Abstract
Background
Bacteriocins are protein antimicrobial agents that are produced by all prokaryotic lineages. Escherichia coli strains frequently produce the bacteriocins known as colicins. One of the most prevalent colicins, colicin M, can kill susceptible cells by hydrolyzing the peptidoglycan lipid II intermediate, which arrests peptidoglycan polymerization steps and provokes cell lysis. Due to the alarming rise in antibiotic resistance and the lack of novel antimicrobial agents, colicin M has recently received renewed attention as a promising antimicrobial candidate. Here the effects of subinhibitory concentrations of colicin M on whole genome transcription in E. coli were investigated, to gain insight into its ecological role and for purposes related to antimicrobial therapy.
Results
Transcriptome analysis revealed that exposure to subinhibitory concentrations of colicin M altered expression of genes involved in envelope, osmotic and other stresses, including genes of the CreBC two-component system, exopolysaccharide production and cell motility. Nonetheless, there was no induction of biofilm formation or genes involved in mutagenesis.
Conclusion
At subinhibitory concentrations colicin M induces an adaptive response primarily to protect the bacterial cells against envelope stress provoked by peptidoglycan damage. Among the first induced were genes of the CreBC two-component system known to promote increased resistance against colicins M and E2, providing novel insight into the ecology of colicin M production in natural environments. While an adaptive response was induced nevertheless, colicin M application did not increase biofilm formation, nor induce SOS genes, adverse effects that can be provoked by a number of traditional antibiotics, providing support for colicin M as a promising antimicrobial agent.
Keywords: Antimicrobial agent, Bacteriocin, Colicin M, Escherichia coli, Gene expression, Peptidoglycan
Background
In their natural environments, bacteria are frequently exposed to various stresses, including antimicrobials. It has been generally assumed that the role of antibiotics in nonclinical environments is the inhibition of competitors. Nevertheless, antibiotic concentrations in natural habitats can be variable, with high concentrations only in the vicinity of the producer. Recent studies have shown that antibiotics can act in a concentration-dependent manner that exhibits dual ecological roles: (i) at high concentrations they can destroy microorganisms; while (ii) at low concentrations they can modulate bacterial gene expression to promote ecological adaptation [1,2].
Bacteriocins are ribosomally synthesized antimicrobial agents that are produced by all prokaryotic lineages and they are generally active against closely related species. Among the best characterized bacteriocins are those produced by Escherichia coli, which are known as colicins. The majority of colicins act by membrane permeabilization, followed by nuclease activity, while one colicin, colicin M, inhibits peptidoglycan synthesis.
Uptake of colicin M proceeds by binding to the FhuA outer membrane receptor followed by energy-dependent translocation into the periplasm through the TonB system (TonB, ExbB and ExbD) and the proton motive force of the inner membrane [3]. Colicin M is a phosphotase that cleaves the undecaprenyl-phosphate-linked peptidoglycan precursor, lipid II producing free undecaprenol and 1-pyrophospho-Mur-GlCNAc-pentapeptide. In the periplasm, hydrolysis of peptidoglycan lipid precursors results in arrest of polymerization steps and cell lysis [4]. Operons that encode colicin M and B are tightly linked on large conjugative plasmids [5,6], and these are among the most abundant colicins produced by E. coli strains [7].
A number of studies have been aimed at defining the function of colicins in microbial communities. They might serve to enable invasion or defense of an ecological niche [8]. They have been shown to mediate population and community level interactions, promoting microbial diversity within E. coli populations in the mammalian colon [9]. To obtain more insight into the ecological roles of one of the most prevalent colicins, the effects of subinhibitory concentrations of colicin M on genome wide transcription in E. coli was studied.
Antibiotic resistance currently represents one of the greatest worldwide threats to human health therefore, novel antibiotics are urgently needed. Antibiotic resistance among the Enterobacteriaceae represents a particular threat [10,11]. As colicin M promotes the irreversible hydrolysis of lipid II, a peptidoglycan lipid intermediate that is common to all bacteria, it is also a promising candidate for development of a novel antimicrobial agent [12]. Analysis of the gene expression profile was thus also undertaken, to acquire insight into adaptive responses to colicin M that might be detrimental during antimicrobial therapy.
Results and discussion
Transcriptome analysis of E. coli MG1655 exposed to subinhibitory concentrations of colicin M
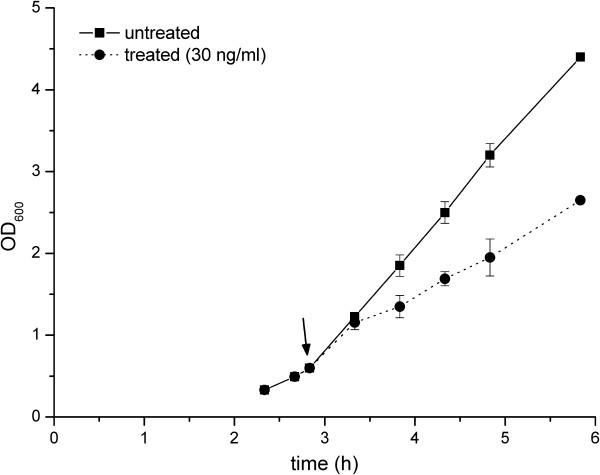
The effects of colicin M on whole genome transcription of E. coli MG1655, a laboratory strain with minimal genetic manipulation that approximates the wild type [13], was investigated by microarray analysis. To choose the appropriate conditions for determing the colicin M induced transcriptome, mid-exponential phase cultures of strain MG1665 were exposed to various concentrations of colicin M and the growth response was monitored. On the basis of these results a concentration of 30 ng/ml was determined as subinhibitory and chosen for transcriptome analysis. To focus on stress responses provoked by colicin M, samples were collected as growth slowed, 30 min following exposure to colicin M and upon regrowth, 60 min following exposure to colicin M, Figure 1 (see also Additional file 1: Figure S1 and Additional file 2: Figure S2).
Figure 1.
Growth of MG1655 without and with colicin M. The arrow denotes the time of addition of colicin M at subinhibitory concentrations (30 ng/ml). The experiment was performed three times, and the means ± standard errors of the means (error bars) are shown.
The 30 min exposure up-regulated the expression of 49 genes, with 2 genes down-regulated (log2 fold change >1 and < −1, P ≤0.05). On the other hand, the 60-min exposure to colicin M significantly up-regulated the expression of 210 genes, with expression of 51 genes down-regulated (log2 fold change >1 and < −1, P ≤0.05). Time course analysis showed that 46 genes were differentially expressed following 30 and 60 min colicin M treatment while 5 were differentially expressed only after 30 min treatment, (Figure 2). Whereas 30 min exposure provoked differential expression of a limited number of genes across several gene groups, more genes were altered in their expression (extensive transcriptional changes were observed) following 60 min treatment. Among the first significantly induced genes were those of two component sensory systems and several genes encoding membrane proteins.
Figure 2.
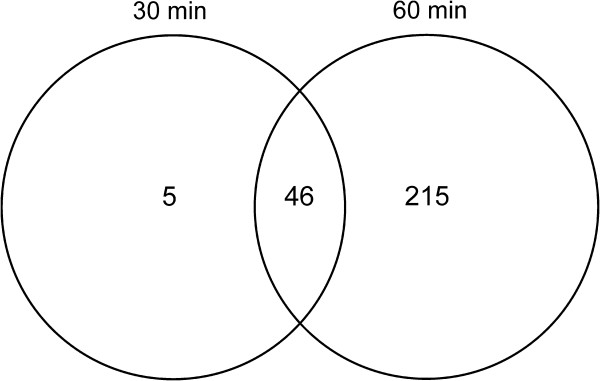
Venn diagram of gene expression in 30 min and 60 min treated E. coli MG1655. Time course analysis of differentially expressed genes, reveals number of genes induced following 30 min and 60 min exposure to subinhibitory concentrations of colicin M.
Time course analysis of differential gene expression, after 30 and 60 min treatment, is presented in Additional file 3: Table S1 (log2 fold change >1 and < −1, P ≤0.05). Genes considered for interpretation are presented in Table 1 and are described below.
