Abstract
Ribosomally synthesized posttranslationally modified peptides (RiPPs) are a rapidly growing class of natural products with diverse structures and activities. In recent years, a great deal of progress has been made in elucidating the biosynthesis of various RiPP family members. As with the study of nonribosomal peptide and polyketide biosynthetic enzymes, these investigations have led to the discovery of entirely new biological chemistry. With each unique enzyme investigated, a more complex picture of Nature’s synthetic potential is revealed. This review focuses on recent reports (since 2008) that have changed the way that we think about ribosomal natural product biosynthesis and the enzymology of complex bond-forming reactions.
Introduction
How does one measure the impact that natural products have had on human health and science as a whole? Natural products, or the derivatives thereof, encompass the lion’s share of drugs in therapeutic use.1–3 The exquisite complexity of these compounds has served as a source of inspiration for organic chemists for over a century, leading to the development of reaction methodologies too numerous to list.3, 4 Synthetic organic chemistry as we know it today would likely not exist had the study of natural products been neglected. While the impact on human therapeutics and synthesis has been immense, natural products research also enabled the study of fundamental biological processes. For example, research into the mode of action of antibiotics continues to provide insight into bacterial biochemistry and physiology,5–7 while compounds such as phalloidin (filamentous actin stabilizer), rapamycin (mTOR inhibitor), and tunicamycin (N-glycosylation inhibitor) have become invaluable tools for probing eukaryotic cell biology.8–10 Furthermore, the study of natural products has provided a window through which the chemical toolbox of Nature can be explored, revolutionizing the way we think about the chemistry of biological systems. With the vast majority of Nature’s biosynthetic potential remaining to be discovered,11, 12 the study of natural products will likely dominate the endeavors of future generations of scientists.
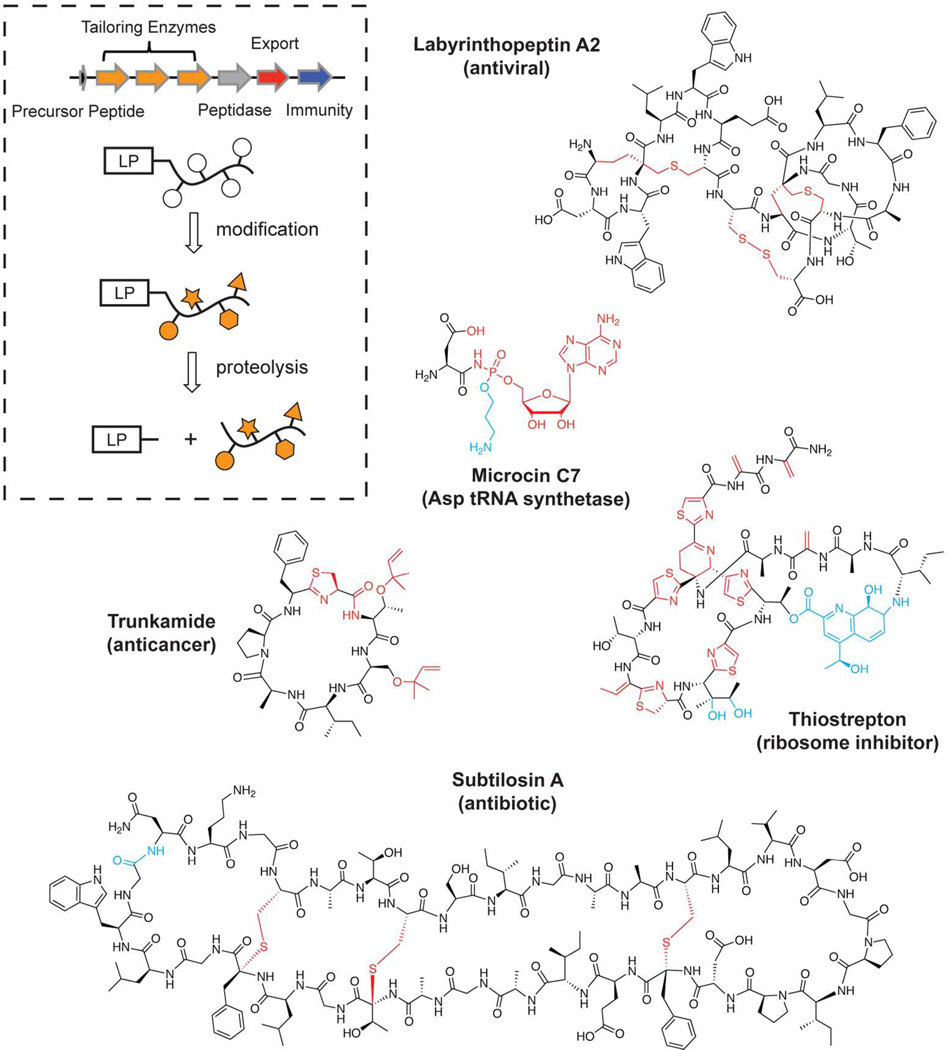
A class of natural products receiving significant recent attention is the ribosomally synthesized posttranslationally modified peptides (RiPPs).13, 14 Comprised of the lantipeptides, microcins and the thiazole/oxazole-modified microcins (TOMMs) among others, RiPPs occupy a large chemical, genetic, and functional space, but remain linked by a common mechanism of biosynthesis. In all cases, a ribosomally synthesized precursor peptide undergoes modification by a set of tailoring enzymes usually found in the local genomic region (i.e. within the biosynthetic gene cluster, Figure 1). These modifications include, but are not limited to, varied cyclizations, dehydrations, and rearrangements, all of which are responsible for endowing the peptide with a rigidified structure and biological activity. The tailoring enzymes encoded in the biosynthetic gene cluster govern the posttranslational modifications received and, in some cases, the structure of the final product can be accurately predicted.15–17 In most cases, modification is followed by the proteolytic removal of a N-terminal leader peptide required for recognition by the tailoring enzymes, and the mature compound is exported.18 Using this simple strategy, a varied compound library can be constructed from a minimal amount of genetic space. In fact, the investigation of RiPPs has overturned the longstanding paradigm that large genomes are required for an organism to produce architecturally complex natural products.19, 20
Figure 1.
Example of RiPP natural product diversity. The general strategy for RiPP biosynthesis is illustrated in the generic gene cluster and schematic below. The functional assignment for each of the open reading frames is displayed. An array of ribosomal natural products generated from this strategy is displayed along with their molecular target. In all figures within this review, the red moieties represent posttranslational modifications where the tailoring enzyme(s) responsible have been characterized. The modification mechanisms for the blue moieties are unknown and are shown to highlight additional chemistry that remains unexplored in RiPP modification enzymes.
Adding to the allure of this class of natural products is the gene-encoded nature of the precursor peptide coupled with the often high level of promiscuity of the biosynthetic enzymes, allowing for the facile generation of unnatural compound derivatives.13, 17, 21–23 This approach has garnered a great deal of excitement due to the potential to develop therapeutically relevant derivatives of this underutilized compound class.24, 25 The profound interest in these natural products has fueled extensive studies into biosynthetic mechanisms. This review will focus on the recent successes in the field and discuss the advancement that these discoveries have made in the understanding of biological chemistry. RiPP biosynthetic enzymes are classically categorized by the subfamily into which their natural product falls (lantipeptide, TOMM, etc.). However, for the purposes of this review, they will be grouped based on the chemical transformations they catalyze to both highlight the interrelationships of these disparate tailoring enzymes and better illustrate Nature’s remarkable synthetic abilities.
ATP-Driven Cyclizations
Given the ubiquity of ATP as a biological energy currency and its status as an enzymatic cofactor/cosubstrate, it is perhaps not surprising that ATP is heavily employed in the posttranslational modification of ribosomal natural products. In recent years, there have been multiple biosynthetic reports illustrating the use of ATP by enzymes to catalyze varied cyclizations on ribosomal peptide backbones. Below, three examples are discussed.
Cyclodehydration in thiazole/oxazole biogenesis
Thiostrepton was the first azole/azolinecontaining RiPP natural product to be structurally characterized.26 Although the structures of other thiazol(in)e- and oxazol(in)e-containing natural products were solved over the following 25 years, it was not until the mid-1990s that the first insights into azole biosynthesis were revealed with the study of microcin B17.27 Following this discovery, just two additional biosynthetic gene clusters of this type were identified over the next decade (streptolysin S28 and patellamide A/C29), and the unification of the TOMM family as a whole was only recently realized.20 The seminal paper characterizing the TOMM synthetase demonstrated that azole heterocycles were installed on the precursor peptide in a two-step process through the action of a heterotrimeric enzymatic complex. First, the ATP-dependent cyclodehydration of select Ser, Thr and Cys residues affords the azoline heterocycles. Subsequently, in clusters containing a dehydrogenase, select azoline rings are oxidized to the aromatic azoles.27 Studies conducted on various TOMM biosynthetic complexes provided insights into the mechanism of substrate handling, the dehydrogenase activity assigned to a flavin mononucleotide-binding protein (B protein), and the mechanism of select ancillary tailoring enzymes from thiopeptide- and cyanobactin-type clusters was elucidated, the more recent of which will be described in greater detail below.30–37 However, the mechanism of ATP utilization and the exact role of the other two proteins in the heterotrimeric complex (C and D) remained enigmatic due to the relative experimental intractability of the enzymes responsible for microcin B17, streptolysin S, and patellamide A/C biosynthesis.
