Abstract
Background
A nitrilase-mediated pathway has significant advantages in the production of optically pure (R)-(−)-mandelic acid. However, unwanted byproduct, low enantioselectivity, and specific activity reduce its value in practical applications. An ideal nitrilase that can efficiently hydrolyze mandelonitrile to optically pure (R)-(−)-mandelic acid without the unwanted byproduct is needed.
Results
A novel nitrilase (BCJ2315) was discovered from Burkholderia cenocepacia J2315 through phylogeny-based enzymatic substrate specificity prediction (PESSP). This nitrilase is a mandelonitrile hydrolase that could efficiently hydrolyze mandelonitrile to (R)-(−)-mandelic acid, with a high enantiomeric excess of 98.4%. No byproduct was observed in this hydrolysis process. BCJ2315 showed the highest identity of 71% compared with other nitrilases in the amino acid sequence. BCJ2315 possessed the highest activity toward mandelonitrile and took mandelonitrile as the optimal substrate based on the analysis of substrate specificity. The kinetic parameters Vmax, Km, Kcat, and Kcat/Km toward mandelonitrile were 45.4 μmol/min/mg, 0.14 mM, 15.4 s-1, and 1.1×105 M-1s-1, respectively. The recombinant Escherichia coli M15/BCJ2315 had a strong substrate tolerance and could completely hydrolyze mandelonitrile (100 mM) with fewer amounts of wet cells (10 mg/ml) within 1 h.
Conclusions
PESSP is an efficient method for discovering an ideal mandelonitrile hydrolase. BCJ2315 has high affinity and catalytic efficiency toward mandelonitrile. This nitrilase has great advantages in the production of optically pure (R)-(−)-mandelic acid because of its high activity and enantioselectivity, strong substrate tolerance, and having no unwanted byproduct. Thus, BCJ2315 has great potential in the practical production of optically pure (R)-(−)-mandelic acid in the industry.
Keywords: (R)-(−)-mandelic acid, Nitrilase, Burkholderia cenocepacia J2315, Substrate specificity prediction, Enantioselective hydrolysis
Background
Optically pure 2-hydroxycarboxylic acids are important intermediates in the pharmaceutical and fine chemical industries [1-4]. (R)-(−)-mandelic acid is one of the important 2-hydroxycarboxylic acids, which is widely used for the production of semisynthetic cephalosporins [5], penicillins [6], antitumor agents [7], and antiobesity agents [8]. It is also used as a universal acidic chiral resolving agent for the resolution of racemic alcohols and amines [9].
Several methods have been proposed for the production of optically pure (R)-(−)-mandelic acid [4,10]. Among these methods, nitrilase-mediated pathway is increasingly popular because of its lack of cofactor involvement, cheap starting material in the form of mandelonitrile, high enantioselectivity, and theoretically 100% of the product [10-15]. However, these reported nitrilases either have low enantioselectivity or low specific activity toward mandelonitrile [16]. Furthermore, some also produce a byproduct in the form of mandelamide [16,17]. Therefore, an ideal nitrilase that can efficiently hydrolyze mandelonitrile to optically pure (R)-(−)-mandelic acid without the unwanted byproduct is needed.
Several approaches have been developed to discover novel nitrilases toward mandelonitrile [18-22]. Among these approaches, an enrichment culture [19] and the metagenome approach [20] have been used successfully. However, these methods require screening a large number of clones, and are thereby time consuming. Considering that the number of genes increases exponentially based on an automated genome annotation in the database, genome mining has become increasingly popular in the recent years. Researchers can easily find many genes with a defined function, such as nitrilase, from databases, such as GenBank, Pfam, and Brenda. Nitrilases of interest can be discovered more efficiently by combining the existing methods with substrate specificity prediction. Zhu et al. [21] discovered a mandelonitrile hydrolase (nitrilase) by combining traditional mining with the functional analysis of the flanking genes around this nitrilase. This nitrilase was organized in a mandelonitrile metabolic pathway and displayed high activity toward mandelonitrile. Seffernick et al. [22] also discovered a nitrilase and another mandelonitrile hydrolase from Burkholderia xenovorans LB400 using computational methods. However, these two nitrilases exhibited no or only slight enantioselectivity in producing (R)-(−)-mandelic acid.
In our study, phylogeny-based enzymatic substrate specificity prediction (PESSP) was introduced for the efficient discovery of an ideal nitrilase to solve the problems of unwanted byproduct production, low enantioselectivity, and specific activity. A novel nitrilase (BCJ2315) was discovered from Burkholderia cenocepacia J2315. BCJ2315 could efficiently hydrolyze mandelonitrile to (R)-(−)-mandelic acid with high enantioselectivity. No byproduct was observed in the hydrolysis process. BCJ2315 was cloned and overexpressed in Escherichia coli M15, and its catalytic properties were investigated by analyzing its substrate specificity and kinetic parameters. The catalytic efficiency of the recombinant E. coli M15/BCJ2315 was also tested in the hydrolyzing mandelonitrile biotransformation to (R)-(−)-mandelic acid to investigate the potential of BCJ2315 further.
Results and discussion
Discovery of a predicted mandelonitrile hydrolase subgroup through PESSP
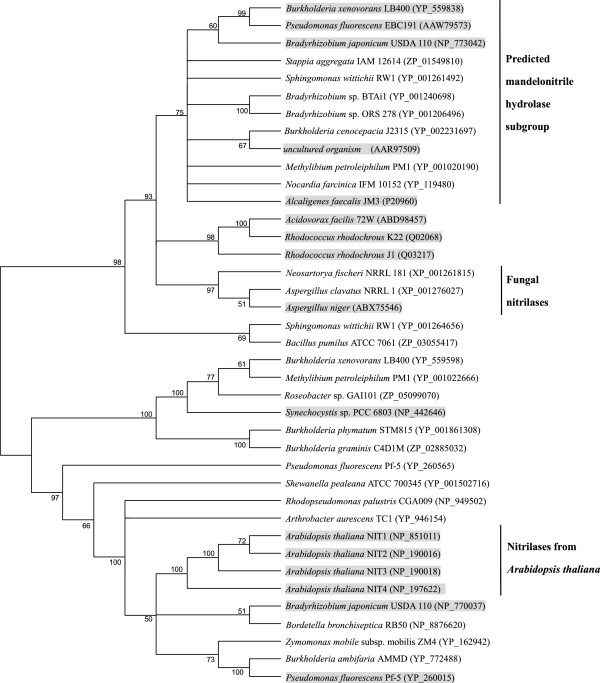
Based on the screening criteria mentioned in Database mining and sequence analysis section, a total of 39 proteins were chosen for the mandelonitrile hydrolase activity assay (Table 1). These proteins were annotated as nitrilase, putative nitrilase, aliphatic nitrilase, and unnamed protein products. Among the 39 proteins, 16 were experimentally determined to have a nitrilase activity with different substrate specificities. For example, the nitrilase from Rhodococcus rhodochrous J1 [23] was designated as an aromatic nitrilase. The nitrilases from Synechocystis sp. PCC6803 [24] and Acidovorax facilis 72W [25] were specific to aliphatic (di)nitrile. The nitrilase from Pseudomonas fluorescens Pf-5 [26] had a regioselective activity toward aliphatic dinitrile. The nitrilases from Alcaligenes faecalis JM3 [13], Pseudomonas fluorescens EBC191 [17], Bradyrhizobium japonicum USDA110 [21], Burkholderia xenovorans LB400 [22], and an uncultured organism (nitrilase I, 2A6) [20] were characterized as mandelonitrile hydrolases. Finally, a cluster containing all the defined mandelonitrile hydrolases was found based on the phylogenetic analysis (Figure 1). Seven proteins were not characterized experimentally within this cluster, and their functions remained unclear. Based on the substrate specificities of the defined nitrilases, this cluster was designated as the predicted mandelonitrile hydrolase subgroup. The uncharacterized seven proteins in this subgroup were further studied.