Table 1.
Genes with modulated expression after exposure to colicin M over time, 30 and 60 min
| Category/Gene symbol | Gene accession No. | Gene description | 30 min log2 ratio | 60 min log2 ratio |
|---|---|---|---|---|
|
Envelope stress regulators/systems |
||||
|
rcsA |
946467 |
DNA-binding transcriptional activator, co-regulator with RcsB |
3.38 |
6.13 |
|
cpxP |
2847688 |
inhibitor of the cpx response; periplasmic adaptor protein |
1.57 |
2.61 |
|
pspA |
945887 |
regulatory protein for phage-shock-protein operon |
1.35 |
1.18 |
|
pspB |
945893 |
DNA-binding transcriptional regulator of psp operon |
1.32 |
1.47 |
|
pspC |
945499 |
DNA-binding transcriptional activator |
1.14 |
1.52 |
|
pspD |
945635 |
peripheral inner membrane phage-shock protein |
0.83 |
1.78 |
|
pspG |
948557 |
phage shock protein G |
1.55 |
2.29 |
|
Colanic acid biosynthetic process |
||||
|
wza |
946558 |
lipoprotein required for capsular polysaccharide translocation through the outer membrane |
3.59 |
7.12 |
|
wzb |
946564 |
protein-tyrosine phosphatase |
2.44 |
6.33 |
|
wzc |
946567 |
protein-tyrosine kinase |
1.52 |
6.72 |
|
wcaA |
946570 |
predicted glycosyl transferase |
0.93 |
5.7 |
|
wcaB |
946573 |
predicted acyl transferase |
0.69 |
5.73 |
|
wcaC |
946579 |
predicted glycosyl transferase |
0.56 |
5.47 |
|
wcaD |
946550 |
predicted colanic acid polymerase |
0.78 |
7.23 |
|
wcaE |
946543 |
predicted glycosyl transferase |
1.25 |
7.26 |
|
wcaF |
946578 |
predicted acyl transferase |
0.97 |
7.21 |
|
gmd |
946562 |
GDP-D-mannose dehydratase, NAD(P)-binding |
0.71 |
6.65 |
|
fcl |
946563 |
bifunctional GDP-fucose synthetase: GDP-4-dehydro-6-deoxy-D-mannose epimerase/GDP-4-dehydro-6-L-deoxygalactose reductase |
0.32 |
6.57 |
|
gmm |
946559 |
GDP-mannose mannosyl hydrolase |
0.3 |
6.15 |
|
wcaI |
946588 |
predicted glycosyl transferase |
0.3 |
5.92 |
|
cpsG |
946574 |
phosphomannomutase |
0.09 |
5.15 |
|
cpsB |
946580 |
mannose-1-phosphate guanyltransferase |
0.26 |
5.1 |
|
wcaJ |
946583 |
predicted UDP-glucose lipid carrier transferase |
0.11 |
4.82 |
|
wzxC |
946581 |
predicted colanic acid exporter |
0.1 |
4.45 |
|
wcaK |
946569 |
Colanic acid biosynthesis protein |
−0.12 |
4.45 |
|
wcaL |
946565 |
predicted glycosyl transferase |
−0.13 |
3.63 |
|
manA |
944840 |
mannose-6-phosphate isomerase |
0.19 |
1.05 |
|
ugd |
946571 |
UDP-glucose 6-dehydrogenase |
0.46 |
4.36 |
|
wcaM |
946561 |
colanic acid biosynthesis protein |
−0.01 |
2.71 |
|
galU |
945730 |
glucose-1-phosphate uridylyltransferase |
0.44 |
1.4 |
|
Extracellular polysaccharide distinct from colanic acid |
||||
|
yjbE |
948534 |
predicted protein |
1.55 |
5.74 |
|
yjbF |
948533 |
predicted lipoprotein |
1.73 |
5.67 |
|
yjbG |
948526 |
conserved protein |
0.67 |
4.29 |
|
yjbH |
948527 |
predicted porin |
0.66 |
5.23 |
|
Peptidoglycan synthesis |
||||
|
anmK |
946810 |
anhydro-N-acetylmuramic acid kinase |
0.16 |
1.17 |
|
mrcB |
944843 |
fused glycosyl transferase and transpeptidase |
0.47 |
1.01 |
|
ycfS |
945666 |
L,D-transpeptidase linking Lpp to murein |
0.77 |
2 |
|
Osmotic stress response |
||||
|
osmB |
945866 |
lipoprotein |
2.41 |
2.95 |
|
osmC |
946043 |
osmotically inducible, stress-inducible membrane protein |
0.44 |
1.15 |
|
opgB |
948888 |
phosphoglycerol transferases I and II |
0.12 |
1.27 |
|
opgC |
946944 |
membrane protein required for succinylation of osmoregulated periplasmic glucans (OPGs) |
0.31 |
1.85 |
|
ivy |
946530 |
inhibitor of vertebrate C-lysozyme |
1.55 |
1.26 |
|
mliC |
946811 |
inhibitor of C-lysozyme, membrane-bound; predicted lipoprotein |
2.17 |
3.92 |
|
ybdG |
946243 |
predicted mechanosensitive channel |
0.69 |
1.26 |
|
dppB |
948063 |
dipeptide/heme transporter |
−0.29 |
3.29 |
|
dppF |
948056 |
dipeptide transporter |
−0.1 |
2.33 |
|
dppC |
948064 |
dipeptide/heme transporter |
−0.09 |
2.33 |
|
dppD |
948065 |
dipeptide/heme transporter |
−0.09 |
2.1 |
|
dppA |
948062 |
dipeptide transporter |
0.02 |
1.13 |
|
Other stress responses |
||||
|
ydeI |
946068 |
conserved protein |
1.99 |
3.96 |
|
treR |
948760 |
DNA-binding transcriptional repressor |
0.65 |
1.88 |
|
ibpA |
948200 |
heat shock chaperone |
−0.01 |
1.78 |
|
ibpB |
948192 |
heat shock chaperone |
0.02 |
2.9 |
|
hslJ |
946525 |
heat-inducible lipoprotein involved in novobiocin resistance |
2.33 |
3.32 |
|
yhbO |
947666 |
predicted intracellular protease |
2.29 |
2.67 |
|
iraM |
945729 |
RpoS stabilizer during Mg starvation, anti-RssB factor |
0.33 |
1.6 |
|
creD |
948868 |
inner membrane protein |
5.66 |
4.96 |
|
cbrB |
948231 |
inner membrane protein, creBC regulon |
5.2 |
4.29 |
|
cbrA |
948197 |
predicted oxidoreductase with FAD/NAD(P)-binding domain |
4.3 |
3.35 |
|
cbrC |
948230 |
conserved protein, UPF0167 family |
3.77 |
2.8 |
|
spy |
946253 |
envelope stress induced periplasmic protein |
1.71 |
2.99 |
|
htpX |
946076 |
predicted endopeptidase |
0.27 |
1.01 |
|
yggG |
945173 |
heat shock protein binding to Era protein; predicted peptidase |
1.01 |
1.82 |
|
Biofilm formation |
||||
|
ycfJ |
945977 |
predicted protein |
4.77 |
5.8 |
|
rprA |
2847671 |
ncRNA |
3.86 |
4.85 |
|
omrA |
2847746 |
ncRNA |
0.36 |
1.76 |
|
omrB |
2847747 |
ncRNA |
0.77 |
1.74 |
|
bdm |
946041 |
biofilm-dependent modulation protein |
4.49 |
4.21 |
|
ydeH |
946075 |
diguanylate cyclase, required for pgaD induction |
1.38 |
1.68 |
|
Cell motility |
||||
|
fliZ |
946833 |
RpoS antagonist; putative regulator of FliA activity |
−0.41 |
−1.05 |
|
fliE |
946446 |
flagellar basal-body component |
−0.66 |
−1.07 |
|
fliG |
946451 |
flagellar motor switching and energizing component |
−0.28 |
−1.07 |
|
flgN |
945634 |
export chaperone for FlgK and FlgL |
−0.29 |
−1.12 |
|
flgA |
946300 |
assembly protein for flagellar basal-body periplasmic P ring |
−0.09 |
−1.17 |
|
flgF |
945639 |
flagellar component of cell-proximal portion of basal-body rod |
−0.29 |
−1.21 |
|
flgM |
946684 |
anti-sigma factor for FliA (sigma 28) |
−0.27 |
−1.23 |
|
fliA |
948824 |
RNA polymerase, sigma 28 (sigma F) factor |
−0.21 |
−1.45 |
|
flgD |
945813 |
flagellar hook assembly protein |
−0.33 |
−1.61 |
|
flgE |
945636 |
flagellar hook protein |
−0.05 |
−1.72 |
|
flgC |
946687 |
flagellar component of cell-proximal portion of basal-body rod |
−0.04 |
−2.14 |
|
flgB |
945678 |
flagellar component of cell-proximal portion of basal-body rod |
−0.19 |
−2.4 |
|
flhC |
947280 |
DNA-binding transcriptional dual regulator with FlhD |
−0.76 |
−2.54 |
|
flhD |
945442 |
DNA-binding transcriptional dual regulator with FlhC |
−0.76 |
−2.54 |
|
Amino acid transport/acid resistance |
||||
|
glnP |
945621 |
glutamine transporter subunit |
−0.23 |
−1.17 |
|
gadB |
946058 |
glutamate decarboxylase B, PLP-dependent |
0.03 |
−1.18 |
|
glnQ |
945435 |