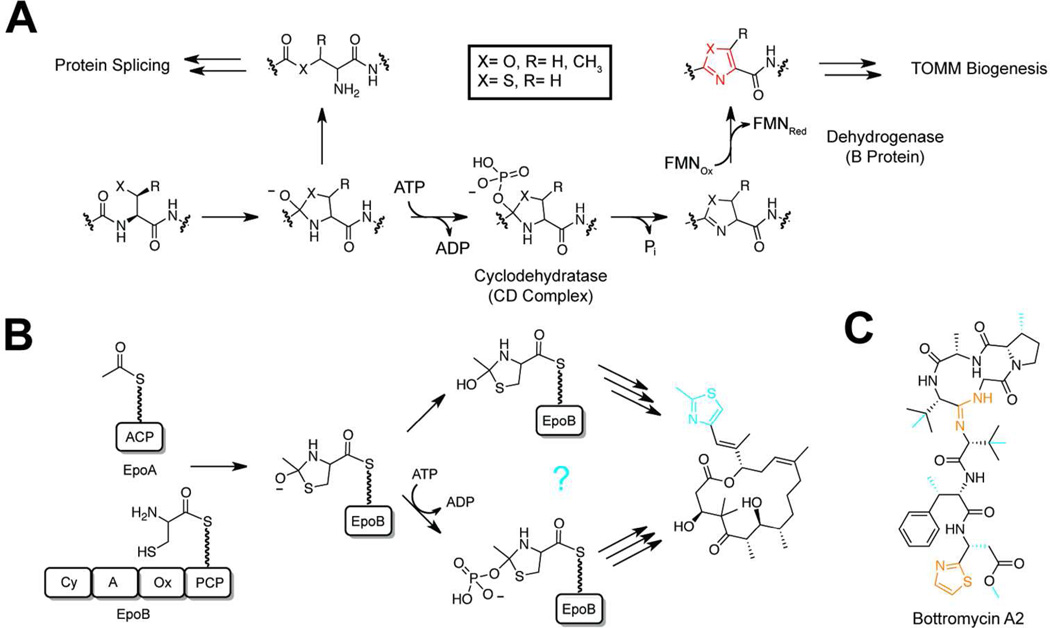
Recently, our group reported the characterization of a novel TOMM synthetase complex from Bacillus sp. Al Hakam.23 In vitro reconstitution of this tractable enzyme complex facilitated the discovery that ATP is directly used to phosphorylate the amide backbone of the peptide substrate.38 This information was used in conjunction with a recent report of azoline formation in engineered intein domains39 and the discovery of depsipeptide bonds in naturally produced microcin B1740 to propose a mechanism of azoline biosynthesis (Figure 2A). In this mechanism, phosphorylation of the peptide backbone drives the tetrahedral intermediate towards oxygen elimination and azoline formation rather than N-protonation and ester formation, as seen in autoprocessing enzymes.41 This innovative use of ATP resembles the chemistry employed by the purine biosynthetic enzymes PurL and PurM,42 but was unprecedented in the study of ribosomal natural products. Moreover, single turnover experiments assigned this phosphorylation activity to the YcaO protein homolog in the cyclodehydratase complex, providing the first definitive activity for a member of this poorly understood protein family.38 Since this initial report, two bottromycin biosynthetic clusters encoding YcaO homologs have been identified that are predicted to catalyze cyclodehydration in the absence of a C protein. This indicates that select YcaO members may have evolved to act as stand-alone cyclodehydratases.43–46 Furthermore, a second YcaO homolog in the bottromycin clusters has been predicted to catalyze the formation of the highly unusual macrolactamidine ring via direct activation of the carbonyl backbone (Figure 2C). Additional studies are required to validate these activities. Finally, because many of the bioinformatically identifiable YcaO homologs do not appear in recognizable TOMM clusters, the possibility remains that this mechanism of amidine formation is more widespread than previously thought.
Figure 2.
Cyclizations via backbone activation. a) A mechanistic proposal for the biosynthesis of azoline heterocycles in TOMM natural products. The first step is shared with the mechanism of protein splicing. Following phosphorylation and elimination of phosphate, a FMN-dependent dehydrogenation affords the aromatic azole (red). b) Two possible strategies are presented for the biosynthesis of azoline heterocycles in PKS/NRPS natural products (e.g. epothilone). The top route is ATP-independent while the bottom follows the mechanism of the TOMM cyclodehydratase complex. As the mechanism for the Cy-dependent azoline formation is unknown the moiety is shown in blue. c) The structure of bottromycin A2 is shown with the putative YcaO-installed modifications highlighted in orange. Modifications installed by uncharacterized mechanisms are shown in blue.
The strategy used in RiPP natural product biosynthesis represents the fourth known strategy for the construction of azole heterocycles. Two other examples are found in thiamin biogenesis,47 the most recently characterized of which involves a bona fide suicide enzyme.48 The remaining azole-synthesizing pathway comes from nonribosomal peptide synthetases (NRPS). Similar to TOMM biogenesis, these megasynthases are predicted to catalyze azole formation through sequential cyclodehydration (catalyzed by a Cy domain) and oxidation (Ox domain) reactions; however, the mechanistic details of these transformations remain ill-defined.49, 50 Currently, it is believed that these proteins do not require an energy source, such as ATP, to catalyze cyclodehydration (Figure 2B). However, to our knowledge, no study on a NRPS Cy domain has been conducted in the absence of ATP, as the adenylation domains in these modules require ATP for substrate loading. Consequently, the possibility remains that Cy domains utilize a strategy similar to TOMM cyclodehydratases.
Cyclodehydration as a reaction intermediate
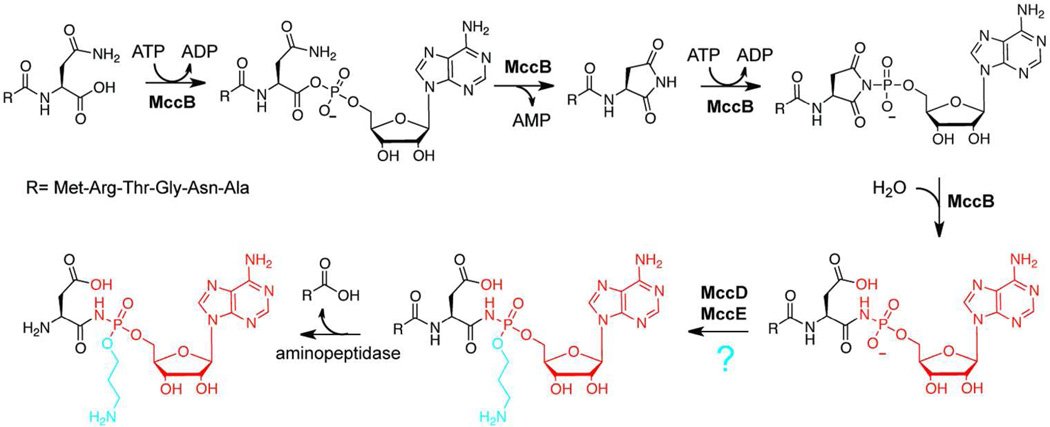
Microcin C7 (MccC7) is a plasmid-encoded RiPP produced by some strains of Escherichia coli to inhibit the growth of closely related species.51 MccC7 contains an unusual phosphoramidate (N-P) linkage between a C-terminal aspartate and AMP, as well as an aminopropylation of the phosphoramidate moiety (Figure 3).52 The sequence of the gene cluster demonstrated that a third posttranslational modification occurred during MccC7 maturation, as the naturally encoded C-terminal asparagine residue is converted to isoasparagine (isoAsn) by an unknown mechanism.53, 54 Subsequently, it was discovered that the 7-mer peptide was not the bioactive form of MccC7. Rather, the six N-terminal amino acids facilitate uptake of the antibiotic into target cells. After entry, cellular proteases cleave off these N-terminal residues, releasing the adenylated warhead, which disrupts protein synthesis through aspartyl-tRNA synthetase inhibition.55, 56 As such, MccC7 is commonly referred to as a “Trojan horse” antibiotic.
Figure 3.
Microcin C7 biosynthesis. The mechanism of the MccB-catalyzed phosphoramidate bond installation is shown with the final posttranslational modification of the precursor peptide displayed in red. Addition of the aminopropyl group (blue) to the phosphoramidate moiety is proposed to be catalyzed by MccD and/or MccE enzymes. Upon entering a target cell, the N-terminal six amino acids are proteolyically removed to afford the bioactive compound.
