greenfeder S. A., Ong L. Y., Zhu J. G., Broach J. R., Newlon C. S., Huberman J. A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol Cell Biol. 1991 Oct;11(10):5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman J. B., Hicks J. B., Broach J. R. Identification of sites required for repression of a silent mating type locus in yeast. J Mol Biol. 1984 Oct 5;178(4):815–834. doi: 10.1016/0022-2836(84)90313-9. [DOI] [PubMed] [Google Scholar]
- Floor E. Interaction of morphogenetic genes of bacteriophage T4. J Mol Biol. 1970 Feb 14;47(3):293–306. doi: 10.1016/0022-2836(70)90303-7. [DOI] [PubMed] [Google Scholar]
- Giesman D., Best L., Tatchell K. The role of RAP1 in the regulation of the MAT alpha locus. Mol Cell Biol. 1991 Feb;11(2):1069–1079. doi: 10.1128/mcb.11.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990 Nov 16;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Hardy C. F., Sussel L., Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992 May;6(5):801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985 Oct;42(3):913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Ivy J. M., Klar A. J., Hicks J. B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne P. S., Kim U. J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Kimmerly W. J., Rine J. Replication and segregation of plasmids containing cis-acting regulatory sites of silent mating-type genes in Saccharomyces cerevisiae are controlled by the SIR genes. Mol Cell Biol. 1987 Dec;7(12):4225–4237. doi: 10.1128/mcb.7.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988 Jul;7(7):2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991 Apr;5(4):616–628. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- Laurenson P., Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992 Dec;56(4):543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Wilson N. M., Petracek M. E., Berman J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet. 1989 Oct;16(4):225–239. doi: 10.1007/BF00422108. [DOI] [PubMed] [Google Scholar]
- Mahoney D. J., Broach J. R. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol Cell Biol. 1989 Nov;9(11):4621–4630. doi: 10.1128/mcb.9.11.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney D. J., Marquardt R., Shei G. J., Rose A. B., Broach J. R. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 1991 Apr;5(4):605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- Marshall M., Mahoney D., Rose A., Hicks J. B., Broach J. R. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Dec;7(12):4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D., Hartwell L. H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986 Jan 17;44(1):43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- Megee P. C., Morgan B. A., Mittman B. A., Smith M. M. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990 Feb 16;247(4944):841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Miller A. M., Nasmyth K. A. Role of DNA replication in the repression of silent mating type loci in yeast. Nature. 1984 Nov 15;312(5991):247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- Mullen J. R., Kayne P. S., Moerschell R. P., Tsunasawa S., Gribskov M., Colavito-Shepanski M., Grunstein M., Sherman F., Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989 Jul;8(7):2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. C., Szostak J. W. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990 Sep;10(9):4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L., Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989 Nov 17;59(4):637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Rine J., Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987 May;116(1):9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier D. H., Rine J. An origin of DNA replication and a transcription silencer require a common element. Science. 1992 May 1;256(5057):659–663. doi: 10.1126/science.1585179. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E. M., Swanson M. J., Romeo A. M., Hicks J. B., Sternglanz R. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol Cell Biol. 1991 Apr;11(4):2253–2262. doi: 10.1128/mcb.11.4.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L., Shore D. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Freedman R., Van Arsdell S., Szostak J. W., Thorner J. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol Cell Biol. 1987 Oct;7(10):3713–3722. doi: 10.1128/mcb.7.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]