glutamine transporter subunit |
−0.15 |
−1.25 |
|
glnG |
948361 |
fused DNA-binding response regulator in two-component regulatory system with GlnL: response regulator/sigma54 interaction protein |
−0.15 |
−1.32 |
|
gadA |
948027 |
glutamate decarboxylase A, PLP-dependent |
−0.23 |
−1.64 |
|
gadE |
948023 |
DNA-binding transcriptional activator |
0.13 |
−1.38 |
|
slp |
948022 |
outer membrane lipoprotein |
−0.18 |
−1.91 |
|
hdeB |
948026 |
acid-resistance protein |
0.13 |
−1.17 |
|
hdeD |
948024 |
acid-resistance membrane protein |
−0.01 |
−1.04 |
|
Poorly characterized |
||||
|
ymgD |
945732 |
predicted protein |
3.45 |
3.65 |
|
ymgG |
945728 |
conserved protein, UPF0757 family |
3.87 |
3.55 |
|
yfdC |
944801 |
predicted inner membrane protein |
1.02 |
2.25 |
|
yjbJ |
948553 |
conserved protein, UPF0337 family |
0.97 |
1.19 |
|
yaaX |
944747 |
predicted protein |
1.59 |
4.12 |
|
yegS |
946626 |
phosphatidylglycerol kinase, metal-dependent |
0.81 |
1.65 |
| yaiY | 945223 | inner membrane protein, DUF2755 family | 3.94 | 5.22 |
(bold indicates genes with gene expression log2 ratio (fold change) ≥1 and ≤−1, denoting fold change ≥2 or ≤−2, and with p≤0.05, and italic indicates genes with p≥0.05).
Colicin M treatment affects signal transduction pathways
The bacterial envelope protects the bacterial cell from external stress and performs essential functions such as, transport of nutrients and waste, as well as respiration and adhesion. In Gram-negative bacteria the outer membrane acts as a permeability barrier, with porins for transport and protection against phagocytosis [14] while processes central for energy generation and nutrient transport occur in the inner membrane [15]. In the periplasmic space peptidoglycan ensures the structural integrity of the cell by preventing osmolysis. To sense and rapidly respond to environmental signals, bacteria primarily use two-component signal transduction systems, composed of an inner membrane histidine kinase and a cytoplasmic response regulator [16].
Peptidoglycan is the major structural component of the bacterial cell wall. It provides the bacterial cell structural strength and protects the osmotically sensitive membrane [17]. As expected, by targeting peptidoglycan synthesis, colicin M induced an envelope stress response. In E. coli envelope homeostasis is monitored by several stress responses; namely, Rcs, Cpx, Psp, σE, Bae and vesicle release responses [18-20]. Our microarray analysis revealed that the Rcs regulon has a major role in the response to envelope stress induced by colicin M. RcsC and RcsD are inner-membrane-bound proteins, with RcsC as the sensor kinase that autophosphorylates upon sensing the appropriate signal, while RcsD transfers the phosphoryl group to the transcriptional regulator RcsB. Certain promoters require the RcsA protein to act as an auxiliary regulatory protein, apparently exerting its effects by forming a heterodimer with RcsB [21]. The Rcs system controls the production of exopolysaccharides [22], biofilm formation [23,24], cell motility, and chemotaxis [25]. We observed induction of rcsA and a number of other Rcs-regulated genes: the exopolysaccharide operons wca for colanic acid synthesis, and yjbEFGH, as well as genes osmB, ymgG and ymgD, ivy, yfbR, ugd, yfdC, yjbJ, galU, yaaX, yggG, yegS, spy, rprA, bdm and yaiY (Table 1). Recently, perturbations to peptidoglycan by several ß-lactam antibiotics were shown to elicit shared as well as unique responses with all activating the Rcs system [26] indicating that, the Rcs pathway elicits a global response to peptidoglycan stress [27].
Colicin M treatment also induced the expression of cpxP, which encodes the periplasmic inhibitor of the Cpx envelope stress response. The Cpx system appears to sense misfolded proteins that are synthesized for the periplasm, and it is controlled by the sensor kinase CpxA, the response regulator CpxR, and the periplasmic inhibitor CpxP. CpxP has been assumed to fine tune Cpx activation during envelope stress, by preventing its incorrect activation and enabling its rapid shut-down following envelope stress relief [28].
Treatment with colicin M also up-regulated a third cell envelope stress system, the psp genes that encode the membrane-bound phage shock proteins: PspA, PspB, PspC, PspD and PspG. The psp regulon consists of the psp operon with genes pspA, pspB, pspC, pspD and pspE, as well as genes pspF and pspG. Proteins PspB, PspC and PspD are located in the inner membrane, while PspA is on the cytoplasmic side of the inner membrane. In the absence of stress, PspA binds to protein PspF, thus inhibiting transcription of the psp operon. The psp genes are induced by filamentous phage infection and other stresses that impair cell membrane function [29,30]. This system is responsible for repair of inner membrane damage and maintenance of the proton motive force across the inner membrane [31,32]. Peptidoglycan damage provoked by colicin M exposes the sensitive inner membrane to osmotic damage requiring activation of membrane repair mechanisms.
Colicin M induces expression of exopolysaccharide genes
Among the most strongly up-regulated genes, were those of the wca operon, which encodes the production of the exopolysaccharide, colanic acid [33]. The highly viscous colanic acid [34] is secreted into the extracellular environment to protect cells from osmotic stress such as provoked by cell envelope perturbations, including peptidoglycan damage or dessication [35]. In addition, colanic acid is involved in the later stages of biofilm formation; namely, the maturation and development of complex three-dimensional biofilm structures [24]. The wca operon is comprised of 19 genes that are involved in colanic acid synthesis from the nucleoside diphosphate sugars: GDP-L-fucose, UDP-d-glucose, UDP-d-galactose and UDP-D-glucuronate [36]. Colicin M treatment induced the expression of all 19 of the wca genes.
Exposure to colicin M also up-regulated the D-galactose transporter galP, as well as galU, which encodes the glucose-1-phosphate uridylyltransferase that is needed for UDP-glucose, an intermediate involved in the synthesis of colanic acid, trehalose, lipopolysaccharide and membrane-derived oligosaccharides [37].
Furthermore, our studies revealed strong induction of the yjbEFGH operon that is involved in the production of another, as-yet-unidentified, exopolysaccharide [38]. Recent studies have shown that the yjbEFGH operon is also induced by osmotic stress, and that the wca and yjbEFGH operons are negatively regulated by the general stress response sigma factor RpoS (σ38) [39]. Both the wca and the yjbEFGH operons are induced by the activated Rcs pathway to protect the bacterial cell from osmolysis.