Table 1.
| Organism | GenBank Accession no. | Predicted function | Defined functionb | Identitya(%) | Reference |
|---|---|---|---|---|---|
|
Bradyrhizobium japonicum USDA 110 |
NP_773042 |
|
nitrilase |
100 |
[21] |
|
Burkholderia xenovorans LB400 |
YP_559838 |
|
nitrilase |
60 |
[22] |
|
Pseudomonas fluorescens EBC191 |
AAW79573 |
|
nitrilase |
60 |
[17] |
|
Sphingomonas wittichii RW1 |
YP_001261492 |
unnamed protein product |
|
58 |
This study |
|
Bradyrhizobium sp. ORS 278 |
YP_001206496 |
aliphatic nitrilase |
|
58 |
This study |
|
Bradyrhizobium sp. BTAi1 |
YP_001240698 |
aliphatic nitrilase |
|
57 |
This study |
|
uncultured organism |
AAR97509 |
|
nitrilase |
56 |
[20] |
|
Burkholderia cenocepacia J2315 |
YP_002231697 |
putative nitrilase |
|
56 |
This study |
|
Methylibium petroleiphilum PM1 |
YP_001020190 |
aliphatic nitrilase |
|
55 |
This study |
|
Nocardia farcinica IFM 10152 |
YP_119480 |
nitrilase |
|
55 |
This study |
|
Stappia aggregata IAM 12614 |
ZP_01549810 |
nitrilase |
|
53 |
This study |
|
Rhodococcus rhodochrous K22 |
Q02068 |
|
nitrilase |
51 |
[27] |
|
Rhodococcus rhodochrous J1 |
Q03217 |
|
nitrilase |
50 |
[23] |
|
Acidovorax facilis 72W |
ABD98457 |
|
nitrilase |
49 |
[25] |
|
Alcaligenes faecalis JM3 |
P20960 |
|
nitrilase |
47 |
[13] |
|
Sphingomonas wittichii RW1 |
YP_001264656 |
unnamed protein product |
|
46 |
This study |
|
Neosartorya fischeri NRRL 181 |
XP_001261815 |
nitrilase, putative |
|
45 |
This study |
|
Aspergillus clavatus NRRL 1 |
XP_001276027 |
nitrilase, putative |
|
41 |
This study |
|
Aspergillus niger |
ABX75546 |
|
nitrilase |
38 |
[28] |
|
Bordetella bronchiseptica RB50 |
NP_887662 |
unnamed protein product |
|
37 |
This study |
|
Arthrobacter aurescens TC1 |
YP_946154 |
nitrilase |
|
36 |
This study |
|
Bacillus pumilus ATCC 7061 |
ZP_03055417 |
nitrilase |
|
36 |
This study |
|
Rhodopseudomonas palustris CGA009 |
NP_949502 |
nitrilase |
|
36 |
This study |
|
Pseudomonas fluorescens Pf-5 |
YP_260565 |
carbon-nitrogen family hydrolase |
|
36 |
This study |
|
Arabidopsis thaliana |
NP_190018 |
|
nitrilase |
35 |
[29] |
|
Burkholderia graminis C4D1M |
ZP_02885032 |
nitrilase |
|
35 |
This study |
|
Bradyrhizobium japonicum USDA 110 |
NP_770037 |
|
nitrilase |
35 |
[30] |
|
Arabidopsis thaliana |
NP_197622 |
|
nitrilase |
35 |
[31] |
|
Methylibium petroleiphilum PM1 |
YP_001022666 |
nitrilase |
|
34 |
This study |
|
Synechocystis sp. PCC 6803 |
NP_442646 |
|
nitrilase |
34 |
[24] |
|
Burkholderia phymatum STM815 |
YP_001861308 |
unnamed protein product |
|
34 |
This study |
|
Arabidopsis thaliana |
NP_190016 |
|
nitrilase |
34 |
[29] |
|
Arabidopsis thaliana |
NP_851011 |
|
nitrilase |
34 |
[29] |
|
Shewanella pealeana ATCC 700345 |
YP_001502716 |
nitrilase |
|
33 |
This study |
|
Burkholderia xenovorans LB400 |
YP_559598 |
nitrilase |
|
32 |
This study |
|
Roseobacter sp. GAI101 |
ZP_05099070 |
aliphatic nitrilase |
|
32 |
This study |
|
Burkholderia ambifaria AMMD |
YP_772488 |
nitrilase/cyanide hydratase hydratase and apolipoprotein N-acyltransferase |
|
31 |
This study |
|
Pseudomonas fluorescens Pf-5 |
YP_260015 |
|
nitrilase |
31 |
[26] |
| Zymomonas mobilis ZM4 | YP_162942 | unnamed protein product | 30 | This study |
a BLASTP was performed by comparing the amino acid sequence with that of the mandelonitrile hydrolase (bll6042, NP_773042) from Bradyrhizobium japonicum USDA 110.
b The enzymes were experimentally characterized to harbor nitrilase activity.
Figure 1.
Unrooted neighbor-joining tree based on the amino acid sequences of the organisms from Table 1(accession numbers are in parentheses). A consensus tree was constructed using a bootstrap test with 1000 replications. Bootstrap values greater than 50% are shown at the branch points. The organisms harboring defined nitrilase activity are shadowed.
Identification of BCJ2315 from the predicted mandelonitrile hydrolase subgroup
The respective genes of the seven proteins were cloned and overexpressed in E. coli to verify whether these uncharacterized proteins in the predicted mandelonitrile hydrolase subgroup have a mandelonitrile hydrolase activity. The resulting recombinant histidine (His)-tagged proteins were all soluble and purified to homogeneity for the catalytic activity assay. All the seven enzymes were active toward mandelonitrile and have relatively high enantioselectivity (Table 2). Among these enzymes, BCJ2315 from Burkholderia cenocepacia J2315 exhibited the highest specific activity (27.79 U/mg) and enantioselectivity (98.4%). Only the SA12614 produced amide as a reaction byproduct. The sequence analysis of BCJ2315 showed 52%, 59%, 56%, 60%, and 66% identities with the nitrilases from Alcaligenes faecalis JM3 [13], Pseudomonas fluorescens EBC191 [17], Bradyrhizobium japonicum USDA110 [21], Burkholderia xenovorans LB400 [22], and an uncultured organism (nitrilase I, 2A6) [20], respectively. The highest identity was 71% compared with nitrilase (2A12) from an uncultured organism discovered by Robertson et al. [20]. To the best of our knowledge, the current study is the first report of nitrilase BCJ2315. BCJ2315 was chosen for further study because it had the highest activity and enantioselectivity toward mandelonitrile.