The discovery of the “Trojan horse” mode of action revitalized interest in MccC7 biosynthesis. As is the case for many RiPPs, bioinformatic analysis provided the first insights into how biosynthesis might occur. Of the three genes required for MccC7 maturation (mccB, mccDand mccE), mccB was homologous to E1 ubiquitin ligases and thus, was the most likely choice for the formation of the N-P bond.57 Indeed, the characterization of MccB validated this functional assignment and demonstrated that two equivalents of ATP were consumed during N-P bond formation. The use of 15N-Asn7 labeled MccC7 precursor peptide generated a product that had two 15N labels, indicating that the nitrogen of the asparagine sidechain was incorporated into the phosphoramidate linkage. Additionally, a kinetically competent succinimide intermediate was identified during substrate processing. Based on these observations, the mechanism displayed in Figure 3 was proposed.57 In agreement with other adenylating enzymes, the first proposed step is an adenylation of the C-terminal carboxylate. This modification activates the C-terminus of the peptide and facilitates an intramolecular cyclization with Asn7generating the succinimide ring identified in the aforementioned experiments. A second equivalent of ATP is then used to adenylate the nitrogen on the succinimide ring, and subsequent hydrolysis of the activated heterocycle with water affords the phosphoramidate linkage and the isoAsn residue. Importantly, the observation that the isoAsn7 derivative of MccA was not a substrate for MccB provided strong support for a mechanism involving direct adenylation of the succinimide nitrogen.
The reaction carried out by MccB is notable for several reasons: (1) a formal cyclodehydration is employed to generate an intermediate that is subject to further tailoring, (2) MccB adenylates two weak nucleophiles in succession in a single active site, (3) it represented the first instance of an E1 ubiquitin ligase homolog that modifies a small peptide and (4) MccB was the first adenylating enzyme shown to create an N-P bond. The latter two aspects of MccB catalysis are of particular interest, as they expand the known chemistry of adenylating enzymes, and can potentially provide a platform to better understand their evolution. In a follow up study, the crystal structure of MccB was solved and the peptide specificity of the adenylation enzyme was established.58 The crystal structure had the typical E1 superfamily fold but had a large N-terminal extension domain that served as a “peptide clamp” for binding MccA. Removal of this clamp abolished MccA processing, but activity could be partially restored by adding the extension in trans. Notably, it is the strong interactions of this clamp with the substrate that allow the peptide to undergo multiple adenylations in a single active site by preventing intermediate release. Such a mechanism is also used by the macrocyclase involved in cyanobactin biosynthesis (vide infra).59
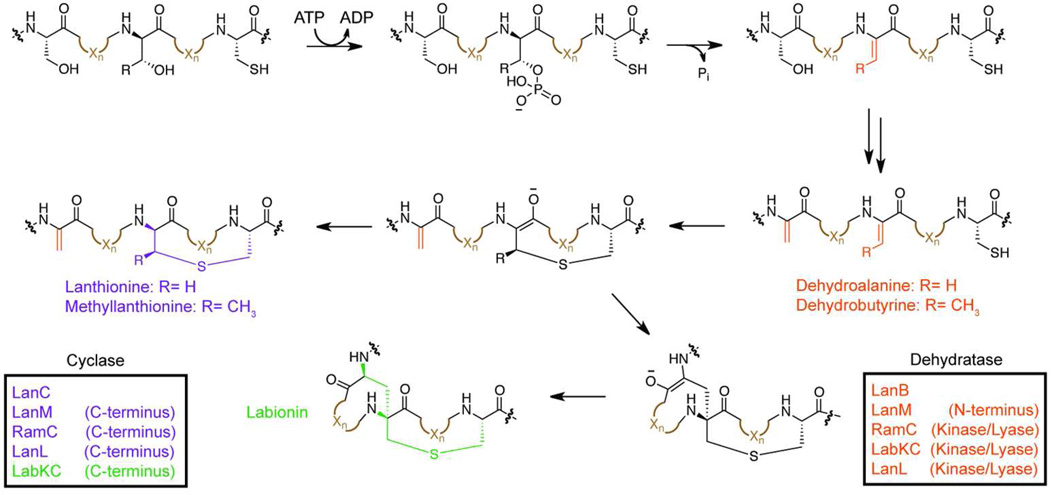
ATP-dependent dehydration and cyclization in lantipeptide biosynthesis
Lantipeptides are the archetypical members of the RiPP natural product family and are the only RiPPs to see widespread commercial use. Nisin, the first characterized lantipeptide, is an inhibitor of Gram-positive bacteria and has been utilized as a food preservative for over 50 years without reported resistance.60 As such, a substantial amount of work has been done to both understand the biosynthesis of the eponymous lanthionine rings and engineer novel lantipeptide derivatives.13, 24, 60, 61 The β-thioether linkages in lantipeptides are assembled via a two-step process. Firstly, a dehydratase catalyzes the ATPdependent dehydration of Ser/Thr residues to afford dehydroalanine/dehydrobutyrine moieties (Figure 4). Subsequently, a lantipeptide cyclase catalyzes the regioselective Michael-type addition of a Cys thiol to a particular unsaturated position. For all characterized lantipeptide cyclases, this step is ATP-independent but requires an ATP-dependent dehydration to generate a suitable electrophile. As the enzymes responsible for these transformations have been thoroughly reviewed,61 they will not be discussed further. However, the recent discovery of a novel lantipeptide cyclase, LabKC, capable of catalyzing C-C bond formation deserves a brief mention.62, 63 Whereas canonical lantipeptide cyclases resolve the enolate intermediate formed after Cys thiol attack through protonation, the LabKC enzyme quenches the enolate through a second Michael addition to afford a labionin ring (Figure 4). Further characterization of the labionin cyclases is required to understand how these enzymes catalyze tandem macrocycle formation. Moreover, additional posttranslational modifications continue to be identified within this RiPP subfamily, promising a wealth of biological chemistry still to be uncovered.60, 61
Figure 4.
Lanthionine ring formation in lantipeptides. The mechanism of dehydration of Ser/Thr residues to generate dehydroalanine/dehydrobutyrine moieties (orange) in lantipeptides is displayed. In all studied cases, dehydration is ATP-dependent and proceeds through a phosphorylated intermediate. Subsequently, a cyclase domain catalyzes a Michael-type addition to form the lanthionine rings (purple). In a subclass of lantipeptides, a second Michael-addition occurs to generate a labionin ring (green). In all cases, the enzymes responsible for each transformation are displayed in the same color as the modification they install.
ATP-Independent Cyclizations
As discussed above, an energy source is often required during the cyclization of RiPPs to both accelerate leaving group departure and overcome the entropic barrier to cyclization. Even lantipeptide thioether biogenesis requires ATP to generate the electrophile for cyclization (vide supra). Cyclases that do not require any external energy source for catalysis are of particular interest, as these proteins have evolved unique strategies to overcome cyclization barriers. Below, we will discuss two recently described cases of such enzymes involved in RiPP biosynthesis.
Cyanobactin macrocyclization
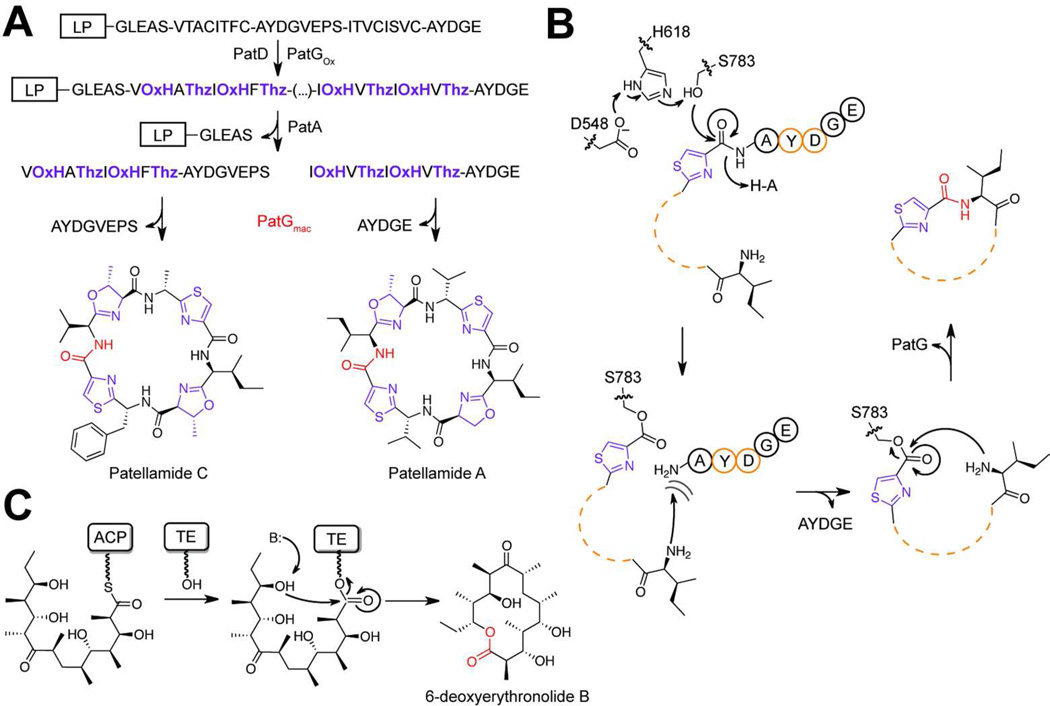
Cyanobactins are a diverse class of macrocyclized RiPPs produced by cyanobacteria isolated from soil, marine and freshwater environments.15, 17, 64 With over 100 identified members, these often azol(in)e-containing peptides have become a prime example for RiPP diversity.16, 21, 65 Despite the heterogeneity of this compound class, the associated posttranslational tailoring enzymes display remarkable sequence similarity, suggesting that the enzymes possess a high degree of promiscuity.15, 22, 66 As a result, cyanobactin biosynthetic enzymes have gained attention as suitable targets for bioengineering efforts.22, 25, 67 In several notable cases, cyanobactin precursor peptides encode for more than one natural product (e.g. PatE encodes for patellamide A and C).29 PatE modification begins with the installation of azole(in)e heterocycles by PatD, a TOMM synthetase (Figure 5A).68 Next, a subtilisin-like protease, PatA, removes the leader peptide and separates the heterocyclized patellamide A/C peptides, each bearing an AYDG sequence at their C-termini.33 This tetrapeptide motif serves as a recognition sequence for another subtilisinlike protease, PatG. In vitro studies demonstrated that PatG removes the C-terminal AYDG tag and facilitates cyanobactin N-to-C macrocyclization.33 Given the homology to subtilisin, a mechanism was proposed where the acyl-enzyme intermediate was resolved with the N-terminus, rather than water, to generate the macrolactam.