Colicin M induced additional osmotic and other stress responses
By inhibiting peptidoglycan synthesis, colicin M weakens membrane protection, provoking osmotic stress. Interestingly, genes creD, cbrA, cbrB and cbrC of the CreB/CreC regulon were strongly induced already 30 min after exposure to colicin M. The Cre system was previously found to be involved in the switch from aerobic to anaerobic conditions. CreC is the sensor that also senses changes in the growth medium and/or metabolite pool levels, while CreB is a transcriptional regulator [40]. The two-component CreBC system positively controls transcription of cbrA. Recently, the CbrA protein was shown to protect against colicin M and osmotic shock, implying a function of CbrA in outer membrane structure [41]. Thus, it was proposed that in the natural environment with a limited supply of nutrients, CbrA protects sensitive cells from colicin M synthesized by competitors. Furthermore, CbrC, which we also found to be induced by colicin M treatment, has been shown to protect against colicin E2 and also seems to be involved in alteration of outer membrane structure [41]. Our results indicate that subinhibitory concentrations of colicin M could induce protection against colicins. Thus, in the natural environment, both colicin synthesis and the CreBC system are induced upon nutrient limitation [42,43]. Colicin produced in microbial communities by colicinogenic bacteria could in colicin sensitive community members induce protective responses. Moreover, activation of the CreBC two component regulator system was recently shown to play a major role in the ß-lactam resistance response [44] indicating that, subinhibitory concentrations of colicin M might elicit broader antimicrobial protection.
It can also be noted that more than 100 of the open reading frames up-regulated by colicin M treatment are classified as poorly characterized or with predicted functions. Among these, many are predicted membrane proteins and lipoproteins indicating that, to protect cells against peptidoglycan damage provoked by colicin M, an adaptive response to strengthen/stabilize the osmosensitive membrane is induced.
To resist the effects of colicin M treatment, other genes involved in the response to hyperosmotic stress were up-regulated; namely, osmB and osmC[45] as well as two inhibitors of C-lysozyme, ivy and a membrane bound and predicted lipoprotein mliC, were also induced by the Rcs system. Antibiotic-mediated peptidoglycan stress has also been shown to trigger expression of both of these genes [27].
Colicin M also induced other stress response genes, including ydeI, which is involved in hydrogen peroxide stress [46], as well as the ibpA and ibpB heat shock genes, which encode chaperones that can cooperate to prevent irreversible aggregation of proteins [47].
Colicin M induces biofilm associated genes
In natural environments, bacteria often form biofilms, microbial communities in which bacteria adhere to an abiotic or biotic surface via surface charges as well as production of pili, fimbriae and exopolysaccharides. Microbial cells in biofilms show distinct properties, particularly resistance to antibiotics, disinfectants, shear stress and the immune system [48]. Biofilm formation proceeds in several tightly regulated steps: initial attachment, three-dimensional development by microcolony formation, biofilm maturation and the final step dispersal or cellular detachment to colonize other surfaces. Initially, flagella promote motility toward a surface; subsequently, flagella are lost and adhesive organelles such as curli fimbria enable attachment; and finally, colanic acid production promotes maturation into the three dimensional biofilm structure [49,50].
Colicin M treatment upregulated several genes involved in biofilm production. Thus, in addition to up-regulation of colanic acid, colicin M treatment up-regulated rprA, which encodes a small regulatory RNA affecting target mRNA translation. Expression of rprA has been shown to be activated by the Rsc system. RprA has been shown to repress csgD. This latter encodes the master transcriptional regulator that activates curli fimbriae and cellulose synthesis, both of which are involved in the initial stage of biofilm formation [51]. It has been postulated that by interfering with csgD mRNA translation, RprA might prevent the undesired co-expression of curli/cellulose and colanic acid [52]. In accordance, as we found upregulation of the colanic acid operon genes we also determined upregulation of the omrA and omrB genes, which encode two redundant small/antisense RNAs that have recently been shown to inhibit CsgD translation [53].
Colicin M exposure up-regulated another biofilm-associated gene, bdm, which encodes the biofilm-dependent modulation protein. Bdm expression is positively regulated by RcsB in response to osmotic shock [25], and the Bdm protein has been recently shown to enhance biofilm formation [54].
The exposure to colicin M also upregulated ydeH, which codes for a diguanylate cyclase that can synthesize the second messenger bis-(3′-5′) cyclic di-guanosine monophosphate (c-di-GMP) [55-57]. ydeH is positively regulated by CpxR, and has been shown to inhibit motility as well as to promote adhesin and biofilm formation. In E. coli, c-di-GMP controls the synthesis of two exopolysaccharides: cellulose and poly-GlcNaC (PGA), a virulence factor of uropathogenic E. coli[58]. Our study thus showed that colicin M induced an envelope stress response which could provoke switching from a planktonic to a sessile lifestyle. Nevertheless, in crystal violet assays no induction of biofilm formation was observed (data not shown).
Colicin M treatment downregulates flagellar biosynthesis genes
Not unexpectedly, among the down-regulated genes, there were in particular the genes involved in flagellar motility and in glutamine biosynthetic processes. In E. coli, flagellar expression and motility is controlled by the FlhDC complex that comprises >60 genes. Flagellar synthesis and function are processes that demand high energy consumption, and therefore, expression of the flagellar genes is tightly regulated [59]. In contrast to exopolysaccharide production, expression of the flhDC operon has been shown to be down-regulated by numerous environmental signals, such as high temperature, high osmolarity (concentrations of salts, in the presence of carbohydrates or low-molecular alcohols) and extreme pH [60,61].
Both the exopolysaccharide synthesis operons, wca and yjbEFGH, and the flagellar flhDC operon genes are controlled by the Rcs phosphorelay system. However, while the activated Rcs phosphorelay system induces exopolysaccharide synthesis, it down-regulates the flhDC operon due to repression by the RcsB cofactor RcsA.
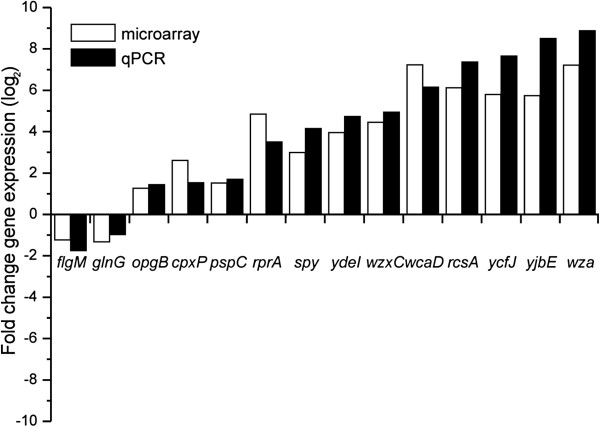
Validation of microarray results by real-time PCR
The gene expression results from the microarray analyses of total RNA isolated from cultures exposed to colicin M for 60 min were verified using qPCR. For this purpose, 14 genes differentially expressed upon colicin M treatment and from different functional groups, were selected: ydeI, pspC, opgB, rprA, cpxP, ycfJ, rcsA, yjbE, wcaD, spy, wzxC, wza, glnG and wza. For this comparison, the fold-changes of mRNA abundance of selected genes after 60 min colicin M exposure were plotted as those determined by qPCR versus those seen in the microarray analysis. The qPCR results confirmed differential gene expression observed by microarray analysis of the selected genes (Figure 3).
Figure 3.
Validation of the microarray results by qPCR. Expression analysis of the 14 selected genes determined by microarray (open bars) and validated by qPCR (solid bars). The fold-changes for the microarray and qPCR were calculated as described (Materials and Methods) and represent gene expression levels following 60 min exposure to colicin M.
Colicin M treatment does not promote significantly increased exopolysaccharide production
As the microarray data showed that the colanic acid operon genes were among the most strongly induced, a functional assay was performed to address whether the amount of colanic acid was changed accordingly. Production of colanic acid was quantified following exposure of E. coli to colicin M. Colanic acid was extracted from bacterial cultures treated with subinhibitory concentrations of colicin M for 60 min, 90 min and 120 min, as well as from an untreated control. While at the mRNA level there was significant induction of the wca operon genes, only a slight, 1.3-fold, increase in the production of colanic acid was seen at all sampling times. As an additional control, colanic acid was quantified from a culture overexpressing the wca operon encoded by a multicopy plasmid, pATC400 [62]. A 6-fold increase in colanic acid production was seen in comparison with an isogenic strain that did not overexpress the wca operon genes.