Table 2.
Specific activity and enantioselectivity of the nitrilases in predicted mandelonitrile hydrolase subgroup
| Nitrilase | Organism | Specific activity (U/mg) | ee value of acid (%) | Amide formation (% of total products) |
|---|---|---|---|---|
| BCJ2315 |
Burkholderia cenocepacia J2315 |
27.79±1 |
98.4 |
N.D. |
| SWRW1 |
Sphingomonas wittichii RW1 |
4.60±0.3 |
96.4 |
N.D. |
| MPPM1 |
Methylibium petroleiphilum PM1 |
7.67±0.5 |
93.4 |
N.D. |
| SA12614 |
Stappia aggregata IAM 12614 |
3.87±0.3 |
92.8 |
8 |
| NF10152 |
Nocardia farcinica IFM 10152 |
10.11±0.7 |
91.5 |
N.D. |
| BSBTAi1 |
Bradyrhizobium sp. BTAi1 |
5.82±0.5 |
90.9 |
N.D. |
| BS278 | Bradyrhizobium sp. ORS 278 | 1.33±0.06 | 89.7 | N.D. |
N.D. Not detected.
The rest of the uncharacterized genes outside the predicted mandelonitrile hydrolase subgroup in the phylogenetic tree were also cloned and overexpressed in E. coli (Additional file 1: Table S1) so that any other nitrilases with good characteristics toward mandelonitrile would not be missed. After optimizing the expression conditions (induction temperature, Isopropyl-β-D-thiogalactopyranoside (IPTG) concentration, and expression vector/host), all the enzymes were expressed in a soluble form and showed activity toward at least one of the four assayed nitrile substrates (benzonitrile, phenylacetonitrile, acrylonitrile, and succinonitrile). The recombinant His-tagged proteins were purified for the mandelonitrile hydrolase activity assay. Little to no activity was observed in the high-performance liquid chromatography (HPLC) analysis after 12 h of hydrolysis (Additional file 1: Table S1). This result further proved the accuracy of the prediction based on phylogenetic analysis.
Properties of the purified BCJ2315
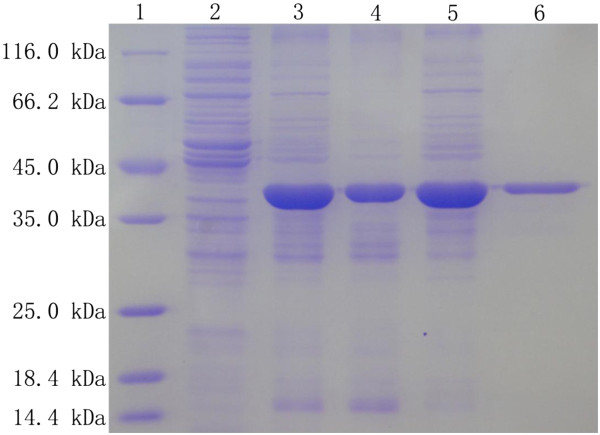
The molecular weight of the purified native BCJ2315 estimated by conducting a gel filtration chromatography was about 450 kDa. BCJ2315 showed one single band on the SDS-PAGE with a molecular weight of 37 kDa (Figure 2). This result indicated that the native BCJ2315 consisted of 12 subunits with identical sizes, which is in agreement with most nitrilases reported with 6 to 26 identical subunits that self-aggregated to form active enzymes [32].
Figure 2.
SDS-PAGE analysis of the nitrilase BCJ2315. Lane 1: protein marker; Lane 2; whole cell lysates of E. coli M15/pQE30; Lane 3: whole cell lysates of E. coli M15/BCJ2315; Lane 4: insoluble fractions of the whole cell lysates of E. coli M15/BCJ2315; Lane 5: soluble fractions of the whole cell lysates of E. coli M15/BCJ2315; Lane 6: purified BCJ2315.
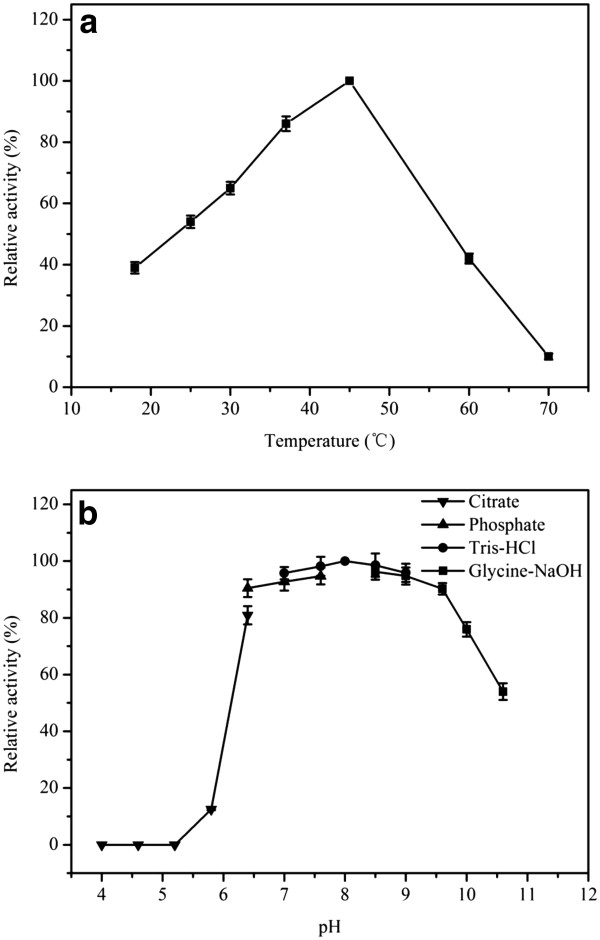
The optimum temperature and pH of the purified BCJ2315 were determined. The optimum temperature was 45°C, as shown in Figure 3a. When the temperature was above 45°C, the activity of BCJ2315 decreased sharply. This behavior is similar to the nitrilases reported from mesophilic organisms, having optimum temperatures ranging from 30°C to 50°C [1,11,12,17,33]. BCJ2315 showed the highest activity at pH 8.0 (Figure 3b). Only small changes in activity were observed between pH 6.4 and 9.6. These variations suggested that BCJ2315 had a relatively broad optimum pH in contrast to other arylacetonitrilases that have a rather narrow optimum pH at neutral or slightly alkaline pH values [11].
Figure 3.