Figure 5.
Strategies for natural product macrocyclization. a) Cyanobactin biosynthetic scheme. Oxazoline (OxH; purple) and thiazole (Thz; purple) heterocycles are installed on cyanobactin precursor peptides by PatD (cyclodehydratase) and the N-terminal oxidation domain of PatG. After heterocycle installation, the peptide is proteolyzed by PatA and PatG, the latter also catalyzing macrocyclization (red). b) The PatG macrocyclization mechanism is displayed with the catalytic triad shown. The residues important for recognition in the C-terminal AYDG recognition sequence are colored orange. c) As a comparison, a mechanism for the DEBS thioesterase domain (non-RiPP) is displayed. This mechanism is conserved in a majority of the characterized PKS and NRPS thioesterases.
Recently, the crystal structure of the central PatG macrocyclase domain, PatGmacwas solved.59 As expected, PatGmac adopts the subtilisin-like fold. However, PatG contains an additional loop positioned over the active site. The PatG-type protease/macrocyclase proteins all contain a 30– 50 amino acid insertion between two of the three residues involved in subtilisin catalytic triad, providing a bioinformatic handle for identifying cyanobactin macrocylases.33 An analysis of PatGmac co-crystallized with a peptide substrate demonstrated that this insertion was responsible for shielding the active site from water and also binding of the AYDG sequence.59 Using both site-directed mutagenesis and selective truncations of this insertion sequence, it was determined that any disruption of AYDG recognition or water shielding resulted in a loss of macrocyclase, but not protease, activity. To further demonstrate the importance of the insertion in the macrocyclization reaction, Lys598, a residue that forms a pivotal salt bridge with the aspartate of the AYDG motif, was mutated. As expected, the K598D mutant was able to catalyze AYDG cleavage, but not macrocyclization. However, if a PatE substrate analog bearing a complementary AYKG sequence was supplied to K598D, macrocyclization activity was restored. Combined with the observation that the acyl-enzyme intermediate is remarkably long-lived, the mechanism in Figure 5B was proposed. In this mechanism, the insertion loop prevents AYDG peptide dissociation after cleavage until the N-terminus displaces it and resolves the covalent intermediate.
The PatG-catalyzed transformation is extraordinary for three reasons: (1) unlike the thioesterase domains in polyketide and non-ribosomal peptide synthetases (Figure 5C), PatG macrocyclization occurs on an unactivated substrate without an external energy source, (2) the prime side specificity (recognition is C-terminal to the scissile bond) of PatG facilitates the processing of hypervariable substrates with a minimal sequence tag is in stark contrast to other subtilisin-like proteases and (3) the strong sequence similarity to subtilisin proteases suggests that these proteins have been naturally evolved to catalyze a variety of chemical transformations.33 Importantly, all three of these properties are enabled by the addition of a short insertion to the subtilisin fold. Altering protein function by such a simple and elegant means begs the question as to whether such insertions are a general strategy utilized by Nature to expand the catalytic versatility of biosynthetic enzymes.
Thiopeptide macrocyclization: The discovery of a Diels-Alderase?
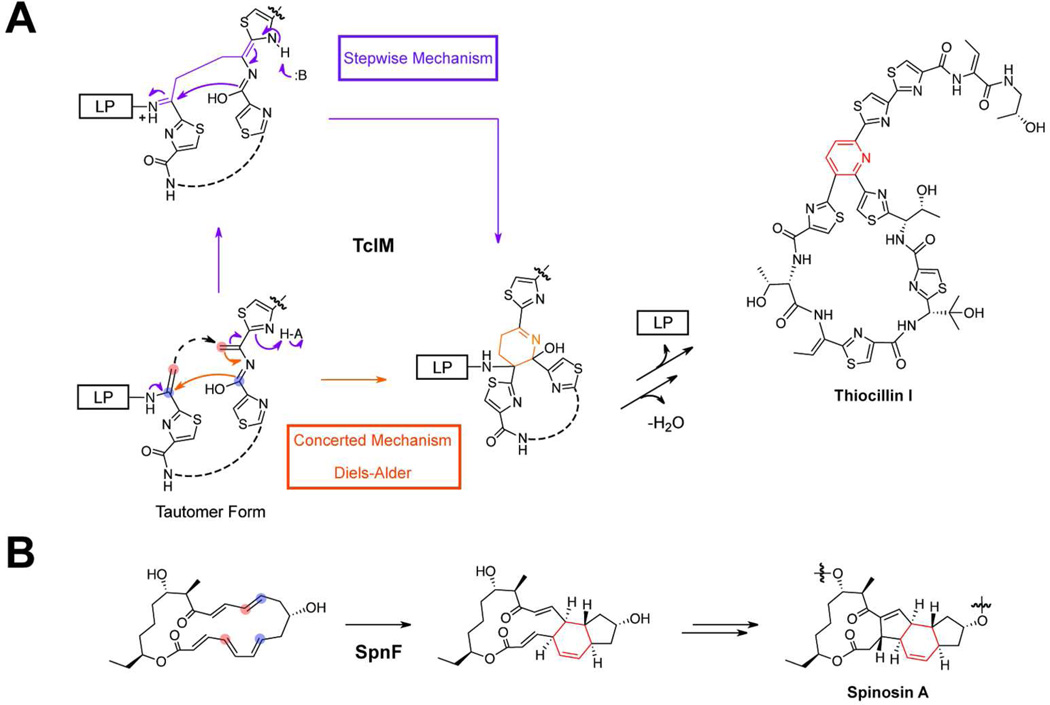
While cyanobactins are head-totail macrolactams, the central macrocycle found in all thiopeptides is formed through a cycloaddition process that forms two carbon-carbon linkages.69 This macrocyclization forms the ubiquitous thiopeptide (dehydro)pyridine ring, which is critical for target recognition.70 Recent investigations into thiocillin biosynthesis have demonstrated that the pyridine ring is assembled via the TclMcatalyzed trans-annular heteroannulation of two dehydroalanine residues.71, 72 This formal [4+2] cycloaddition has received a great deal of attention, as it has been postulated to proceed through an aza-Diels-Alder mechanism. Although the Diels-Alder is one of the most widely employed reactions in organic synthesis, no naturally occurring catalyst has been definitively shown to carry out a concerted [4+2] cycloaddition.73 Theoretically, 13C primary kinetic isotope effects (KIEs) could be used to determine if the TclM-catalyzed cycloaddition occurs via a stepwise or concerted mechanism (Figure 6A; see reference 74 for an in-depth discussion of KIEs and how they are used to support enzymatic mechanisms). To perform this experiment, the two differentially (dual) labeled substrates shown in Figure 6A would be required. If the mechanism were concerted, a primary 13C KIE would be observed with both substrates; however, if cycloaddition occurred via a stepwise mechanism, a KIE would only be observed for one of the dual-labeled substrates. While this experiment may seem relatively straightforward, in practice it will be difficult. TclM has never been reconstituted in vitrothe heterocyclized and dehydrated substrate is not readily accessible, and the expected primary 13C KIEs require a highly sensitive kinetic assay to measure. A more tractable alternative for the discovery of a bona fide Diels-Alderase may be to examine the enzyme responsible for the [4+2] cycloaddition in spinosin A biosynthesis (Figure 6B).75 While SpnF is not responsible for producing a RiPP, it has been reconstituted in vitrothe reaction allows for the use of 2H secondary KIEs (which are typically 30% larger than 13C primary KIEs),74 and the substrate is more amenable to chemical synthesis.
Figure 6.
[4+2] cycloadditions in natural product biosynthesis. a) Two potential mechanisms for installing the pyridine moiety of thiocillin I (red) are displayed. Starting from the tautomer form, the orange arrows demonstrate a concerted Diels-Alder mechanism, while the purple arrows follow a stepwise, polar mechanism. The shared step of each mechanism is displayed as a dashed black arrow. After the [4+2] cycloaddition is complete, the elimination of water and the leader peptide (LP) affords the central pyridine ring. The colored circles on the tautomer form of the non-cyclized intermediate indicate positions where dual 13C-labeling could distinguish between stepwise and concerted cyclization mechanisms. b) The SpnF-catalyzed cycloaddition of spinosin A (red) is displayed. The colored circles indicate positions where dual 2H-labeling could be used for establishing the precise mechanism.