Treatment of E. coli with colicin M promotes the hydrolysis of the peptidoglycan lipid precursors, which results in the arrest of the polymerization steps and exposes the bacterial cells to envelope stress, which activates the Rcs and Cpx phosphorelay systems. Subsequently, cell motility is down-regulated, with induction of the expression of the exopolysaccharide wca and the yjbEFGH operon genes. Colicin M promoted hydrolysis of lipid II which prevents recycling of the lipid carrier for peptidoglycan synthesis and also limits its availability for exopolysaccharide biosynthesis, including colanic acid.
Following an initial growth stagnation (Figure 1 and also see Additional file 1: Figure S1), regrowth of cultures treated with these subinhibitory concentrations of colicin M indicate an adaptive response to the stress through the activation of the envelope and other stress responses. While neither an increase in colanic acid or biofilm production was detected, in the natural environment, activation of the various above-described stress responses might allow rapid adaptation, including mature biofilm formation, in the event of a sudden decrease/absence in antimicrobial concentrations.
Genome-wide transcriptional analysis and other antimicrobials
A number of studies have shown that traditional antibiotics affect bacterial gene expression and physiology [1,2,63,64]. Thus, some β-lactam antibiotics that can also inhibit peptidoglycan synthesis have been shown to induce the production of colanic acid in E. coli, which indicates that these might exacerbate biofilm formation [65]. Investigation of the E. coli transcription profile in response to bactericidal concentrations of ampicillin also showed induction of the colanic acid biosynthetic pathway, as well as rcsA, the transcriptional activator of colanic acid synthesis and other stress responses [66]. However, the authors did not detect induction of the additional exopolysaccharide operon yjbEFGH, distinct from colanic acid. In Staphylococcus aureus, subinhibitory concentrations of β-lactams have been shown to up-regulate some virulence genes [67]. Moreover, the aminoglycoside tobramycin has been shown to induce biofilm formation in E. coli and in Pseudomonas aeruginosa, due to alterations in the levels of c-di-GMP [68]. Biofilm formation was also induced following exposure of P. aeruginosa to subinhibitory concentrations of tetracycline and norfloxacin [69].
Further to this, a number of studies have investigated the effects of antibiotics on the expression of the SOS regulon genes. Thus, the β-lactam antibiotic, ceftazidime, which is an inhibitor of a protein involved in cell wall biosynthesis, PBP3, has been shown to induce transcription of the dinB gene, which encodes the error-prone DNA polymerase Pol IV [70]. Subsequently, subinhibitory concentrations of ampicillin, norfloxacin and kanamycin were shown to induce mutagenesis due to antibiotic-mediated increases in reactive oxygen species, which results in SOS-induced mutagenesis that might lead to multidrug resistance [71]. An additional study showed that a number of antibiotics can promote an increase in mutation frequency; namely, ampicillin, ceftazidime, imipenem, ciprofloxacin, trimethoprim, sulfamethoxazole and tetracycline [72]. With the exception of imipenem, fosfomycin and tetracycline, the antibiotics tested were shown to induce recA expression, while inactivation of recA abolished the mutagenic effects.
In the present study, subinhibitory concentrations of colicin M did not induce the expression of dinB or recA. To further confirm that colicin M does not induce the SOS response, induction of the sulA gene following colicin M treatment was investigated. SOS-regulated SulA inhibits cell division by binding to FtsZ, which is required for septum formation. For this purpose, expression of the chromosomal sulA-lacZ fusion was studied in the ENZ1257 strain [73] without and with colicin M: no induction was detected (data not shown).
While colicins have been assumed to have roles in defense or invasion of an ecological niche, previous studies have shown that colicins are also involved in the promotion of microbial diversity within E. coli populations in the mammalian colon [9,74]. Furthermore, the nuclease colicins, E9 and E3, have been shown to have the potential to promote microbial genetic diversity via induction of the SOS response or via increased transcription of laterally acquired mobile elements, respectively [75].
Another study showed that colicins from one producer can induce production in another producer, thus resulting in colicin-mediated colicin induction [74]. Here, we show that subinhibitory concentrations of colicin M induced an envelope and other stress responses including the two component CreBC system connected with increased resistance to colicins M and E2. In natural environments, subinhibitory concentrations of colicin M could thus affect E. coli bacterial communities by promoting ecological adaptation enabling noncolicinogenic cells to survive and compete with colicin producers. The above-described phenomena might also be relevant in the natural settings of other bacterial species, as colicin M homologous proteins have been identified recently in human and plant pathogenic Pseudomonas species that have hydrolytic activity against peptidoglycan precursors [76]. Further, activation of the P. aeruginosa CreBC system has been shown to play a major role in the ß-lactam resistance response [44].
Resistance of pathogens to traditional antibiotics represents one of the greatest health care threats. The present lack of novel antibiotics is also of great concern. Colicin M has been recently shown to hydrolyse lipid II intermediates of Gram-negative and Gram-positive bacteria [12]. In addition, as the isolated colicin M catalytic domain displays full enzymatic activity, protein engineering can be used to allow binding and translocation in various Gram-negative and Gram-positive species [77,78]. Furthermore, low concentrations and low protein-to-bacteria ratios suffice for colicin M to kill E. coli. Targeting of lipid II has been indicated as a potential antibacterial strategy [79].
Conclusion
In conclusion, subinhibitory concentrations of colicin M induced genes involved in adaptive responses to protect the population against envelope and other stresses, including the two component CreBC system associated with increased resistance to some colicins. Our study of the global transcriptional response to colicin M thus provides novel insight into the ecology of colicin M production in natural environments. While an adaptive response was provoked by colicin M treatment there was no induction of biofilm formation, SOS response genes, or other genes involved in mutagenesis, adverse effects shown to be promoted by a number of clinically significant traditional antibiotics. Thus, our study also corroborates the potential of colicin M as a promising antimicrobial and shows the value of microarrays for investigation of novel antimicrobials.
Methods
Colicin M expression and isolation
Prior to isolation of colicin M, the cma colicin M structural and cmi immunity genes were PCR amplified from the natural colicin M coding plasmid pCHAP1 using the primers ColM1 5′-TCACTCGAGCATGGAAACCTTAACTGTTCATGCA-3′ and ColM2 5′-CCACGCGTCCACTTCACAGTATGCTCACATTG-3′. The amplified fragment was digested with the XhoI and MluI restriction enzymes and cloned into the pET8c expression vector, also cut with the same two enzymes [80]. The isolated plasmid was designated pColM-imm Cloning of cma into the pET8c vector introduced an N-terminal histidine tag with expression under the control of a T7 promotor. Colicin M and the immunity protein were subsequently expressed in E. coli BL21 (DE3)pLysS and colicin M was purified using nickel affinity chromatography [80,81].
For large scale isolation an overnight culture of BL21 (DE3)pLysS, with plasmid pColM-imm was diluted 100 fold in 500 ml LB with ampicillin (120 μg/ml) and grown at 37°C to an OD600 0.6-0.8, when chromosomal T7 polymerase production was induced by addition of 0.8 mM IPTG and incubated for a further 4 h. Subsequently, cells were harvested and resuspended in 50 mM phosphate, 300 mM NaCl buffer, pH 8, containing RNaseA (20 μg/ml), DNAse (10 μg/ml), lysozyme (1 mg/ml), 10 mM imidazole as well as protein inhibitors and incubated for 1 h at 4°C with shaking. The cells were then lysed with 3 min sonification, 40% amplitude and the supernatant obtained by centrifugation at 17000×g for 1 h at 4°C. The histidine-tag enabled Ni-NTA affinity column purification according to the user’s manual (Qiagen). Nonspecifically bound proteins were washed off the column with 50 mM phosphate, 300 mM NaCl, pH 8.0 buffer, containing 50 mM imidazole while colicin M was subsequently eluted with the same buffer containing 300 mM imidazole.
