Abstract
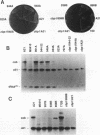
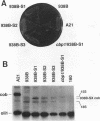
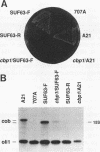
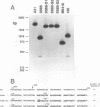
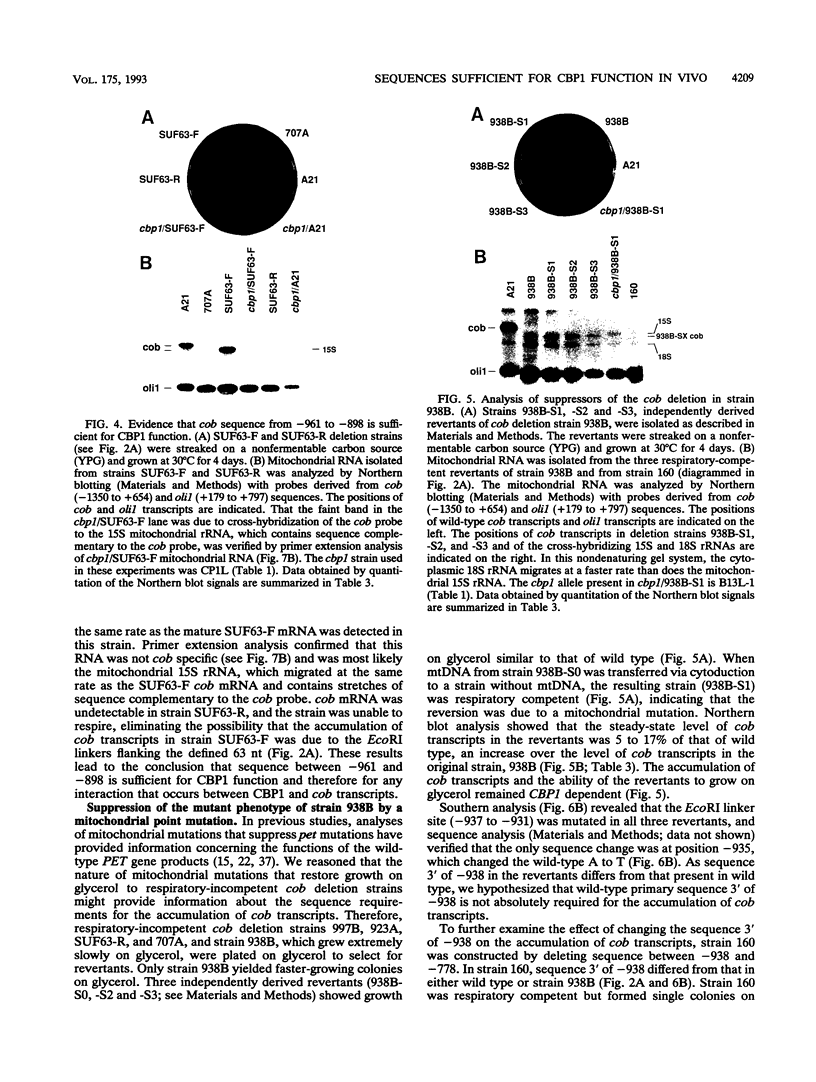
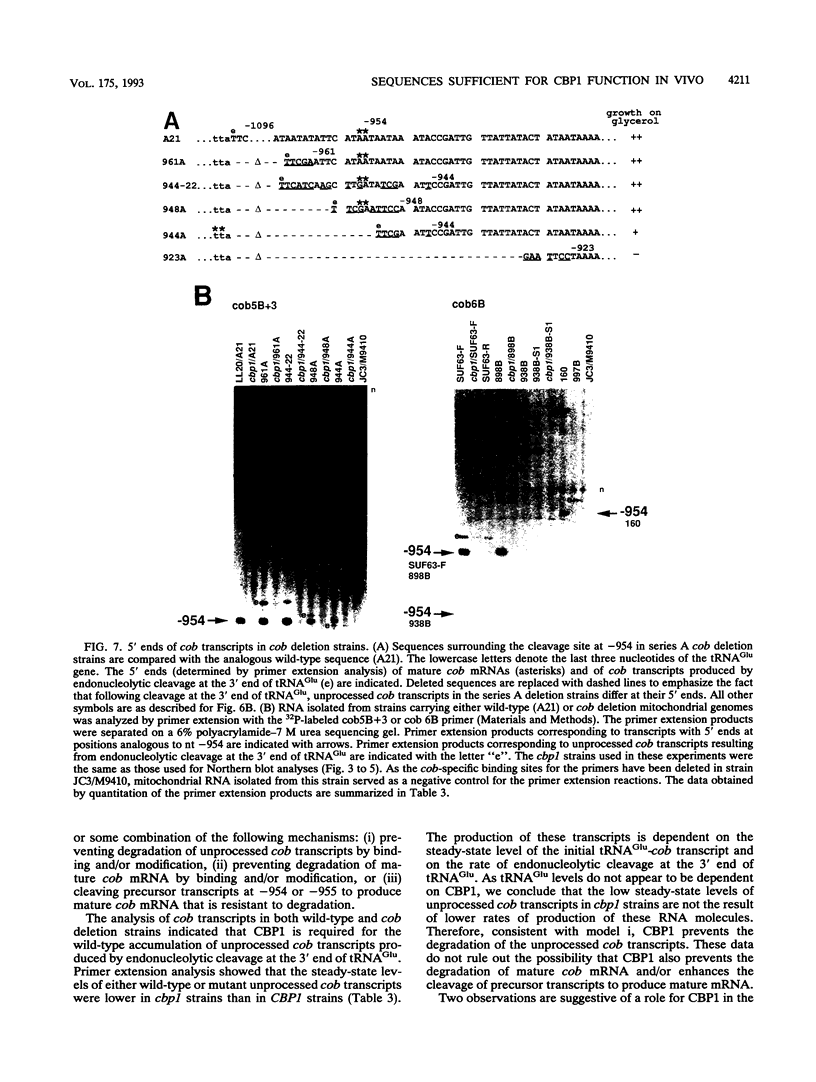
In Saccharomyces cerevisiae, cytochrome b, an essential component of the respiratory chain, is encoded by the mitochondrial gene cob. The cob transcription unit includes the tRNA(Glu) gene from positions -1170 to -1099 relative to the cob ATG at +1. The initial tRNA(Glu)-cob transcript undergoes several processing events, including removal of tRNA(Glu) and production of the mature 5' end of cob mRNA at nucleotide -954. The nuclear gene product CBP1 is specifically required for the accumulation of cob mRNA. In cbp1 mutant strains, cob transcripts are not detectable by Northern (RNA) blot analysis, but the steady-state level of tRNA(Glu) is similar to that of wild type. The results of a previous study led to the conclusion that a 400-nucleotide region just downstream of tRNA(Glu) is sufficient for CBP1 function. In the present study, the microprojectile bombardment method of mitochondrial transformation was used to introduce deletions within this region of cob. The analysis of cob transcripts in strains carrying the mitochondrial deletion genomes indicates that a 63-nucleotide sequence that encompasses the cleavage site at -954 is sufficient both for CBP1 function and for correct positioning of the cleavage. Furthermore, the data indicate that CBP1 prevents the degradation of unprocessed cob transcripts produced by endonucleolytic cleavage at the 3' end of tRNA(Glu).