Effect of temperature and pH on the activity of purified BCJ2315 toward mandelonitrile. (a) Optimum temperature: enzyme activity was measured at various temperatures (18°C to 70°C) in 100 mM of sodium phosphate buffer (pH 7.0). (b) Optimum pH: enzyme activity was measured in different buffers (pH 4.0 to 10.6) at 30°C. The relative activity was expressed as a percentage of the maximum activity under the experimental condition used.
The catalytic activity of BCJ2315 toward 24 different nitriles with structural diversity was investigated. Table 3 lists the relative activities determined by quantifying the amount of ammonia released during the hydrolysis. A clear preference of BCJ2315 for arylacetonitriles as substrates indicated that this enzyme is an arylacetonitrilase. A lower activity was also observed with the aliphatic and heterocyclic nitriles. No detectable activities were observed with the aromatic nitriles. BCJ2315 showed the highest activity toward mandelonitrile (8.8 times more than that of phenylacetonitrile), indicating that BCJ2315 is a highly active mandelonitrile hydrolase. Moreover, the activities of other arylacetonitrilases, such as Alcaligenes faecalis ATCC 8750 [10], Alcaligenes sp. ECU0401 [11], Alcaligenes faecalis ZJUTB10 [12], Alcaligenes faecalis JM3 [13], and Pseudomonas putida[33], toward mandelonitrile are only 12% to 50% of that for phenylacetonitrile.
Table 3.
Substrate specificity of the purified BCJ2315
| Entry | Substrate | Relative activitya(%) |
|---|---|---|
| 1 |
Iminodiacetonitrile |
21±1 |
| 2 |
acrylonitrile |
17±1 |
| 3 |
2-Methylglutaronitrile |
25±3 |
| 4 |
Succinonitrile |
5±1 |
| 5 |
Fumaronitrile |
78±3 |
| 6 |
Malononitrile |
17±1 |
| 7 |
Valeronitrile |
4±1 |
| 8 |
3-Hydroxyglutaronitrile |
N.D. |
| 9 |
4-Chlorobutyronitrile |
11±1 |
| 10 |
Glycolonitrile |
69±4 |
| 11 |
3-Phenylpropionitrile |
N.D. |
| 12 |
Benzonitrile |
N.D. |
| 13 |
Cinnamonitrile |
N.D. |
| 14 |
Phenylacetonitrile |
100±4 |
| 15 |
2-Chloromandelonitrile |
298±7 |
| 16 |
1,2-Phenylenediacetonitrile |
37±5 |
| 17 |
2-Phenylbutyronitrile |
N.D. |
| 18 |
alpha-Methylphenylacetonitrile |
N.D. |
| 19 |
mandelonitrile |
884±7 |
| 20 |
3-Cyanopyridine |
N.D. |
| 21 |
Thiophene-3-carbonitrile |
1±1 |
| 22 |
Indole-3-acetonitrile |
20±1 |
| 23 |
2-Cyanopyridine |
7±1 |
| 24 | 4-Cyanopyridine | 7±1 |
a Nitrilase activity against various nitriles was measured under standard assay conditions. The activity toward phenylacetonitrile was defined as 100%.
N.D. not detected.
The kinetic parameters of BCJ2315 were determined using mandelonitrile as substrate. The obtained Km and Vmax were 0.14 mM and 45.4 μmol/min/mg, respectively. The low value of Km indicated that BCJ2315 had high affinity toward mandelonitrile. The Km-values of other highly enantioselective mandelonitrile hydrolases were one to three orders of magnitude higher (above 3.4 mM) than that of BCJ2315 (Table 4) [11,12,16,33]. The Kcat and Kcat/Km were 15.4 s-1 and 1.1×105 M-1s-1, respectively. BCJ2315 also had a high catalytic efficiency, comparable with that of the nitrilase (bll6402) from Bradyrhizobium japonicum USDA110 (1.04×105 M-1s-1) [21].
Table 4.
Kinetic parameters, specific activity, enantioselectivity, and specificity of BCJ2315 compared with other mandelonitrile hydrolases[33-35]
| Organism |
Kinetic parameters |
Specific activity (U/mg) | ee value of acid (%) | Amide formation (% of total products) | Reference | |
|---|---|---|---|---|---|---|
| Km(mM) | Vmax(μmol/min/mg) | |||||
|
uncultured organisma |
N.A. |
N.A. |
50 |
98 |
N.D. |
[34] |
|
Pseudomonas fluorescens EBC191 |
N.A. |
N.A. |
32.8 |
31 |
19 |
[19] |
|
Burkholderia cenocepacia J2315 |
0.14 |
45.4 |
27.79 |
98.4 |
N.D. |
This study |
|
Bradyrhizobium japonicum USDA110b |
0.26 |
44.7 |
24.38 |
- |
N.D. |
[21] |
|
Alcaligenes sp. ECU0401 |
21.8 |
27.9 |
19 |
97 |
N.D. |
[11] |
|
Alcaligenes faecalis JM3 |
N.A. |
N.A. |
12.4 |
99 |
N.D. |
[13] |
|
Aspergillus niger CBS 513.88 |
11.4 |
12.4 |
7.7 |
99 |
N.D. |
[16], [35] |
|
Neurospora crassa OR74A |
3.4 |
9.9 |
6.1 |
99 |
15 |
[16], [35] |
|
Pseudomonas putida |
13.39 |
16.5 |
3.26 |
99.9 |
N.D. |
[33] |
|
Alcaligenes faecalis ATCC8750 |
N.A. |
N.A. |
3.1 |
100 |
N.D. |
[10] |
|
Burkholderia xenovorans LB400 |
0.084 |
N.A. |
1.523 |
- |
N.D. |
[22] |
| Alcaligenes faecalis ZJUTB10 | 4.74 | 15.85 | N.A. | 99 | N.D. | [12] |
N.A. data not available.
N.D. not detected.
- no enantioselectivity was observed.
a nitrilase I (2A6) was referred here.
b nitrilase (bll6402) was referred here.
BCJ2315 exhibited a relatively high specific activity (27.79 U/mg) among the reported mandelonitrile hydrolases. It has the 2nd highest activity of all known highly enantioselective nitrilases in literature (Table 4). The highest specific activity toward mandelonitrile was observed with nitrilase I (50 U/mg) from an uncultured organism discovered by Robertson et al. Nitrilase I also has an excellent enantioselectivity toward mandelonitrile (enantiomeric excess (ee), 98%) and mandelonitrile derivatives [20,34]. Strangely, these two nitrilases only shared a 66% identity in the amino acid sequence, probably because of the method we used (PESSP). The PESSP method discovered new enzymes based on their substrate specificity toward mandelonitrile, other than the sequence identity. Therefore, despite the low identity between these two nitrilases, they were successfully clustered together into the predicted mandelonitrile hydrolase subgroup. PESSP exhibited an advantage in searching for enzymes with similar characteristics, although these enzymes may be quite different in the amino acid sequence. A higher specific activity was also observed with the nitrilases from Pseudomonas fluorescens EBC191 (32.8 U/mg) and Bradyrhizobium japonicum USDA110 (24.38 U/mg). However, these two nitrilases had lower ees, and the nitrilase from Pseudomonas fluorescens EBC191 also produced amide as a byproduct. The other highly enantioselective mandelonitrile hydrolases from Alcaligenes sp. ECU0401, Alcaligenes faecalis JM3, Aspergillus niger CBS 513.88, Neurospora crassa OR74A, Pseudomonas putida and Alcaligenes faecalis ATCC8750 exhibited a relatively low specific activity (Table 4). Thus, when the specific activity, enantioselectivity, and production of unwanted byproducts were taken into account, BCJ2315 demonstrated a great potential for the industrial production of optically pure (R)-(−)-mandelic acid.