Radical Chemistry In RiPP Maturation
The radical S-adenosylmethionine (rSAM) family of proteins is responsible for carrying out some of the most difficult biological transformations. Using a 4Fe-4S cluster ligated by three cysteine residues, typically in an easily identifiable CxxxCxxC motif, these proteins use the Fe-S cluster to perform a reductive cleavage of the labile sulfonium moiety of SAM to afford methionine and a 5’-deoxyadenosine (5’-Ado) radical.76 This radical is then used to carry out a variety of synthetically challenging reactions.76–79 The functionality of the rSAM proteins continues to expand as more members of the family are characterized. In just the last three years, a large role for rSAM enzymes in RiPP biosynthesis has emerged. The following section highlights recently elucidated chemistry performed by these enzymes.
Radical rearrangements in thiopeptide biosynthesis
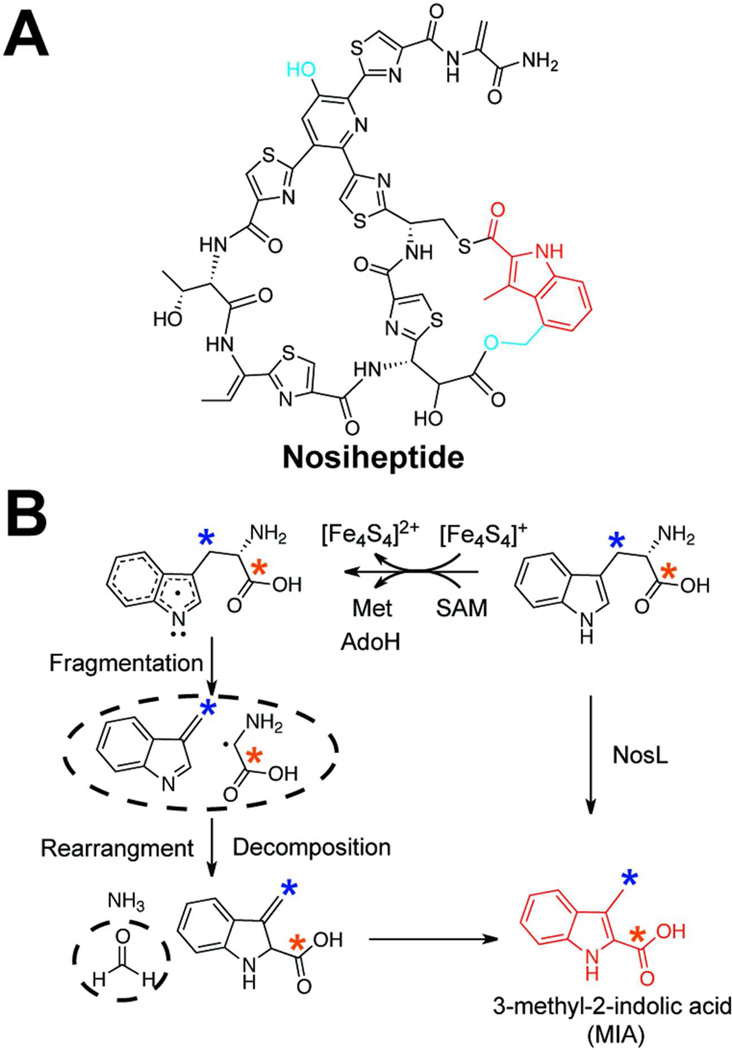
While the presence of the core macrocycle is seen in all thiopeptides, select family members also contain a second macrocycle.69 This ring is not formed from the gene-encoded peptide sequence but rather occurs through the posttranslational attachment of a modified tryptophan.80–84 Currently, two flavors of this auxiliary macrocycle have been identified. In thiostrepton, the macrocycle is comprised of a L-tryptophan-derived quinaldic acid moiety, whose biosynthesis occurs via a complex and poorly understood pathway and thus will not be discussed further.81, 82, 85 In contrast, nosiheptide contains an indolic acid derivative, which is assembled via the action of two rSAM enzymes, NosL and NosN (Figure 7A).80 Through genetic studies, it was discovered that deletion of nosL abolished nosiheptide production, but that supplementation of 3-methyl-2-indolic acid (MIA) rescued product formation. Moreover, a nosN deletion formed a nosiheptide derivative with MIA attached via a single thioester linkage rather than the expected 3,4-dimethyl indolic acid derivative. These results were used to putatively assign NosL and NosN as the enzymes involved in the rearrangement of L-tryptophan to MIA and the methylation of the unactivated C4 position, respectively.80
Figure 7.
The mechanism of MIA formation in nosiheptide biosynthesis. a) The structure of nosiheptide is displayed with the indolic acid ring highlighted in red. The uncharacterized radical SAM methyltransferase, NosA, and an as of yet unidentified protein, methylate and oxidize MIA to afford the second attachment point (blue moiety). b) The proposed mechanism of indolic acid formation by the radical SAM protein NosL is shown. The structures of the intermediates/products enclosed in the dotted circles were detected in reaction mixtures. The colored asterisks denote 13C-labeled positions that were used to demonstrate the fate of the carbon backbone during the rearrangement.
To better understand the transformation of L-tryptophan to MIA, NosL was recently heterologously expressed and investigated in vitro.34 Support for assigning NosL as the sole protein responsible for MIA formation came initially from the detection of MIA in NosL-expressing E. coli. Further characterization demonstrated that NosL reductively cleaved SAM and used the 5’-Ado radical to form MIA. Reactions conducted with [1-13C]- and [3-13C]-labeled L-tryptophan showed that both carbons were retained during the transformation, while reactions conducted with [2H8]-L-tryptophan established that initial hydrogen abstraction does not occur from any of the C-H bonds in tryptophan. Fortuitously, a detailed analysis of the NosL product profile identified four side products: 3-methylindole, glyoxate, glycine and formaldehyde. This led to the mechanistic proposal presented in Figure 7B. Based on the [2H8]-L-tryptophan experiment, the 5’-Ado radical is proposed to abstract the N-H hydrogen to generate a stabilized indole radical. This radical then fragments to form 3-methylene indole and a glycine radical, derivatives of which were identified as shunt products. Subsequent radical attack on the C2 position of the indole ring, and decomposition of the glycine subunit to form formaldehyde and ammonium, affords MIA. A subsequent report on NocL, a NosL homolog in nocathiacin MIA biogenesis, directly detected the glycine radical by EPR, supporting the proposed mechanism.86
Prior to the study of NosL, no member of the rSAM superfamily had been shown to catalyze both the fragmentation and rearrangement of a substrate.79 While this chemistry is noteworthy, glutamate mutase, a cobalamin-dependent enzyme, catalyzes a similar fragmentation-rearrangement reaction.87 Glutamate mutase uses a cobalamin-generated 5’-Ado radical to perform C-H abstraction. Akin to the NosL mechanism, the resulting radical decomposes into a glycine radical and acrylate. In a final step, these recombine to afford 2-methylaspartate. However, unlike glutamate mutase, NosL catalyzes a fragmentation-recombination reaction with elimination of a portion of the molecule during the transformation. The discovery of this complex radical transformation adds to the ever-expanding list of reactions catalyzed by rSAM proteins and serves as another demonstration of how Nature adjusts a general strategy to accomplish a specific reaction.
A tale of two Fe-S clusters: subtilosin thioether biogenesis
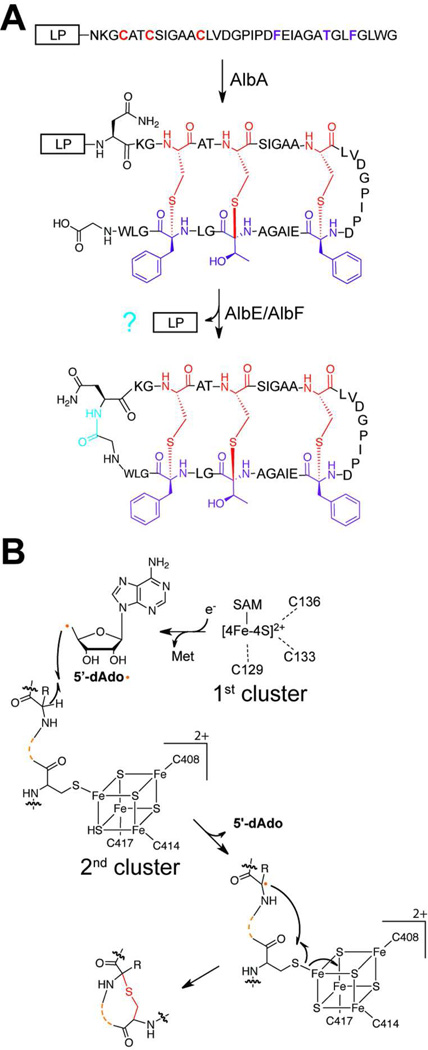
Subtilosin A is a broad spectrum antimicrobial RiPP produced by Bacillus subtilis.88–89 In addition to a head-to-tail macrocyclization, subtilosin A contains unusual thioether linkages connecting the sulfurs of Cys4, Cys7 and Cys13 to the α-carbons of Phe31, Thr28 and Phe22, respectively.90 Since the structure of subtilosin A was determined, similar linkages have been discovered in only four other RiPPs, although genome mining suggests more exist.91–94 Genetic studies of subtilosin A biosynthesis demonstrated that three genes, albA, albEand albFwere responsible for the maturation of the linear precursor peptide, SobA.95, 96 AlbE and AlbF are homologous to proteases and are predicted to be involved in leader peptide cleavage and macrocyclization. AlbA, on the other hand, shares homology with rSAM proteins and was predicted to install the thioether linkages on subtilosin A (Figure 8A).90 Indeed, AlbA was demonstrated to be necessary and sufficient to catalyze thioether formation on SobA via the reductive cleavage of SAM.97 By subjecting both wild-type AlbA and a Fe-S cluster deficient mutant (CxxxCxxC triple mutant) to spectroscopic interrogation, it was discovered that AlbA contained two 4Fe-4S clusters. The first, ligated by Cys129, Cys133, and Cys136, was responsible for SAM coordination and 5’-Ado radical generation, while the function of the second cluster was unknown. As expected from studies on other rSAM enzymes with two Fe-S clusters,98–100 mutation of the cysteine residues within the second cluster (Cys408, Cys414 and Cys414) abolished activity.94, 97 Studies to assign a precise functional role for this auxiliary 4Fe-4S cluster utilized an AlbA mutant with the first Fe-S cluster removed. UV-Vis spectra showed an absorbance shift to the remaining 4Fe-4S cluster upon the addition of SobA. This shift was leader peptide-dependent, suggesting that proper substrate binding is required to place the peptide in close proximity of the second Fe-S cluster within the protein.