Full text
PDF
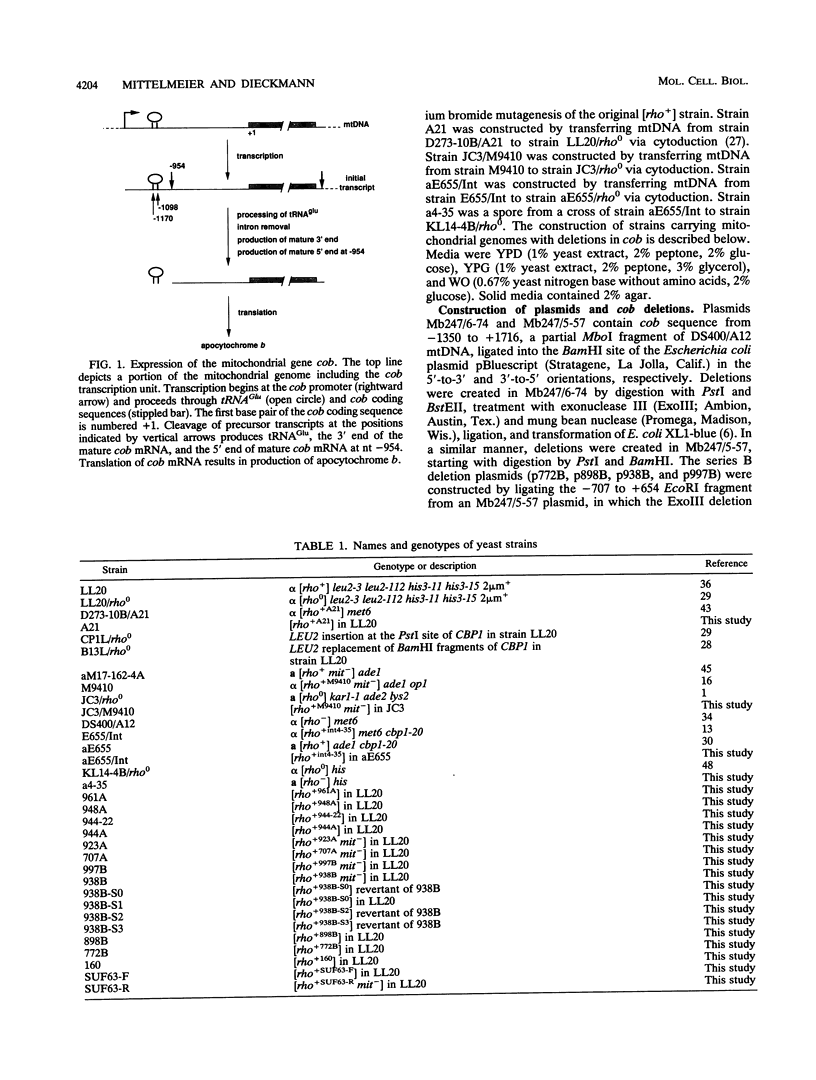


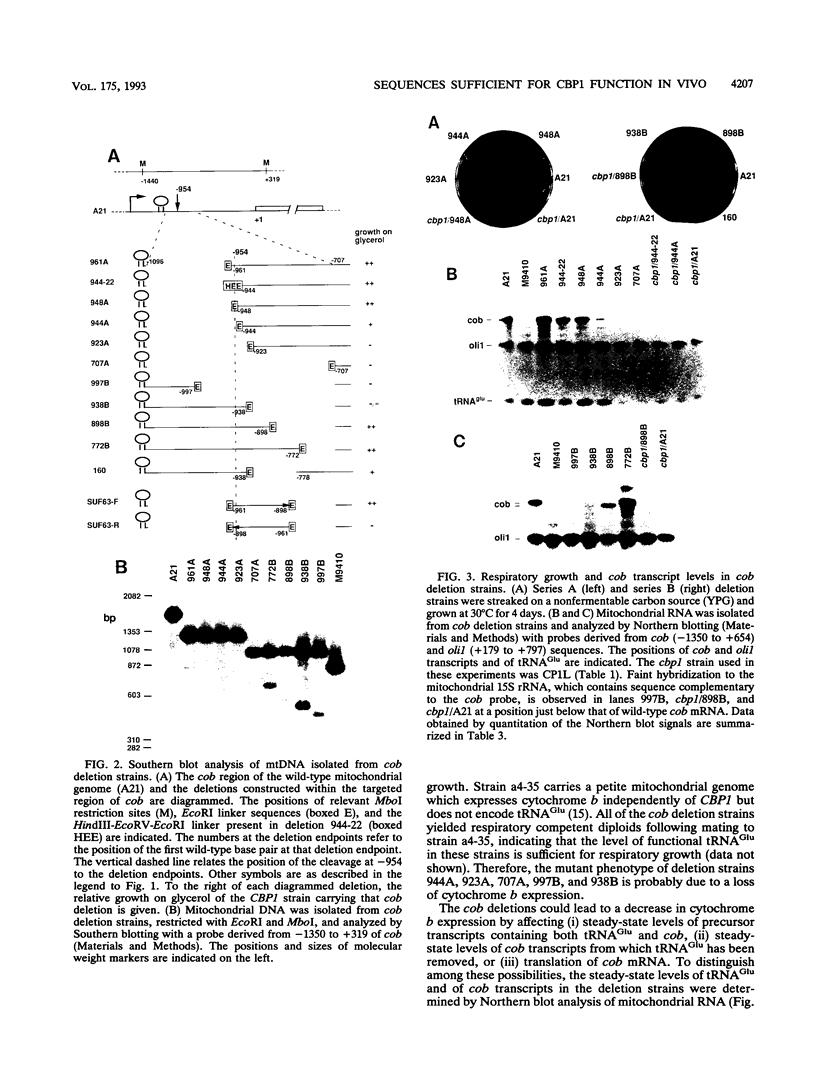
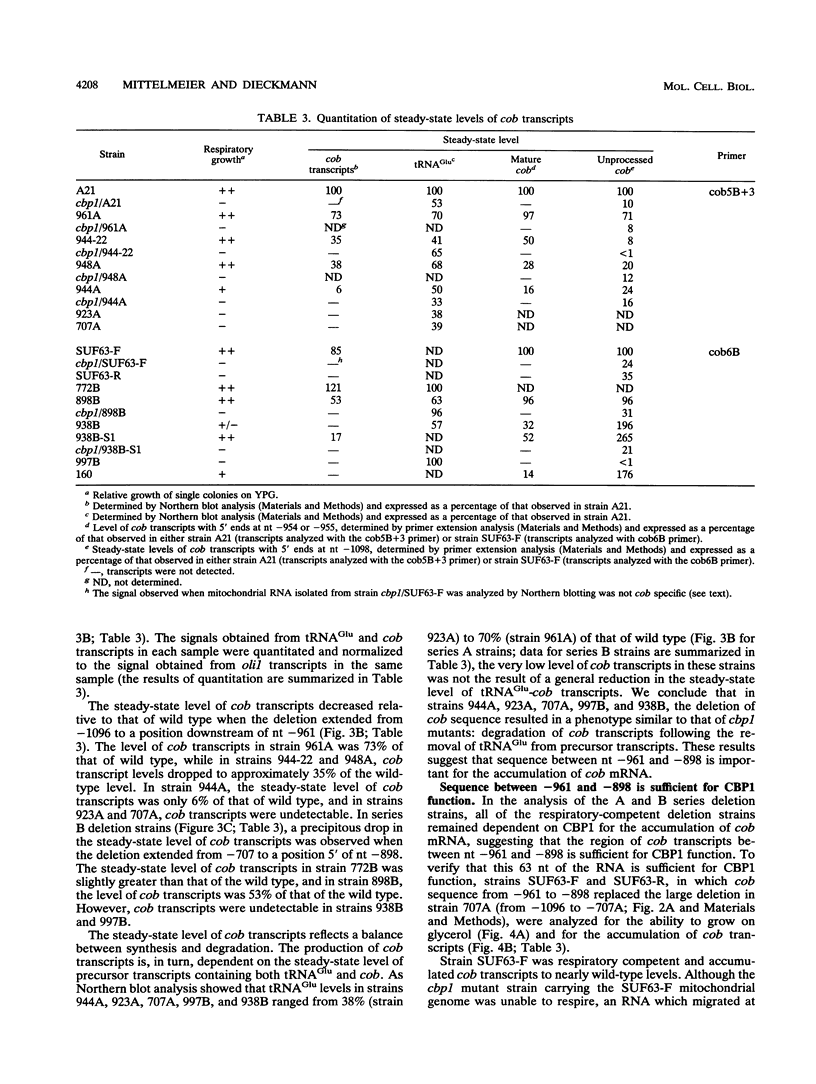



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander N. J., Periman P. S., Hanson D. K., Mahler H. R. Mosaic organization of a mitochondrial gene: evidence from double mutants in the cytochrome b region of Saccharomyces cerevisiae. Cell. 1980 May;20(1):199–206. doi: 10.1016/0092-8674(80)90247-0. [DOI] [PubMed] [Google Scholar]
- Armaleo D., Ye G. N., Klein T. M., Shark K. B., Sanford J. C., Johnston S. A. Biolistic nuclear transformation of Saccharomyces cerevisiae and other fungi. Curr Genet. 1990 Feb;17(2):97–103. doi: 10.1007/BF00312852. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Bonitz S. G., Homison G., Thalenfeld B. E., Tzagoloff A., Nobrega F. G. Assembly of the mitochondrial membrane system. Processing of the apocytochrome b precursor RNAs in Saccharomyces cerevisiae D273-10B. J Biol Chem. 1982 Jun 10;257(11):6268–6274. [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Chen J. Y., Martin N. C. Biosynthesis of tRNA in yeast mitochondria. An endonuclease is responsible for the 3'-processing of tRNA precursors. J Biol Chem. 1988 Sep 25;263(27):13677–13682. [PubMed] [Google Scholar]
- Chen L. H., Emory S. A., Bricker A. L., Bouvet P., Belasco J. G. Structure and function of a bacterial mRNA stabilizer: analysis of the 5' untranslated region of ompA mRNA. J Bacteriol. 1991 Aug;173(15):4578–4586. doi: 10.1128/jb.173.15.4578-4586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T., Edwards J. C., Mueller D. M., Rabinowitz M. Identification of a single transcriptional initiation site for the glutamic tRNA and COB genes in yeast mitochondria. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5564–5568. doi: 10.1073/pnas.80.18.5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T., Rabinowitz M. Identification of multiple transcriptional initiation sites on the yeast mitochondrial genome by in vitro capping with guanylyltransferase. J Biol Chem. 1983 Nov 25;258(22):14025–14033. [PubMed] [Google Scholar]
- Dieckmann C. L., Gandy B. Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J. 1987 Dec 20;6(13):4197–4203. doi: 10.1002/j.1460-2075.1987.tb02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C. L., Homison G., Tzagoloff A. Assembly of the mitochondrial membrane system. Nucleotide sequence of a yeast nuclear gene (CBP1) involved in 5' end processing of cytochrome b pre-mRNA. J Biol Chem. 1984 Apr 25;259(8):4732–4738. [PubMed] [Google Scholar]