Conversion of mandelonitrile by the recombinant E. coli M15/BCJ2315
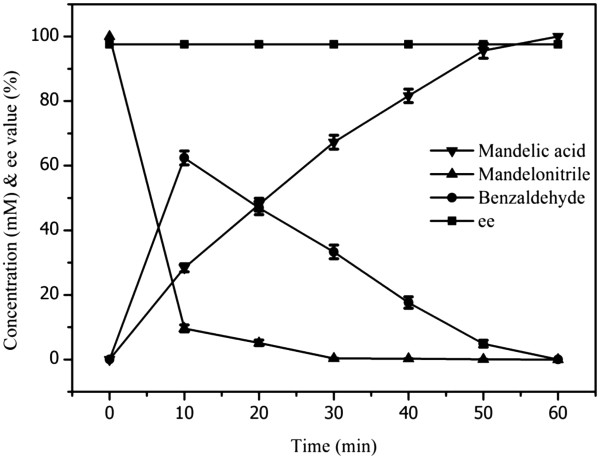
Whole cell biocatalysis was performed using the recombinant E. coli M15/BCJ2315 as biocatalyst to estimate the potential of BCJ2315 for mandelonitrile hydrolysis further. An optimization of the reaction conditions for M15/BCJ2315 was performed by evaluating the effect of pH, temperature, cell concentration, and mandelonitrile concentration in a reaction system of 10 ml. The optimal reaction system consisted of wet cells (100 mg) and mandelonitrile (100 mM) in 10 ml of phosphate buffer (100 mM, pH 8.0) (data not shown). Although the optimal temperature was 45°C, we conducted the reaction at 30°C to consider the thermal deactivation of the enzyme. The reaction process is shown in Figure 4. Considering that mandelonitrile can be decomposed into benzaldehyde and hydrogen cyanide in aqueous solution at pH 7.0 and above, the benzaldehyde concentration was also plotted into the figure to help understand the mechanism of the mandelonitrile hydrolysis mediated by M15/BCJ2315. The mandelonitrile (100 mM) could be hydrolyzed completely by M15/BCJ2315 within 1 h. No mandelamide was detected during the reaction. The ee value for the (R)-(−)-mandelic acid was constant at 97.6% during the whole hydrolysis process. Finally, the (R)-(−)-mandelic acid was recovered with a total yield of 93.5%. The ee of the production was determined as 99.8% after the recrystallization in benzene through HPLC. The product was characterized as follows: [α]D25 = −152.7 (c 1.0, H2O) (literature: [α]D25 = −155 (c 1.0, H2O) [10]; 1H NMR (400 MHz, DMSO): δ12.58 (brs, 1H), 7.45–7.39 (m, 2H), 7.58 (d, J = 7.2 Hz, 2H), 7.32–7.26 (m, 1H), 5.84 (brs, 1H), 5.03 (s, 1H) ppm.
Figure 4.
Time course of the mandelonitrile hydrolysis mediated by the recombinant E. coli M15/BCJ2315. The reaction mixture (10 ml) containing wet cells (100 mg) and mandelonitrile (100 mM) suspended in phosphate buffer (100 mM, pH 8.0) was incubated in a rotary shaker (30°C, 200 rpm). Samples were taken every 10 min and quenched by the addition of 10% (v/v) 2 M HCl.
Among the reported nitrilase-mediated hydrolysis of mandelonitrile, only the nitrilase from Alcaligenes sp. ECU0401 could enantioselectively hydrolyze mandelonitrile in a high substrate concentration [36]. When 100 mM of mandelonitrile was used, the yield of the (R)-(−)-mandelic acid reached 100% with wet cells (100 mg/ml) in 2 h and the ee was as high as 99%. In the current study, we used less biocatalysts (10 mg/ml of wet cells) to realize the hydrolysis of the same mandelonitrile concentration (100 mM) with a high enantioselectivity (97.6%) in 1 h of reaction time. This result may be due to the high specific activity of BCJ2315 and the high soluble expression in E. coli. The biocatalyst preparation accounts for a great part of the total cost of an enzymatic process. Therefore, using M15/BCJ2315 for the production of (R)-(−)-mandelic acid is beneficial in two ways, i.e., the processing time is shorter and require less amount of biocatalyst (resting cells), thereby significantly reducing the production cost compared with other reported systems.
Conclusions
A novel mandelonitrile hydrolase BCJ2315 was discovered from Burkholderia cenocepacia J2315 through PESSP. BCJ2315 took mandelonitrile as its optimal substrate and exhibited great advantages in the production of optically pure (R)-(−)-mandelic acid. These advantages include a high enantioselectivity and activity, strong substrate tolerance, and having no byproduct. These advantages make BCJ2315 more efficient in the hydrolysis of mandelonitrile. Thus, BCJ2315 has a great potential in the practical production of optically pure (R)-(−)-mandelic acid.
Methods
Materials
All the bacterial strains used for the mandelonitrile hydrolase analysis were obtained from the China General Microbiological Culture Collection Center, German collection of Microorganisms and Cell Cultures, and American Type Culture Collection (ATCC). E. coli BL21/pET28a(+) and M15/pQE30 were used for expressing the nitrilases. The nitrile substrates and carboxylic acids were purchased from Sigma–Aldrich (Milwaukee, USA).
Database mining and sequence analysis
The database searches of the sequence data were performed using the BLASTP program (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins&PROGRAM=blastp&BLAST_PROGRAMSblastp&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome). The nitrilase (bll6402, NP_773042) from Bradyrhizobium japonicum USDA110 [21] was chosen as the identifier to detect mandelonitrile-specific nitrilase (mandelonitrile hydrolase). Bll6042 was the most active toward mandelonitrile and took mandelonitrile as its optimal substrate. The identity of the amino acid sequence was used as the first criterion to screen the BLASTP result. Sequences with identities greater than 90% and less than 30% were removed to filter the enzymes with the same characteristics or with more distinct characteristics. No more than two sequences with the same identity were kept for analysis. The second criterion was the source of the sequence. Sequences from the same species were chosen once because these sequences usually share high identities and the same characteristics with one another. Two or more different sequences from the same strain were kept. The third criterion was the availability of the organisms that harbor nitrilase. Regardless of these three criteria, the sequences with experimentally defined nitrilase substrate specificity were chosen as a priority to refine the phylogenetic analysis. The sequences annotated as unnamed protein products were checked for the Glu-Lys-Cys catalytic triad [32,37] using the ScanProsite tool in the ExPASy proteomic server.