The colicin-M-containing fractions, as established by 10% SDS-PAGE, were then dialyzed against 5 mM phosphate buffer, pH 7.3, centrifuged at 17,000× g at 4°C for 30 min, and stored at −80°C. Colicin M purity was verified by SDS-PAGE (see Additional file 4: Figure S3), and (a concentration of 3.4 mg/ml) protein concentrations were determined using bicinchoninic acid protein assay kits (Pierce) and a Nanodrop ND 1000 spectrophotometer (Thermo Scientific). Finally colicin M was stored at −80°C.
Growth conditions
The agar dilution method (National Committee for Clinical Laboratory Standards, 2000) was used to determine the minimal inhibitory concentration (MIC) of colicin M 50 ng/ml. For this purpose, an overnight culture of E. coli MG1655 [13] was diluted 1:625 in LB broth and grown at 37°C with aeration to an OD600 0.6 when the culture was divided into several parts. One part served as a control while the other parts were treated with various concentrations of colicin M. The subinhibitory concentration (30 ng/ml) provoked temporary growth stagnation while growth decline was observed at higher concentrations, presumably due to cell lysis (see Additional file 1: Figure S1 in Additional file 2: Figure S2).
To investigate the effects of colicin M on the whole genome expression profile, an overnight culture of the E. coli strain MG1655 (F-lambda-ilvG-rfb-50 rph-1) was grown as described above. One part was treated with colicin M (30 ng/ml), while the untreated part served as the control. For gene expression analysis by microarray and qPCR, total RNA was isolated from 2-ml samples removed from each flask following 30 min and 60 min incubations at 37°C. The experiments were repeated at least two times.
For quantification of colanic acid, the growth conditions and the application of subinhibitory concentrations of colicin M were as described above. Colanic acid was purified from 50 ml cultures treated with colicin M for 60 min, 90 min and 120 min at 37°C, with aeration. The experiment was repeated at least two times.
RNA isolation
Total RNA was extracted using the RNAProtect Bacteria Reagent (Qiagen) and RNeasy Mini kits (Qiagen), according to the manufacturer instructions. To remove residual DNA, on-column DNase digestion was performed during the RNA purification using the RNase-Free DNase Set (Qiagen). A Nanodrop ND 1000 spectrophotometer (Thermo Scientific) was used to confirm total RNA concentrations, while an Agilent 2100 Bioanalyser (Agilent Technologies, CA, USA) was used to evaluate the RNA quality. The isolated RNA was stored at −80°C until use.
Microarray procedures
Gene expression analysis was performed using Affymetrix GeneChip® E. coli Genome 2.0 arrays. Target preparation, hybridization, washing, staining and scanning were performed as recommended by the Affymetrix GeneChip® Expression Analysis Technical Manual. The experiment was repeated at least two times.
The acquisition of array images and the data quality assessment were performed using an Affymetrix GeneChip Command Console. The GeneChip data was processed using several different R/Bioconductor packages. The Affymetrix raw data were normalized using the RMA algorithm from the XPS package. The data have been deposited in the NCBI Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo) under GEO series accession number GSE37026.
Annotation of the genes and the data representation was managed using the ANNAFFY and AFFYCORETOOLS packages. The normalized data, converted to log2 values, were first limited to the ENTREZ-annotated probes from strain K12 (10208 probes). The remaining data were tested for differential expression, which was performed using the LIMMA package for the 30-min treated versus the 30-min untreated control and for the 60-min treated versus the 60-min untreated control bacterial culture. Differential expression was assessed using the 2-way factorial ANOVA model constructed using LIMMA package. Differential expression was assessed using the false discovery rate multiple test correction [82] and controlling type I error at α = 0.05. Differential expression of genes is represented as log2 ratios between the treated and untreated samples averaged over the two biological replicates. Important differentially expressed genes with log2 (fold change) greater than 1 or less than -1 denoting 2-fold up-regulated or down-regulated genes over time were considered for interpretation and are presented in Table 1. The expression of a subset of selected genes was validated by quantitative real-time PCR (qPCR) (see Additional file 5: Table S2).
Real-time PCR
qPCR was performed for 14 genes that showed significant differential expression in the microarray analysis. Samples of 1 μg total RNA were reverse transcribed to synthesize cDNA using High Capacity cDNA Reverse Transcription kits (Applied Biosystems), according to the manufacturer instructions. qPCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems) with an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). The qPCR amplifications were performed as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, and a final dissociation curve analysis step from 60°C to 95°C. Two negative reverse transcription controls were used to show no reverse transcription contamination. qPCR validation was performed on four biological replicates.
Publicly available sequences of the transcripts from the NetAffyx Analysis Centre (http://www.affymetrix.com/analysis/netaffx/index.affx) were analyzed to select target sequences, and the Primer3 software [83] and Primer Express 3.0 software (Applied Biosystems) were used for the design of the specific primers (Sigma). The primer sequences are listed in Additional file 5: Table S2.
Raw data were acquired using the Sequence Detection System software, version 2.3 (Applied Biosystems), and gene expression levels were analyzed using the 2-δδCT method [84], as the efficiency of the qPCR amplifications for all of the genes tested was >90%. geNorm [85] (available from medgen.ugent.be/~jvdesomp/genorm) was used to select the most stable genes, and out of the seven housekeeping genes tested, lpp, aroE, gapA were used as the reference genes, with their geometric mean used for normalization. The results are presented as log2 ratios between gene expression of treated and untreated cultures of four replicates, and they are presented as a comparison with the microarray data (Figure 3).
Colanic acid quantification
Colanic acid was extracted from cultures grown and treated with colicin M as described above, and from untreated control cultures incubated under the same conditions. Colanic acid extraction and quantification was performed as described previously [86]. Briefly, for quantification, the amount of nondialyzable methylpentose ω-deoxyhexose (L-fucose), a component of colanic acid, was measured using a colorimetric reaction with authentic L-fucose (Sigma) as standard, and with concentrations ranging from 5 μg/ml to 100 μg/ml. Colanic acid levels were calculated as μg of nondialyzable ω-deoxyhexose/unit A600 nm/ml of culture.
Biofilm production
The ability to form biofilms was investigated using crystal violet assays, as described previously [87]. To assess the induction of biofilm formation, 100 μl of overnight cultures were added to microtiter plates without and with colicin M, and incubated for 24 h at 37°C. The experiments were performed in triplicate.
ß-Galactosidase assay
For quantification, the growth conditions and application of subinhibitory concentrations of colicin M are as described above. The ß-galactosidase activity of the sulA-lacZ gene fusion was measured as described previously [88].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: DŽB. Performed the experiments: SK. Analyzed the data: SK, DŽB. Contributed reagents/materials/analysis tools: SK, DŽB. Wrote the paper: SK, DŽB. Both authors read and approved the final manuscript.
Supplementary Material
Growth of E. coli MG1655 treated with colicin M. The arrow denotes the time of addition of colicin M, at inhibitory (100 ng/ml, 50 ng/ml) and subinhibitory concentrations (30 ng/ml, 20 ng/ml, 10 ng/ml). Growth curves represent E. coli MG1655 cultures treated with different colicin M concentrations.
Effect of subinhibitory concentrations of colicin M on E. coli MG1566 viable counts. Growth curves with viable counts (CFU/ml as a function of time relative to antibiotic addition) are shown for untreated and treated culture (30 ng/ml of colicin M).
Time course analysis of differentially expressed genes after 30 and 60 min exposure to subinhibitory concentrations of colicin M. p≤0.05, log2 FC≥1 and ≤−1, log2 FC≤1, ≥−1; p≥0.05, log2 FC≥1 and ≤−1, log2 FC≤1, ≥−1. Log2 FC values in bold correspond to log2 FC≥1 and ≤−1 when p≤0.05 and in regular type to log2 FC≤1, ≥−1 when p≤0.05. Log2 FC values in italics bold correspond to log2 FC≥1 and ≤−1 when p≥0.05 and in italics regular type to log2 FC≤1, ≥−1 when p≥0.05.