- Dieckmann C. L., Koerner T. J., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP1, a yeast nuclear gene involved in 5' end processing of cytochrome b pre-mRNA. J Biol Chem. 1984 Apr 25;259(8):4722–4731. [PubMed] [Google Scholar]
- Dobres M., Gerbl-Rieger S., Schmelzer C., Mueller M. W., Schweyen R. J. Deletions in the cob gene of yeast mtDNA and their phenotypic effect. Curr Genet. 1985;10(4):283–290. doi: 10.1007/BF00365624. [DOI] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou J., Wollman F. A. Evidence for Nuclear Control of the Expression of the atpA and atpB Chloroplast Genes in Chlamydomonas. Plant Cell. 1992 Mar;4(3):283–295. doi: 10.1105/tpc.4.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Folley L. S., Fox T. D. Site-directed mutagenesis of a Saccharomyces cerevisiae mitochondrial translation initiation codon. Genetics. 1991 Nov;129(3):659–668. doi: 10.1093/genetics/129.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Sanford J. C., McMullin T. W. Plasmids can stably transform yeast mitochondria lacking endogenous mtDNA. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7288–7292. doi: 10.1073/pnas.85.19.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell L. A. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur J Biochem. 1989 Jul 1;182(3):477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hill J., McGraw P., Tzagoloff A. A mutation in yeast mitochondrial DNA results in a precise excision of the terminal intron of the cytochrome b gene. J Biol Chem. 1985 Mar 25;260(6):3235–3238. [PubMed] [Google Scholar]
- Hollingsworth M. J., Martin N. C. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986 Apr;6(4):1058–1064. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Anziano P. Q., Shark K., Sanford J. C., Butow R. A. Mitochondrial transformation in yeast by bombardment with microprojectiles. Science. 1988 Jun 10;240(4858):1538–1541. doi: 10.1126/science.2836954. [DOI] [PubMed] [Google Scholar]
- Kuchka M. R., Goldschmidt-Clermont M., van Dillewijn J., Rochaix J. D. Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell. 1989 Sep 8;58(5):869–876. doi: 10.1016/0092-8674(89)90939-2. [DOI] [PubMed] [Google Scholar]