The alignment of the obtained sequences was conducted using ClustalW [38]. A bootstrap consensus tree was built by using a neighbor-joining method packaged in MEGA version 4.0 [39].
Cloning and expression of nitrilase genes in E. coli
The nitrilase gene primers in the predicted mandelonitrile hydrolase subgroup used in this study are listed in Table 5. Recombinant DNA techniques were performed according to standard protocols [40]. All the recombinant expression plasmids were transformed into E. coli BL21 (DE3) or E. coli M15. The recombinant E. coli cells were cultivated in a Luria–Bertani medium containing antibiotics at 37°C. IPTG was added to a final concentration of 0.1 mM to induce the cultures when the OD600 reached 0.6 to 0.8. The E. coli BL21 (DE3) or E. coli M15 cultures were further incubated for 20 h at 20°C or 30°C, respectively. The induced cells were harvested through centrifugation (12,000 rpm, 10 min) at 4°C and stored at −20°C.
Table 5.
Primers and locus tags of the nitrilase genes in the predicted mandelonitrile hydrolase subgroup used in this study
| Sequence Accession no. | Locus_tag | Source | Primer pair (sequence 5’→ 3’) | Restriction enzymes |
|---|---|---|---|---|
|
CP000699 |
Swit_0987 |
Sphingomonas wittichii RW1a |
Forward: CATATGTCGGACGGTCCGTTCAAGGTTG |
NdeI/HindIII |
| Reverse: AAGCTTTTAGGAACCGGCCTTTTCGGCGA | ||||
|
CU234118 |
BRADO4539 |
Bradyrhizobium sp. ORS 278a |
Forward: GGGAATTCCATATGGGACTGGCACATCCG |
NdeI/HindIII |
| Reverse: GAAAAAAGCTTACCCGCCAGCCGCGACCT | ||||
|
CP000494 |
BBta_4766 |
Bradyrhizobium sp. BTAi1b |
Forward: GGATCCATGGGACTGGCACATCCGAAATAC |
BamHI/HindIII |
| Reverse: AAGCTTCAGATGGATTGATCGGGCCGCGCG | ||||
|
AM747720 |
BCAL2585 |
Burkholderia cenocepacia J2315b |
Forward: GGATCCATGACCATCAATCACCC |
BamHI/HindIII |
| Reverse: AAGCTTAAGCGGGTGTGACGC | ||||
|
CP000555 |
Mpe_A0993 |
Methylibium petroleiphilum PM1a |
Forward: CATATGCCGGTTTCGCACCCCAAG |
NdeI/HindIII |
| Reverse: AAAAAAGCTTAAGCCGTGCGGCGCGCGGTC | ||||
|
BAD58116 |
nfa32690 |
Nocardia farcinica IFM 10152a |
Forward: CATATGAGTCAGCGAGACAGTTTCCG |
NdeI/HindIII |
| Reverse: ATATAAGCTTCCGCACCGCGGGTTCGGCGT | ||||
| NZ_AAUW01000019 | SIAM614_26356 | Stappia aggregata IAM 12614a | Forward: CATATGAAAGCTATCAAGGTTGCCGCCGTTC |
NdeI/BamHI |
| Reverse: GGATCCCTACTCCTCGACCTCAAAAGGCCGT |
a The gene was cloned into a pET28a(+) expression vector using E. coli BL21 (DE3) as host strain.
b The gene was cloned into a pQE30 expression vector using E. coli M15 as host strain.
Protein purification
The obtained cell pellets were resuspended in 10 ml of ice-chilled lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 1 mM dithiothreitol (DTT), pH 8.0). A cell disruption was performed by sonication on ice, and the lysate was centrifuged at 10,000 × g for 30 min to remove the cell debris. The resulting supernatant was passed through a 0.22-μm filter, and then applied to a Ni-NTA Superflow column (1 ml, Qiagen) previously equilibrated with the lysis buffer. The column was subsequently washed with 10 ml of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 1 mM DTT, pH 8.0) to remove the impurity protein. The fusion protein was then eluted with the elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 1 mM DTT, pH 8.0). The eluted protein was desalted and concentrated through ultrafiltration using a 50-ml Amicon Ultra Centrifugal Filter Device with a molecular weight cutoff of 10 KDa (Millipore, USA). The purified enzyme was resuspended in a sodium phosphate buffer (pH 7.0) containing 1 mM of DTT and 20% glycerol, and then stored at −40°C. The crude extract and the pure enzyme were analyzed by SDS-PAGE. Protein concentration was determined using the Bradford method, with bovine serum albumin as the standard. All purification steps were carried out at 4°C.
Enzyme assay
The standard reaction with the purified nitrilase was performed at 30°C in a reaction mixture (1 ml) containing 100 μmol sodium phosphate (pH 7.0), 20 μmol of nitrile substrate, and an appropriate amount of nitrilase. Aliquots (100 μl) were withdrawn at different time intervals, and 10 μl of 2 M HCl was added to quench the reaction. The production and optical purity of the mandelic acid were determined through an HPLC analysis. In some cases, the amount of ammonia formed in the reaction was measured by performing the Berthelot assay [41]. All the experiments were performed in triplicates. One unit of the enzyme activity was defined as the amount of enzyme that produced 1 μmol of mandelic acid or ammonia per min under the standard assay conditions.
Determination of the molecular weight
The molecular weight of the purified BCJ2315 was determined by gel filtration chromatography on a Superdex 200 10/300 GL column (GE Healthcare). The calibration of the column was performed with the HMW-Gel Filtration Calibration Kit (GE Healthcare) containing thyreoglobulin (669 kDa), ferritiin (440 kDa), aldolase (158 kDa), and conalbumin (75 kDa). The void volume was determined using Blue Dextran (2,000 kDa).
Effects of temperature and pH on the purified BCJ2315 activity
The reaction was performed at different temperatures or in buffers of different pH values for 10 min with 20 μg of purified BCJ2315 and 20 mM mandelonitrile in 1 ml of reaction mixture to determination the temperature and pH effects. The optimum temperature of BCJ2315 was determined by incubating the enzyme with mandelonitrile at different temperatures (18°C to 70°C). The optimum pH of BCJ2315 was determined by measuring the enzyme activity in buffers with different pH values (4.0 to 10.6) using mandelonitrile as the substrate. Sodium citrate-citric acid buffer (pH 4.0 to 6.4, 0.1 M), sodium phosphate buffer (pH 6.4 to 7.6, 0.1 M), Tris–HCl buffer (pH 7.0 to 9.0, 0.1 M), and Glycine-sodium hydroxide buffer (pH 8.5 to 10.6, 0.1 M) were used in this process.
Measurement of the kinetic parameters
The kinetic parameters of BCJ2315 were determined over a wide range of mandelonitrile concentrations (0.1 mM to 15 mM) under standard assay conditions. The kinetic constants Vmax and Km were calculated from the Lineweaver– Burk plots using standard linear regression techniques.