Figure 8.
Subtilosin biosynthesis. a) The general biosynthetic scheme for subtilosin is shown. Residues involved in thioether crosslinking are colored (red, donor residue; purple, receiver residue). The protein(s) responsible for leader peptide (LP) cleavage and macrocyclization (blue) have not been characterized. b) A proposed mechanism of thioether formation is presented. The first Fe-S cluster is involved in radical generation while the second cluster serves as a one-electron oxidant of the cysteine thiol.
The above results were used to propose the mechanism displayed in Figure 8B. The 5’-Ado radical generated from the reductive cleavage of SAM by the primary 4Fe-4S cluster is used to abstract the hydrogen from the α-carbon of Phe31, Thr28 and Phe22. This radical then attacks the appropriate cysteine thiol, which is ligated to the ancillary Fe-S cluster. The second 4Fe-4S cluster is proposed to facilitate the second electron oxidation of the thiol to generate the thioether linkage, in a fashion analogous to other rSAM-dependent oxidations.99
Radical SAM-dependent methylation: future work in RiPP biosynthesis
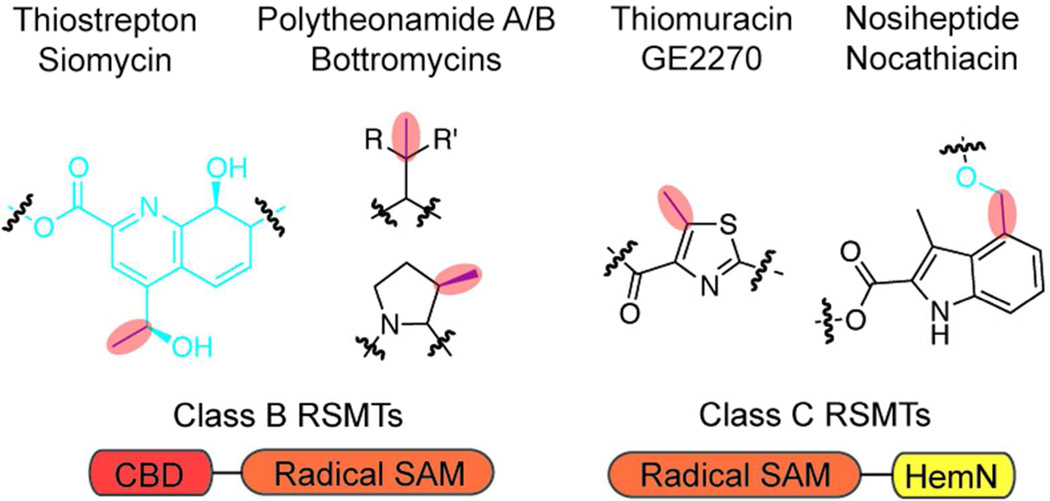
Various positions within thiopeptides are methylated, despite being unactivated and non-nucleophilic, such as the C4 methylation of MIA (Figure 9; vide supra).80, 101 Furthermore, recent efforts have identified similar modifications in other RiPP biosynthetic clusters, including the bottromycins and polytheonamides.43–46, 102 In certain cases, labeling studies indicated that SAM was the source of these methyl groups;103, 104 however, the unactivated nature of the carbon suggested that a radical mechanism would be necessary for modification. Accordingly, recent studies conducted on two rRNA methyltransferases, RlmN and Cfr, provided the first definitive evidence that rSAM enzymes can perform methylation reactions.105, 106 These proteins have become the founding members of the radical SAM methyltransferase (RSMT) protein family, of which there are three bioinformatically-identifiable classes.101 Class A is solely comprised of RlmN and Cfr, contains only a rSAM domain, and is the best characterized class to date. However, as these enzymes are involved in ribonucleotide methylation and not RiPP biosynthesis, they will not be discussed further. Rather, focus will be placed on class B and class C RSMTs because analyses of several RiPP biosynthetic clusters demonstrate that in all cases where the natural product is methylated at an unactivated position, a RSMT of class B or C is present.
Figure 9.
Radical SAM-dependent methylations in RiPPs. Shown are several examples of RSMTdependent RiPP modifications (red highlight). Shown below each structure is the class of RSMT and its domain architecture (CBD, cobalamin-binding domain). The blue moieties are installed by uncharacterized enzymes. R= H, R’= Ph or R= Me, R’= Me/Et.
In addition to the canonical rSAM domain, class B RSMTs contain a N-terminal cobalamin-binding domain. The first class B RSMT to be characterized was TsrM, which is involved in thiostrepton biosynthesis. This enzyme was recently reconstituted in vitro and exhibited a novel strategy for methylation.107 TsrM catalyzed the SAM and cobalamin-dependent methylation of the C2 position of tryptophan (Figure 9), which has been predicted to be the precursor for the quinaldic acid moiety.82 Unlike all other rSAM homologs, TsrM did not carry out the reductive cleavage of SAM despite containing a CxxxCxxC ligated Fe-S cluster.107 Instead, SAM was used as a methyl source for the in situ formation of a methylcobalamin cofactor, which then serves as the methyl source for tryptophan methylation. Although the mechanism of methyltransfer from the methylcobalamin cofactor to tryptophan remains enigmatic, preliminary data suggests that the Fe-S cluster may be pivatol for this transformation. While this initial study of TsrM is interesting from a mechanistic standpoint, it has larger implications for the classification of the rSAM superfamily. TsrM contains the ubiquitous Fe-S cluster ligation motif characteristic of rSAM proteins, and is SAM-dependent, but does not catalyze the reductive cleavage of SAM. This stands in stark contrast to all other rSAMs and prompts the question as to whether TsrM should be considered a true member of the family. Moreover, the characterization of additional class B RSMTs will be required to determine if this unusual utilization of SAM by a rSAM domain is a conserved strategy for methylation of unactivated positions.
To date, no member of the class C RSMTs has been reconstituted. These proteins share significant similarity to the rSAM protein coproporphyrinogen III oxidase (HemN), which is responsible for the oxidative decarboxylation of coproporphyrinogen III to form protoporphyrinogen IX in heme biogenesis.108 Although class C RSMTs contain both a HemN-like rSAM domain and a domain similar to the C-terminus of HemN (Figure 9), they lack the N-terminal “trip-wire” domain of HemN.101 Studies on HemN have demonstrated that the rSAM TIM barrel core binds two molecules of SAM109 and a comparison to class C RSMTs hints that these enzymes may also simultaneously bind two SAM molecules.101 Additional study of these putative methyltransferases is required to fully understand the variety of methods Nature has developed for the methylation of unactivated carbon.
Leader Peptide-Independent Modifications
Apart from the NosL-catalyzed rearrangement of L-tryptophan and the macrocyclization of cyanobactins, all of the modifications discussed above require a leader peptide for recognition by the tailoring enzymes. This broadly employed strategy in RiPP biosynthesis facilitated the evolution of tremendous diversity within the natural product class by separating substrate recognition from the modified regions of the peptides.18 Nonetheless, recent investigations of disparate RiPP biosynthetic pathways have uncovered several posttranslational modification machineries that function in a leader peptide-independent fashion. A few of these unusual cases are discussed below.