- Lancashire W. E., Mattoon J. R. Cytoduction: a tool for mitochondrial genetic studies in yeast. Utilization of the nuclear-fusion mutation kar 1-1 for transfer of drug r and mit genomes in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Mar 5;170(3):333–344. doi: 10.1007/BF00267067. [DOI] [PubMed] [Google Scholar]
- Liu Y. X., Dieckmann C. L. Overproduction of yeast viruslike particles by strains deficient in a mitochondrial nuclease. Mol Cell Biol. 1989 Aug;9(8):3323–3331. doi: 10.1128/mcb.9.8.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer S. A., Dieckmann C. L. The yeast CBP1 gene produces two differentially regulated transcripts by alternative 3'-end formation. Mol Cell Biol. 1989 Oct;9(10):4161–4169. doi: 10.1128/mcb.9.10.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelmeier T. M., Dieckmann C. L. CBP1 function is required for stability of a hybrid cob-oli1 transcript in yeast mitochondria. Curr Genet. 1990 Dec;18(5):421–428. doi: 10.1007/BF00309911. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Prentki P., Belin D., Krisch H. M. Processing of unstable bacteriophage T4 gene 32 mRNAs into a stable species requires Escherichia coli ribonuclease E. EMBO J. 1988 Nov;7(11):3601–3607. doi: 10.1002/j.1460-2075.1988.tb03238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D. M., Getz G. S. Steady state analysis of mitochondrial RNA after growth of yeast Saccharomyces cerevisiae under catabolite repression and derepression. J Biol Chem. 1986 Sep 5;261(25):11816–11822. [PubMed] [Google Scholar]
- Müllner E. W., Neupert B., Kühn L. C. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell. 1989 Jul 28;58(2):373–382. doi: 10.1016/0092-8674(89)90851-9. [DOI] [PubMed] [Google Scholar]
- Nobrega F. G., Tzagoloff A. Assembly of the mitochondrial membrane system. Complete restriction map of the cytochrome b region of mitochondrial DNA in Saccharomyces cerevisiae D273-10B. J Biol Chem. 1980 Oct 25;255(20):9821–9827. [PubMed] [Google Scholar]
- Nobrega F. G., Tzagoloff A. Assembly of the mitochondrial membrane system. DNA sequence and organization of the cytochrome b gene in Saccharomyces cerevisiae D273-10B. J Biol Chem. 1980 Oct 25;255(20):9828–9837. [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödel G., Körte A., Kaudewitz F. Mitochondrial suppression of a yeast nuclear mutation which affects the translation of the mitochondrial apocytochrome b transcript. Curr Genet. 1985;9(8):641–648. doi: 10.1007/BF00449816. [DOI] [PubMed] [Google Scholar]