Substrate specificity
The specific activities of BCJ2315 toward different nitriles with structural diversity were measured under standard conditions. The reaction was incubated at 30°C for 5 min to 240 min. The conversion was determined by measuring the amount of ammonia produced in the reaction using the Berthelot assay, as described previously.
Enantioselective mandelonitrile hydrolysis with the recombinant E. coli M15/BCJ2315
For the recombinant E. coli M15/BCJ2315, a standard reaction mixture (10 ml) containing wet cells (100 mg) and mandelonitrile (100 mM) suspended in phosphate buffer (100 mM, pH 8.0) was incubated in a rotary shaker (30°C, 200 rpm). Aliquots (100 μl) were withdrawn and quenched with 10 μl of 2 M HCl at different time intervals. The production and optical purity of the mandelic acid were determined by HPLC analysis. (R)-(−)-mandelic acid was recovered using an ion-exchange process, as described by Xue et al. [42]. The product was recrystallized using benzene as the solvent.
Analytical methods
The decrease in mandelonitrile and the mandelic acid formation were analyzed by HPLC using a Zorbax SB-Aq column (4.6 mm × 250 mm, 5 μm) (Agilent Technologies, Ltd., USA) at a flow rate of 1 ml/min, with an eluting solvent system of phosphoric acid (0.1%, v/v) and methanol (20:80, v/v). A210 nm was measured.
The optical purity of the mandelic acid was determined by analyzing the enantiomers on a CHIRALCEL-OD-H column (4.6 mm × 250 mm, 5 μm) (Daicel Chemical Industries, Ltd., Japan) at a flow rate of 0.8 ml/min, with an eluting solvent system of hexane, isopropanol, and trifluoroacetic acid (95:5:0.1, v/v). A228 nm was measured.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DW and HW designed the study. HW carried out the bulk of the experiments. HS contributed to the expression and purification of the nitrilases. All authors have read and approved the final manuscript.
Supplementary Material
The respective gene primers and mandelonitrile hydrolase activity of the nitrilases outside the predicted mandelonitrile hydrolase subgroup.
Contributor Information
Hualei Wang, Email: hlwang@mail.ecust.edu.cn.
Huihui Sun, Email: sunhuihui8963@126.com.
Dongzhi Wei, Email: dzhwei@ecust.edu.cn.
Acknowledgements
This work was supported by the National Major Science and Technology Projects of China (2012ZX09304009) and the Fundamental Research Funds for the Central Universities (WF1013008).
References
- Zhang CS, Zhang ZJ, Li CX, Yu HL, Zheng GW, Xu JH. Efficient production of (R)-o-chloromandelic acid by deracemization of o-chloromandelonitrile with a new nitrilase mined from labrenzia aggregata. Appl Microbiol Biotechnol. 2012;95:91–99. doi: 10.1007/s00253-012-3993-4. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng RC, Zheng YG, Shen YC. Microbial transformation of nitriles to high-value acids or amides. Biotechnology in China I. 2009;113:33–77. doi: 10.1007/10_2008_25. [DOI] [PubMed] [Google Scholar]
- Wang MX. Enantioselective biotransformations of nitriles in organic synthesis. Top Catal. 2005;35:117–130. doi: 10.1007/s11244-005-3817-1. [DOI] [PubMed] [Google Scholar]
- Groger H. Enzymatic routes to enantiomerically pure aromatic alpha-hydroxy carboxylic acids: a further example for the diversity of biocatalysis. Adv Synth Catal. 2001;343:547–558. doi: 10.1002/1615-4169(200108)343:6/7<547::AID-ADSC547>3.0.CO;2-A. [DOI] [Google Scholar]
- Terreni M, Pagani G, Ubiali D, Fernandez-Lafuente R, Mateo C, Guisan JM. Modulation of penicillin acylase properties via immobilization techniques: One-pot chemoenzymatic synthesis of cephamandole from cephalosporin C. Bioorg Med Chem Lett. 2001;11:2429–2432. doi: 10.1016/S0960-894X(01)00463-2. [DOI] [PubMed] [Google Scholar]
- Furlenmeier AQP, Volger K, Lanz P. 6-Acyl derivatives of aminopenicillanic acid. US Patent Office 3957758; 1976. [Google Scholar]
- Surivet JP, Vatele JM. Total synthesis of antitumor goniothalamus styryllactones. Tetrahedron. 1999;55:13011–13028. doi: 10.1016/S0040-4020(99)00794-2. [DOI] [Google Scholar]
- Mills JSK, Shaw WN. Phenethanolamines compositions containing the same and method for effecting weight control. US Patent 4391826; 1983. [Google Scholar]
- Yadav GD, Sajgure AD, Dhoot SB. Insight into microwave irradiation and enzyme catalysis in enantioselective resolution of RS-(+/−)-methyl mandelate. J Chem Technol Biotechnol. 2008;83:1145–1153. doi: 10.1002/jctb.1975. [DOI] [Google Scholar]
- Yamamoto K, Oishi K, Fujimatsu I, Komatsu K. Production of R-(−)-mandelic acid from mandelonitrile by alcaligenes faecalis ATCC 8750. Appl Environ Microbiol. 1991;57:3028–3032. doi: 10.1128/aem.57.10.3028-3032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZJ, Xu JH, He YC, Ouyang LM, Liu YY. Cloning and biochemical properties of a highly thermostable and enantioselective nitrilase from alcaligenes sp. ECU0401 And its potential for (R)-(−)-mandelic acid production. Bioprocess Biosyst Eng. 2011;34:315–322. doi: 10.1007/s00449-010-0473-z. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Dong LZ, Cheng F, Xue YP, Wang YS, Ding JN, Zheng YG, Shen YC. Gene cloning, expression, and characterization of a nitrilase from alcaligenes faecalis ZJUTB10. J Agr Food Chem. 2011;59:11560–11570. doi: 10.1021/jf202746a. [DOI] [PubMed] [Google Scholar]
- Nagasawa T, Mauger J, Yamada H. A novel nitrilase, arylacetonitrilase, of alcaligenes faecalis JM3 purification and characterization. Eur J Biochem/FEBS. 1990;194:765–772. doi: 10.1111/j.1432-1033.1990.tb19467.x. [DOI] [PubMed] [Google Scholar]
- Singh R, Banerjee A, Kaul P, Barse B, Banerjee UC. Release of an enantioselective nitrilase from alcaligenes faecalis MTCC 126: a comparative study. Bioprocess Biosyst Eng. 2005;27:415–424. doi: 10.1007/s00449-005-0013-4. [DOI] [PubMed] [Google Scholar]
- Kaul P, Banerjee A, Banerjee UC. Stereoselective nitrile hydrolysis by immobilized whole-cell biocatalyst. Biomacromolecules. 2006;7:1536–1541. doi: 10.1021/bm0507913. [DOI] [PubMed] [Google Scholar]
- Petrickova A, Sosedov O, Baum S, Stolz A, Martinkova L. Influence of point mutations near the active site on the catalytic properties of fungal arylacetonitrilases from aspergillus niger and neurospora crassa. J Mol Catal B: Enzym. 2012;77:74–80. [Google Scholar]