SDS-PAGE gel showing purity of isolated colicin M. Left, Protein ladder Page Ruler (Fermentas); Right, colicin M - 29.5 kDa, colicin M (3.4 mg/ml).
Primer pairs used for qRT-PCR in the present study.
Contributor Information
Simona Kamenšek, Email: simona.kamensek@bf.uni-lj.si.
Darja Žgur-Bertok, Email: darja.zgur.bertok@bf.uni-lj.si.
Acknowledgements
We thank the Centre for Functional Genomics, at the Medical Faculty, University of Ljubljana, Slovenia, for assistance with the microarray procedures. We also thank S. Gottesman for kindly providing strain MG1655 pATC400, P. Moreau for strain ENZ1257 and A. P. Pugsley for pCHAP1. This study was supported by the Slovenian Research Agency (ARRS) grant P1-0198.
References
- Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci USA. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Braun V, Patzer SI, Hantke K. Ton-dependent colicins and microcins: modular design and evolution. Biochimie. 2002;84:365–380. doi: 10.1016/S0300-9084(02)01427-X. [DOI] [PubMed] [Google Scholar]
- El Ghachi M, Bouhss A, Barreteau H, Touzé T, Auger G, Blanot D, Mengin-Lecreulx D. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J Biol Chem. 2006;281:22761–22772. doi: 10.1074/jbc.M602834200. [DOI] [PubMed] [Google Scholar]
- Harkness RE, Olschläger T. The biology of colicin M. FEMS Microbiol Rev. 1991;8:27–41. doi: 10.1111/j.1574-6968.1991.tb04955.x. [DOI] [PubMed] [Google Scholar]
- Sasarman A, Massie B, Zollinger M, Gagnetellier H, Shareck F, Garzon S, Morisset R. Naturally occurring R. ColBM plasmids belonging to the IncfIII incompatibility group. J Genl Microbiol. 1980;119:475–483. doi: 10.1099/00221287-119-2-475. [DOI] [PubMed] [Google Scholar]
- Christenson JK, Gordon DM. Evolution of colicin BM plasmids: the loss of the colicin B activity gene. Microbiology. 2009;155:1645–1655. doi: 10.1099/mic.0.026666-0. [DOI] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- Kirkup BC, Riley MA. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature. 2004;428:412–414. doi: 10.1038/nature02429. [DOI] [PubMed] [Google Scholar]
- Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin Infect Dis. 2001;32:1162–1171. doi: 10.1086/319757. [DOI] [PubMed] [Google Scholar]
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patin D, Barreteau H, Auger G, Magnet S, Crouvoisier M, Bouhss A, Touze T, Arthur M, Mengin-Lecreulx D, Blanot D. Colicin M hydrolyses branched lipids II from Gram-positive bacteria. Biochimie. 2012;94:985–990. doi: 10.1016/j.biochi.2011.12.019. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Nikaido H. In: Escherichia coli and Salmonella: Cellular and Molecular biology. Volume 1. 2. Neidhardt FC, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editor. Washington, DC: ASM Press; 1996. Outer membrane; pp. 29–47. [Google Scholar]
- Kadner RJ. In: Escherichia coli and Salmonella: Cellular and Molecular biology. Volume 1. 2. Neidhardt FC, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editor. Washington, DC: ASM Press; 1996. Cytoplasmic membrane; pp. 58–87. [Google Scholar]
- Casino P, Rubio V, Marina A. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol. 2010;20:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Coyette J, Van Der Ende A. Peptidoglycan: the bacterial Achilles heel. FEMS Microbiol Rev. 2008;32:147–148. doi: 10.1111/j.1574-6976.2008.00108.x. [DOI] [PubMed] [Google Scholar]
- Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009;5:e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Silhavy TJ. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol. 2005;8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Stout V, Torres-Cabassa A, Maurizi MR, Gutnick D, Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991;173:1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn A, Whitfield C. Transcriptional organization and regulation of the Escherichia coli K30 group 1 capsule biosynthesis (cps) gene cluster. Mol Microbiol. 2003;47:1045–1060. doi: 10.1046/j.1365-2958.2003.03354.x. [DOI] [PubMed] [Google Scholar]
- Ferrières L, Clarke DJ. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol. 2003;50:1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Francez-Charlot A, Castanié-Cornet MP, Gutierrez C, Cam K. Osmotic regulation of the Escherichia coli bdm (biofilm-dependent modulation) gene by the RcsCDB His-Asp phosphorelay. J Bacteriol. 2005;187:3873–3877. doi: 10.1128/JB.187.11.3873-3877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubacher ME, Ades SE. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol. 2008;190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert L, Vanoirbeek KGA, Lurquin I, Michiels CW, Aertsen A. The Rcs two-component system regulates expression of lysozyme inhibitors and is induced by exposure to lysozyme. J Bacteriol. 2009;191:1979–1981. doi: 10.1128/JB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL, Popkin DL, Silhavy TJ. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol. 1999;181:5263–5272. doi: 10.1128/jb.181.17.5263-5272.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MPH, Buck M. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev. 2010;34:797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Yamamoto M, Aono R. Appearance of a stress-response protein, phage-shock protein A, in Escherichia coli exposed to hydrophobic organic solvents. Microbiology. 1998;144:353–359. doi: 10.1099/00221287-144-2-353. [DOI] [PubMed] [Google Scholar]
- Jovanovic G, Lloyd LJ, Stumpf MPH, Mayhew AJ, Buck M. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem. 2006;281:21147–21161. doi: 10.1074/jbc.M602323200. [DOI] [PubMed] [Google Scholar]
- Kobayashi R, Suzuki T, Yoshida M. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol. 2007;66:100–109. doi: 10.1111/j.1365-2958.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Stout V. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol Microbiol. 1991;5:1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x. [DOI] [PubMed] [Google Scholar]
- Agladze K, Wang X, Romeo T. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J Bacteriol. 2005;187:8237–8246. doi: 10.1128/JB.187.24.8237-8246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Gottesman S. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1996;178:1204–1206. doi: 10.1128/jb.178.4.1204-1206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhringer J, Fischer D, Mosler G, Henggearonis R. UDP-glucose is a potential intracellular signal molecule in the control of expression of sigma S and sigma S-dependent genes in Escherichia coli. J Bacteriol. 1995;177:413–422. doi: 10.1128/jb.177.2.413-422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrières L, Aslam SN, Cooper RM, Clarke DJ. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology. 2007;153:1070–1080. doi: 10.1099/mic.0.2006/002907-0. [DOI] [PubMed] [Google Scholar]
- Ionescu M, Belkin S. Overproduction of exopolysaccharides by an Escherichia coli K-12 rpoS mutant in response to osmotic stress. Appl Environ Microbiol. 2009;75:483–492. doi: 10.1128/AEM.01616-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariss SJL, Tayler AE, Avison MB. Defining the growth conditions and promoter-proximal DNA sequences required for activation of gene expression by CreBC in Escherichia coli. J Bacteriol. 2008;190:3930–3939. doi: 10.1128/JB.00108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig S, Hantke K, Ammelburg M, Braun V. CbrA is a flavin adenine dinucleotide protein that modifies the Escherichia coli outer membrane and confers specific resistance to colicin M. J Bacteriol. 2012;194:4894–4903. doi: 10.1128/JB.00782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhar I, Žgur-Bertok D. Transcription regulation of the colicin K cka gene reveals induction of colicin synthesis by differential responses to environmental signals. J Bacteriol. 1999;181:7373–7380. doi: 10.1128/jb.181.23.7373-7380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraso JM, Chidambaram M, Weinstock GM. Increased production of colicin E1 in stationary phase. J Bacteriol. 1996;178:1928–1935. doi: 10.1128/jb.178.7.1928-1935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya B, Dötsch A, Juan C, Blázquez J, Zamorano L, Haussler S, Oliver A. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 2009;5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JU, Gutierrez C, Martin F, Ardourel M, Villarejo M. Transcription of osmB, a gene encoding an Escherichia coli lipoprotein, is regulated by dual signals. Osmotic stress and stationary phase. J Biol Chem. 1990;265:10574–10581. [PubMed] [Google Scholar]