C-prenylation through a Claisen rearrangement
Amongst other modifications, select cyanobactins are further decorated by O-prenylation. Deriving from dimethylallyl pyrophosphate (DMAPP), this modification is observed at Ser, Thr and Tyr residues. Ser and Thr prenylation occurs through the C3 position of DMAPP (reverse prenylation). Conversely Tyr prenylation appears to occur exclusively through the C1 position (forward prenylation; Figure 10A).65 This modification is notable, as the only known O-prenylated RiPPs are cyanobactins. Based on comparative genomics, TruF and its homologs were tentatively assigned as O-prenyltransferases.15 However, this assignment was complicated by the fact that TruF-like proteins are not homologous to known prenyltransferases and are essential in the biosynthesis of non-prenylated cyanobactins as well.15, 17
Figure 10.
Cyanobactin prenylation. a) The two forms of O-prenylation are displayed along with examples of natural products containing each modification. b) The LynF prenylation mechanism. After reverse O-prenylation of tyrosine, the intermediate undergoes a Claisen rearrangement to afford the C-prenylated product. As the product of the Lyn cluster has not been identified, the predicted product is displayed without stereochemistry. The DMAPP derived prenyl groups are shown in red.
In 2011, the first insights into cyanobactin prenylation emerged with the in vitro reconstitution of a prenyltransferase from Lyngbya aestuariiLynF.35 While the product of the Lyn cluster remains uncharacterized, initial activity screens with various peptides substrates confirmed that LynF was a tryrosine prenyltransferase. Unexpectedly, LynF displayed a strong selectivity for macrocyclized substrates over linear peptides. Moreover, LynF was not able to prenylate substrates with leader peptides regardless of whether they contained azol(in)e heterocycles. Considering that heterocyclization occurs prior to macrocyclization, this suggested that prenylation is the final step in cyanobactin biosynthesis. Further analysis of the substrate promiscuity demonstrated that LynF displayed relaxed sequence specificity, but a definitive set of modification rules could not be established. This prompted the question as to how substrate recognition was achieved in vivo given that LynF did not utilize the standard leader peptide recognition strategy used by most RiPP tailoring enzymes. Reaction products were analyzed by 2D-NMR to definitively establish the site of Tyr prenylation. Surprisingly, the spectra showed that LynF catalyzed forward C-prenylation ortho to the phenolic oxygen, rather than O-prenylation. Similar transformations have been characterized in other biosynthetic pathways, which were proposed to proceed via electrophilic aromatic substitution.110 To evaluate the mechanism of LynF prenylation, reaction products were analyzed with multiple spectroscopic methods over a reaction time course. The results demonstrated that the substrate initially undergoes reverse O-prenylation, but slowly converts to the forward C-prenylated phenol, most likely through a Claisen rearrangement (Figure 10B).
Although highly similar reaction schemes have been synthetically employed,111, 112 LynF represents the first instance of enzymatic C-prenylation via a reverse O-prenylation/Claisen rearrangement route. Additional experiments concluded that this rearrangement occurs at a slow but measurable rate (0.23 h−1) in the absence of enzyme, demonstrating that reverse O-prenylated phenols are prone to spontaneous rearrangement.35 This result provides a possible explanation as to why reverse O-prenylated phenol rings are not seen in natural products, while reverse O-prenylated hydroxyl moieties are common. It remains uncertain whether the rearrangement is truly uncatalyzed in vivo or if the product of the Lyn cluster is actually C-prenylated. Regardless, LynF remains the only RiPP prenyltransferase characterized to date and demonstrates that C-prenylation in natural product biosynthesis can be achieved via a Claisen rearrangement, as was predicted for over 40 years.113
S-glycosylations in RiPP biogenesis
Sublancin is a RiPP originally discovered in strains of B. subtilis containing SPβ-prophage.114 The initial structural assignment of sublancin reported the presence of two lanthionine rings and a disulfide bridge.115 Recently, the structure of sublancin was revised to contain two disulfide bridges and an S-glycosylated Cys (Figure 11).116 Analysis of the gene cluster and subsequent in vitro reconstitution identified that sunS catalyzed the S-glycosylation reaction. Investigations into the selectivity of SunS showed that the selective S-glycosylation of Cys22 was both leader peptide- and disulfide bond-independent. Additional experiments demonstrated that this selectivity was not from the flanking residues, suggesting that SunS must be recognizing the substrate away from the glycosylated site. Indeed, a follow up study determined that substrate recognition occurred at a N-terminal α-helical stretch in the peptide. This α-helix served as a form of internal leader peptide, granting SunS a high level of substrate promiscuity in the region surrounding Cys22 without the use of a traditional leader peptide.117 Interestingly, this follow up study also assigned a putative role for the glycosyl moiety. Although the modification was not required for activity against sensitive strains of B. subtilisthe aglycone was active against producer strains. This suggested that the glycosylation is part of a novel immunity mechanism; however, further experiments will be necessary to validate this hypothesis.
Figure 11.
The structure of sublancin. The modification installed by SunS (S-glycosyltransferase) is shown in red.
C-terminal amidation through divergent mechanisms
The C-termini of many thiopeptides are amidated. This modification, while easy to overlook when considering the structural complexity of these heavily modified RiPPs, has an important role in biological activity.36 Recently, the tailoring machinery responsible for C-terminal amidation moiety has been characterized in thiostrepton and nosiheptide biosynthesis.36, 37 These studies demonstrated that two divergent strategies have been utilized by Nature to perform the amidation in a leader peptide-independent fashion. In nosiheptide biosynthesis, genetic deletions and in vitro reactions were used to identify nosA as the responsible gene.37 The ΔnosA strain produced a nosiheptide analog with an additional C-terminal dehydroalanine moiety, which is derived from the dehydration of the terminal serine of the precursor peptide. Reactions with purified NosA validated this assignment and revealed that pyruvate was a reaction byproduct. Based on the pH dependence of the reaction, and the cofactor independent activity of NosA, this reaction was predicted to proceed via the enamide de-alkylation mechanism displayed in Figure 12A. Thiostrepton, on the other hand, requires at least three enzymes to perform the same modification.36, 81, 82 An unidentified methyltransferase is hypothesized to generate the C-terminal methylester on the precursor peptide to facilitate the dehydration of the terminal Ser. Subsequently, TsrB (also called TsrU in ref 82) catalyzes the saponification of the methylated thiostrepton intermediate. Finally, the carboxylate is modified by an asparagine synthetase-like amidotransferase, TsrC (also called TsrT in ref 82), which forms the amide linkage, presumably though an adenylated intermediate.36 The proposed mechanism of this modification is displayed in Figure 12B; however, additional studies are necessary to identify the responsible methyltransferase and to establish the mechanism of TsrC. Such disparate strategies employed by Nature to accomplish identical transformations highlight the powerful interplay of convergent evolution and biological activity.
Figure 12.
C-terminal amidation of thiopeptides. The putative mechanism for C-terminal amide formation is displayed for nosiheptide a) and thiostrepton b). The proposed transformation catalyzed by TsrC is boxed. Tables listing the other thiopeptides that utilize each mechanism, along with the pertinent protein, are listed on the right.
Outlook
Recent successes in the study of RiPP biosynthetic enzymes have greatly expanded our view of Nature’s synthetic organic potential. In addition to the discovery of novel biological chemistry, these studies provide a window through which the evolutionary adaptation of enzymes can be observed. However, there are many tailoring enzymes that have yet to be characterized and many posttranslational modifications whose biosynthesis remains enigmatic. The topics covered in this review represent only a small set of the ever-expanding space occupied by RiPPs. Further characterization of RiPPs, and other biosynthetic systems, will be necessary if we are ever to fully understand and intelligently exploit such enzymes.
Acknowledgments
We are grateful to E. Molloy, P. Knerr, and members of Mitchell lab for critical review of this manuscript. This work was supported by funds provided by the National Institutes of Health (1R01 GM097142) to D. Mitchell; K. Dunbar was supported by the University of Illinois Department of Chemistry Harold R. Snyder Fellowship.
Keywords
- RiPP
gene-encoded, peptidic natural products that are ribosomally synthesized and post-translationally modified
- TOMM
RiPP natural products containing one or more azole and/or azoline heterocycles derived from Cys, Ser and Thr residues
- Cycloaddition
a reaction between two or more unsaturated molecules to generate a cyclized adduct. Cycloadditions can be stepwise (dipolar) or concerted (pericyclic). Example: the Diels-Alder reaction is a pericyclic [4+2] cycloaddition that yields a cyclohexene (6-membered) product
- Precursor peptide
the unmodified, ribosomally synthesized peptide that serves as a substrate for RiPP modification enzymes. RiPP precursor peptides are typically comprised of both a leader peptide and the core peptide. One exception to this general rule is that of bottromycin, which contains a follower peptide.
- Leader peptide
the N-terminal portion of a precursor peptide that serves as a recognition sequence for RiPP modification enzymes.
- Core peptide
the portion of the precursor peptide (usually the C-terminal portion) that is post-translationally modified.
- Radical SAM
an enzyme that uses S-adenosylmethionine as a radical source to catalyze a variety of difficult chemical transformations
- Fe-S cluster
an ensemble of iron and sulfur atoms used as a cofactor for enzymatic transformations. Different ratios of iron to sulfur exist depending on the nature of cluster
- Thiopeptide
a RiPP subclass containing a six-membered, nitrogen-containing heterocycle formed via a [4+2]-cycloaddition reaction. All known thiopeptides also contain azole/azoline heterocycles.