- Kiziak C, Conradt D, Stolz A, Mattes R, Klein J. Nitrilase from pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology-Sgm. 2005;151:3639–3648. doi: 10.1099/mic.0.28246-0. [DOI] [PubMed] [Google Scholar]
- Martinkova L, Kren V. Biotransformations with nitrilases. Curr Opin Chem Biol. 2010;14:130–137. doi: 10.1016/j.cbpa.2009.11.018. [DOI] [PubMed] [Google Scholar]
- Kaul P, Banerjee A, Mayilraj S, Banerjee UC. Screening for enantioselective nitrilases: kinetic resolution of racemic mandelonitrile to (R)-(−)-mandelic acid by new bacterial isolates. Tetrahedron-Asymmetry. 2004;15:207–211. doi: 10.1016/j.tetasy.2003.10.041. [DOI] [Google Scholar]
- Robertson DE, Chaplin JA, DeSantis G, Podar M, Madden M, Chi E, Richardson T, Milan A, Miller M, Weiner DP. Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol. 2004;70:2429–2436. doi: 10.1128/AEM.70.4.2429-2436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Mukherjee C, Biehl ER, Hua L. Discovery of a mandelonitrile hydrolase from bradyrhizobium japonicum USDA110 by rational genome mining. J Biotechnol. 2007;129:645–650. doi: 10.1016/j.jbiotec.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Seffernick JL, Samanta SK, Louie TM, Wackett LP, Subramanian M. Investigative mining of sequence data for novel enzymes: a case study with nitrilases. J Biotechnol. 2009;143:17–26. doi: 10.1016/j.jbiotec.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nagasawa T, Yamada H. Nitrilase of rhodococcus rhodochrous J1 purification and characterization. Eur J Biochem/FEBS. 1989;182:349–356. doi: 10.1111/j.1432-1033.1989.tb14837.x. [DOI] [PubMed] [Google Scholar]
- Heinemann U, Engels D, Burger S, Kiziak C, Mattes R, Stolz A. Cloning of a nitrilase gene from the cyanobacterium synechocystis sp. Strain PCC6803 and heterologous expression and characterization of the encoded protein. Appl Environ Microbiol. 2003;69:4359–4366. doi: 10.1128/AEM.69.8.4359-4366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Wu S, Blumerman S, Fallon RD, Gavagan JE, DiCosimo R, Payne MS. Purification, cloning, sequencing and over-expression in escherichia coli of a regioselective aliphatic nitrilase from acidovorax facilis 72W. Appl Microbiol Biotechnol. 2003;61:118–122. doi: 10.1007/s00253-002-1192-4. [DOI] [PubMed] [Google Scholar]
- Kim JS, Tiwari MK, Moon HJ, Jeya M, Ramu T, Oh DK, Kim IW, Lee JK. Identification and characterization of a novel nitrilase from pseudomonas fluorescens Pf-5. Appl Microbiol Biotechnol. 2009;83:273–283. doi: 10.1007/s00253-009-1862-6. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Yanaka N, Nagasawa T, Yamada H. Purification and characterization of a novel nitrilase of rhodococcus rhodochrous K22 that acts on aliphatic nitriles. J Bacteriol. 1990;172:4807–4815. doi: 10.1128/jb.172.9.4807-4815.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan O, Bezouska K, Plihal O, Ettrich R, Kulik N, Vanek O, Kavan D, Benada O, Malandra A, Sveda O. Heterologous expression, purification and characterization of nitrilase from Aspergillus Niger K10. BMC Biotechnol. p. 2. [DOI] [PMC free article] [PubMed] [Retracted]
- Vorwerk S, Biernacki S, Hillebrand H, Janzik I, Muller A, Weiler EW, Piotrowski M. Enzymatic characterization of the recombinant arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta. 2001;212:508–516. doi: 10.1007/s004250000420. [DOI] [PubMed] [Google Scholar]
- Zhu DM, Mukherjee C, Yang Y, Rios BE, Gallagher DT, Smith NN, Biehl ER, Hua L. A new nitrilase from bradyrhizobium japonicum USDA 110 - gene cloning, biochemical characterization and substrate specificity. J Biotechnol. 2008;133:327–333. doi: 10.1016/j.jbiotec.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Piotrowski M, Schonfelder S, Weiler EW. The arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode beta-cyano-L-alanine hydratase/nitrilase. J Biol Chem. 2001;276:2616–2621. doi: 10.1074/jbc.M007890200. [DOI] [PubMed] [Google Scholar]
- O'Reilly C, Turner PD. The nitrilase family of CN hydrolysing enzymes - a comparative study. J Appl Microbiol. 2003;95:1161–1174. doi: 10.1046/j.1365-2672.2003.02123.x. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Kaul P, Banerjee UC. Purification and characterization of an enantioselective arylacetonitrilase from pseudomonas putida. Arch Microbiol. 2006;184:407–418. doi: 10.1007/s00203-005-0061-9. [DOI] [PubMed] [Google Scholar]
- DeSantis G, Zhu Z, Greenberg WA, Wong K, Chaplin J, Hanson SR, Farwell B, Nicholson LW, Rand CL, Weiner DP. An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J Am Chem Soc. 2002;124:9024–9025. doi: 10.1021/ja0259842. [DOI] [PubMed] [Google Scholar]
- Petrickova A, Vesela AB, Kaplan O, Kubac D, Uhnakova B, Malandra A, Felsberg J, Rinagelova A, Weyrauch P, Kren V. Purification and characterization of heterologously expressed nitrilases from filamentous fungi. Appl Microbiol Biotechnol. 2012;93:1553–1561. doi: 10.1007/s00253-011-3525-7. [DOI] [PubMed] [Google Scholar]
- Zhang ZJ, Xu JH, He YC, Ouyang LM, Liu YY, Imanaka T. Efficient production of (R)-(−)-mandelic acid with highly substrate/product tolerant and enantioselective nitrilase of recombinant alcaligenes sp. Process Biochem. 2010;45:887–891. doi: 10.1016/j.procbio.2010.02.011. [DOI] [Google Scholar]
- Brenner C. Catalysis in the nitrilase superfamily. Curr Opin Struct Biol. 2002;12:775–782. doi: 10.1016/S0959-440X(02)00387-1. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fitsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Press: Cold Spring Harbor, New York; 1989. [Google Scholar]
- Weatherburn MW. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. doi: 10.1021/ac60252a045. [DOI] [Google Scholar]
- Xue YP, Liu ZQ, Xu M, Wang YJ, Zheng YG. Efficient separation of (R)-(−)-mandelic acid biosynthesized from (R, S)-mandelonitrile by nitrilase using ion-exchange process. J Chem Technol Biotechnol. 2011;86:391–397. doi: 10.1002/jctb.2528. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The respective gene primers and mandelonitrile hydrolase activity of the nitrilases outside the predicted mandelonitrile hydrolase subgroup.