- Lee J, Hiibel SR, Reardon KF, Wood TK. Identification of stress-related proteins in Escherichia coli using the pollutant cis-dichloroethylene. J Appl Microbiol. 2010;108:2088–2102. doi: 10.1111/j.1365-2672.2009.04611.x. [DOI] [PubMed] [Google Scholar]
- Ratajczak E, Ziętkiewicz S, Liberek K. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J Mol Biol. 2009;386:178–189. doi: 10.1016/j.jmb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Micro. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Danese PN, Pratt LA, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. 2000;182:3593–3596. doi: 10.1128/JB.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A, Vogel J. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol Microbiol. 2012;84:1–5. doi: 10.1111/j.1365-2958.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, Pruteanu M, Hengge R. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol Microbiol. 2012;84:51–65. doi: 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E, Reimegård J, Sterk M, Grantcharova N, Römling U, Wagner EGH. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SH, Yeom JH, Shin C, Song WS, Shin E, Kim HM, Cha CJ, Han SH, Ha NC, Kim SW, Hahn Y, Bae J, Lee K. Escherichia coli ribonuclease III activity is downregulated by osmotic stress: consequences for the degradation of bdm mRNA in biofilm formation. Mol Microbiol. 2010;75:413–425. doi: 10.1111/j.1365-2958.2009.06986.x. [DOI] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors Ö. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NL, Raivio TL. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J Bacteriol. 2009;191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Ishihama A. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci Biotechnol Biochem. 2006;70:1688–1695. doi: 10.1271/bbb.60024. [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- Shi W, Li C, Louise CJ, Adler J. Mechanism of adverse conditions causing lack of flagella in Escherichia coli. J Bacteriol. 1993;175:2236–2240. doi: 10.1128/jb.175.8.2236-2240.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina OA, Krin E, Laurent-Winter C, Hommais F, Danchin A, Bertin PN. Regulation of bacterial motility in response to low pH in Escherichia coli: the role of H-NS protein. Microbiology. 2002;148:1543–1551. doi: 10.1099/00221287-148-5-1543. [DOI] [PubMed] [Google Scholar]
- Torres-Cabassa AS, Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim G, Huimi Wang H, Davies J. The truth about antibiotics. Int J Med Microbiol. 2006;296:163–170. doi: 10.1016/j.ijmm.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Fajardo A, Martinez JL. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Sailer FC, Meberg BM, Young KD. β-Lactam induction of colanic acid gene expression in Escherichia coli. FEMS Microbiol Lett. 2003;226:245–249. doi: 10.1016/S0378-1097(03)00616-5. [DOI] [PubMed] [Google Scholar]
- Kaldalu M, Mei R, Lewis K. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob Agents Chemother. 2004;48:890–896. doi: 10.1128/AAC.48.3.890-896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrt N, Mesak LR, Davies J. Modulation of virulence gene expression by cell wall active antibiotics in Staphylococcus aureus. J Antimicrob Chemother. 2011;66:979–984. doi: 10.1093/jac/dkr043. [DOI] [PubMed] [Google Scholar]
- Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- Linares JF, Gustafsson I, Baquero F, Martinez JL. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci USA. 2006;103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/S1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi DT, López E, Rodríguez-Rojas A, Rodríguez-Beltrán J, Couce A, Guelfo JR, Castañeda-García A, Blázquez J. Effect of recA inactivation on mutagenesis of Escherichia coli exposed to sublethal concentrations of antimicrobials. J Antimicrob Chemother. 2011;66:531–538. doi: 10.1093/jac/dkq496. [DOI] [PubMed] [Google Scholar]
- Moreau PL. Diversion of the metabolic flux from pyruvate dehydrogenase to pyruvate oxidase decreases oxidative stress during glucose metabolism in nongrowing Escherichia coli cells incubated under aerobic, phosphate starvation conditions. J Bacteriol. 2004;186:7364–7368. doi: 10.1128/JB.186.21.7364-7368.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed H, Gillor O, Kerr B, Riley MA. Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J. 2011;5:71–81. doi: 10.1038/ismej.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D, Rolfe M, Thompson A, Moore GR, James R, Hinton JCD, Kleanthous C. Transcriptional profiling of colicin-induced cell death of Escherichia coli MG1655 identifies potential mechanisms by which bacteriocins promote bacterial diversity. J Bacteriol. 2004;186:866–869. doi: 10.1128/JB.186.3.866-869.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreteau H, Bouhss A, Fourgeaud M, Mainardi JL, Touzé T, Gérard F, Blanot D, Arthur M, Mengin-Lecreulx D. Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J Bacteriol. 2009;191:3657–3664. doi: 10.1128/JB.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnéoud-Arnoulet A, Barreteau H, Touzé T, Mengin-Lecreulx D, Lloubès R, Duché D. Toxicity of the colicin M catalytic domain exported to the periplasm is FkpA independent. J Bacteriol. 2010;192:5212–5219. doi: 10.1128/JB.00431-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreteau H, Bouhss A, Gérard F, Duché D, Boussaid B, Blanot D, Lloubès R, Mengin-Lecreulx D, Touzé T. Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II-degrading enzyme. J Biol Chem. 2010;285:12378–12389. doi: 10.1074/jbc.M109.093583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breukink E, de Kruijff B. Lipid II as a target for antibiotics. Nat Rev Drug Discov. 2006;5:321–323. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- Budič M, Rijavec M, Petkovšek Ž, Žgur-Bertok D. Escherichia coli bacteriocins: antimicrobial efficacy and prevalence among isolates from patients with bacteraemia. PLoS One. 2011;6:e28769. doi: 10.1371/journal.pone.0028769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderluh G, Gökçe I, Lakey JH. Expression of proteins using the third domain of the Escherichia coli periplasmic-protein TolA as a fusion partner. Protein Expr Purif. 2003;28:173–181. doi: 10.1016/S1046-5928(02)00681-2. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obadia B, Lacour S, Doublet P, Baubichon-Cortay H, Cozzone AJ, Grangeasse C. Influence of tyrosine-kinase Wzc activity on colanic acid production in Escherichia coli K12 Cells. J Mol Biol. 2007;367:42–53. doi: 10.1016/j.jmb.2006.12.048. [DOI] [PubMed] [Google Scholar]
- Rijavec M, Müller-Premru M, Zakotnik B, Žgur-Bertok D. Virulence factors and biofilm production among Escherichia coli strains causing bacteraemia of urinary tract origin. J Medical Microbiol. 2008;57:1329–1334. doi: 10.1099/jmm.0.2008/002543-0. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics: Assay of β-galactosidase. Cold Spring Harbor: CSH Laboratory Press; 1972. pp. 352–355. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth of E. coli MG1655 treated with colicin M. The arrow denotes the time of addition of colicin M, at inhibitory (100 ng/ml, 50 ng/ml) and subinhibitory concentrations (30 ng/ml, 20 ng/ml, 10 ng/ml). Growth curves represent E. coli MG1655 cultures treated with different colicin M concentrations.
Effect of subinhibitory concentrations of colicin M on E. coli MG1566 viable counts. Growth curves with viable counts (CFU/ml as a function of time relative to antibiotic addition) are shown for untreated and treated culture (30 ng/ml of colicin M).
Time course analysis of differentially expressed genes after 30 and 60 min exposure to subinhibitory concentrations of colicin M. p≤0.05, log2 FC≥1 and ≤−1, log2 FC≤1, ≥−1; p≥0.05, log2 FC≥1 and ≤−1, log2 FC≤1, ≥−1. Log2 FC values in bold correspond to log2 FC≥1 and ≤−1 when p≤0.05 and in regular type to log2 FC≤1, ≥−1 when p≤0.05. Log2 FC values in italics bold correspond to log2 FC≥1 and ≤−1 when p≥0.05 and in italics regular type to log2 FC≤1, ≥−1 when p≥0.05.
SDS-PAGE gel showing purity of isolated colicin M. Left, Protein ladder Page Ruler (Fermentas); Right, colicin M - 29.5 kDa, colicin M (3.4 mg/ml).
Primer pairs used for qRT-PCR in the present study.