- Sieburth L. E., Berry-Lowe S., Schmidt G. W. Chloroplast RNA Stability in Chlamydomonas: Rapid Degradation of psbB and psbC Transcripts in Two Nuclear Mutants. Plant Cell. 1991 Feb;3(2):175–189. doi: 10.1105/tpc.3.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Jones H., Gruissem W. Function of plastid mRNA 3' inverted repeats. RNA stabilization and gene-specific protein binding. J Biol Chem. 1989 Nov 5;264(31):18742–18750. [PubMed] [Google Scholar]
- Stern D. B., Radwanski E. R., Kindle K. L. A 3' stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991 Mar;3(3):285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Foury F. Assembly of the mitochondrial membrane system XVI. Modified form of the ATPase proteolipid in oligomycin-resistant mutants of Saccharomyces cerevisiae. FEBS Lett. 1976 Jun 15;65(3):391–395. doi: 10.1016/0014-5793(76)80154-8. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Dieckmann C. L. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990 Sep;54(3):211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Foury F., Akai A. Assembly of the mitochondrial membrane system. XVIII. Genetic loci on mitochondrial DNA involved in cytochrome b biosynthesis. Mol Gen Genet. 1976 Nov 24;149(1):33–42. doi: 10.1007/BF00275958. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Myers A. M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Weber E. R., Dieckmann C. L. Identification of the CBP1 polypeptide in mitochondrial extracts from Saccharomyces cerevisiae. J Biol Chem. 1990 Jan 25;265(3):1594–1600. [PubMed] [Google Scholar]
- Wolf K., Dujon B., Slonimski P. P. Mitochondrial genetics. V. Multifactorial mitochondrial crosses involving a mutation conferring paromomycin-resistance in Saccharomyces cerevisiae. Mol Gen Genet. 1973 Sep 5;125(1):53–90. doi: 10.1007/BF00292983. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The AT spacers and the var1 genes from the mitochondrial genomes of Saccharomyces cerevisiae and Torulopsis glabrata: evolutionary origin and mechanism of formation. Gene. 1987;54(1):1–22. doi: 10.1016/0378-1119(87)90342-8. [DOI] [PubMed] [Google Scholar]