- Cyanobactin
a RiPP subclass of small, N- to C-macrocyclized peptides produced by cyanobacteria. Often, cyanobactins will harbor azole/azoline heterocycles characteristic of TOMM natural products.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Harvey AL. Natural products in drug discovery. Drug Discov. Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson RM, Danishefsky SJ. Small molecule natural products in the discovery of therapeutic agents: the synthesis connection. J. Org. Chem. 2006;71:8329–8351. doi: 10.1021/jo0610053. [DOI] [PubMed] [Google Scholar]
- 4.Razzak M, De Brabander JK. Lessons and revelations from biomimetic syntheses. Nat. Chem. Biol. 2011;7:865–875. doi: 10.1038/nchembio.709. [DOI] [PubMed] [Google Scholar]
- 5.Falconer SB, Czarny TL, Brown ED. Antibiotics as probes of biological complexity. Nat. Chem. Biol. 2011;7:415–423. doi: 10.1038/nchembio.590. [DOI] [PubMed] [Google Scholar]
- 6.Romero D, Traxler MF, Lopez D, Kolter R. Antibiotics as signal molecules. Chem. Rev. 2011;111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kocaoglu O, Calvo RA, Sham LT, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, Carlson EE. Selective Penicillin-Binding Protein Imaging Probes Reveal Substructure in Bacterial Cell Division. ACS Chem. Biol. 2012 doi: 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 9.Yajima J, Alonso MC, Cross RA, Toyoshima YY. Direct long-term observation of kinesin processivity at low load. Curr. Biol. 2002;12:301–306. doi: 10.1016/s0960-9822(01)00683-2. [DOI] [PubMed] [Google Scholar]
- 10.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, E EM. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2009. Chemical Tools for Inhibiting Glycosylation; pp. 705–719. [PubMed] [Google Scholar]
- 11.Baltz RH. Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 2006;33:507–513. doi: 10.1007/s10295-005-0077-9. [DOI] [PubMed] [Google Scholar]
- 12.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 13.Velasquez JE, van der Donk WA. Genome mining for ribosomally synthesized natural products. Curr. Opin. Chem. Biol. 2011;15:11–21. doi: 10.1016/j.cbpa.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2012;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donia MS, Schmidt EW. Linking chemistry and genetics in the growing cyanobactin natural products family. Chem. Biol. 2011;18:508–519. doi: 10.1016/j.chembiol.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat. Chem. Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat. Chem. Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 18.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haft DH, Basu MK, Mitchell DA. Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family. BMC Biol. 2010;8:70. doi: 10.1186/1741-7007-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melby JO, Nard NJ, Mitchell DA. Thiazole/oxazole-modified microcins: complex natural products from ribosomal templates. Curr. Opin. Chem. Biol. 2011;15:369–378. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tianero MD, Donia MS, Young TS, Schultz PG, Schmidt EW. Ribosomal route to small-molecule diversity. J. Am. Chem. Soc. 2012;134:418–425. doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melby JO, Dunbar KL, Trinh NQ, Mitchell DA. Selectivity, directionality, and promiscuity in peptide processing from a Bacillus sp. Al Hakam cyclodehydratase. J. Am. Chem. Soc. 2012;134:5309–5316. doi: 10.1021/ja211675n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Field D, Hill C, Cotter PD, Ross RP. The dawning of a 'Golden era' in lantibiotic bioengineering. Mol. Microbiol. 2010;78:1077–1087. doi: 10.1111/j.1365-2958.2010.07406.x. [DOI] [PubMed] [Google Scholar]
- 25.Bowers AA. Biochemical and biosynthetic preparation of natural product-like cyclic peptide libraries. Medchemcomm. 2012;3:905–915. [Google Scholar]
- 26.Anderson B, Hodgkin DC, Viswamitra MA. The structure of thiostrepton. Nature. 1970;225:233–235. doi: 10.1038/225233a0. [DOI] [PubMed] [Google Scholar]
- 27.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 28.Nizet V, Beall B, Bast DJ, Datta V, Kilburn L, Low DE, De Azavedo JC. Genetic locus for streptolysin S production by group A streptococcus. Infect. Immun. 2000;68:4245–4254. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milne JC, Roy RS, Eliot AC, Kelleher NL, Wokhlu A, Nickels B, Walsh CT. Cofactor requirements and reconstitution of Microcin B17 synthetase: A multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic Microcin B17. Biochemistry. 1999;38:4768–4781. doi: 10.1021/bi982975q. [DOI] [PubMed] [Google Scholar]
- 31.Kelleher NL, Hendrickson CL, Walsh CT. Posttranslational heterocyclization of cysteine and serine residues in the antibiotic Microcin B17: Distributivity and directionality. Biochemistry. 1999;38:15623–15630. doi: 10.1021/bi9913698. [DOI] [PubMed] [Google Scholar]
- 32.Roy RS, Kim S, Baleja JD, Walsh CT. Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26. Chem. Biol. 1998;5:217–228. doi: 10.1016/s1074-5521(98)90635-4. [DOI] [PubMed] [Google Scholar]
- 33.Lee J, McIntosh J, Hathaway BJ, Schmidt EW. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J. Am. Chem. Soc. 2009;131:2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Li Y, Chen D, Yu Y, Duan L, Shen B, Liu W. Radical-mediated enzymatic carbon chain fragmentation-recombination. Nat. Chem. Biol. 2011;7:154–160. doi: 10.1038/nchembio.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McIntosh JA, Donia MS, Nair SK, Schmidt EW. Enzymatic basis of ribosomal peptide prenylation in cyanobacteria. J. Am. Chem. Soc. 2011;133:13698–13705. doi: 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao R, Liu W. Thiostrepton maturation involving a deesterification-amidation way to process the C-terminally methylated peptide backbone. J. Am. Chem. Soc. 2011;133:2852–2855. doi: 10.1021/ja1111173. [DOI] [PubMed] [Google Scholar]
- 37.Yu Y, Guo H, Zhang Q, Duan L, Ding Y, Liao R, Lei C, Shen B, Liu W. NosA catalyzing carboxyl-terminal amide formation in nosiheptide maturation via an enamine dealkylation on the serine-extended precursor peptide. J. Am. Chem. Soc. 2010;132:16324–16326. doi: 10.1021/ja106571g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunbar KL, Melby JO, Mitchell DA. YcaO domains use ATP to activate amide backbones during peptide cyclodehydrations. Nat. Chem. Biol. 2012;8:569–575. doi: 10.1038/nchembio.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludwig C, Schwarzer D, Mootz HD. Interaction studies and alanine scanning analysis of a semi-synthetic split intein reveal thiazoline ring formation from an intermediate of the protein splicing reaction. J. Biol. Chem. 2008;283:25264–25272. doi: 10.1074/jbc.M802972200. [DOI] [PubMed] [Google Scholar]
- 40.Ghilarov D, Serebryakova M, Shkundina I, Severinov K. A Major Portion of DNA Gyrase Inhibitor Microcin B17 Undergoes an N,O-Peptidyl Shift during Synthesis. J. Biol. Chem. 2011;286:26308–26318. doi: 10.1074/jbc.M111.241315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perler FB, Xu MQ, Paulus H. Protein splicing and autoproteolysis mechanisms. Curr. Opin. Chem. Biol. 1997;1:292–299. doi: 10.1016/s1367-5931(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Morar M, Ealick SE. Structural biology of the purine biosynthetic pathway. Cell. Mol. Life Sci. 2008;65:3699–3724. doi: 10.1007/s00018-008-8295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez-Escribano JP, Song L, Bibb MJ, Challis GL. Chem. Sci. 2012;3 [Google Scholar]
- 44.Crone WJK, Leeper FJ, Truman AW. Chem. Sci. 2012;3 [Google Scholar]
- 45.Hou Y, Tianero MD, Kwan JC, Wyche TP, Michel CR, Ellis GA, Vazquez-Rivera E, Braun DR, Rose WE, Schmidt EW, Bugni TS. Structure and Biosynthesis of the Antibiotic Bottromycin D. Org. Lett. 2012 doi: 10.1021/ol3022758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huo L, Rachid S, Stradler M, Wenzel SC, Müller R. Synthetic Biotechnology to Study and Engineer Ribosomal Bottromycin Biosynthesis. Chem. Biol. 2012;19 doi: 10.1016/j.chembiol.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 47.Begley TP, Ealick SE, McLafferty FW. Thiamin biosynthesis: still yielding fascinating biological chemistry. Biochem. Soc. Trans. 2012;40:555–560. doi: 10.1042/BST20120084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee A, Abeydeera ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, Ealick SE, Begley TP. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature. 2011;478:542–546. doi: 10.1038/nature10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keating TA, Marshall CG, Walsh CT, Keating AE. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat. Struct. Biol. 2002;9:522–526. doi: 10.1038/nsb810. [DOI] [PubMed] [Google Scholar]
- 50.McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat. Prod. Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Bustos JF, Pezzi N, Mendez E. Structure and mode of action of microcin 7, an antibacterial peptide produced by Escherichia coli. Antimicrob. Agents Chemother. 1985;27:791–797. doi: 10.1128/aac.27.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guijarro JI, Gonzalez-Pastor JE, Baleux F, San Millan JL, Castilla MA, Rico M, Moreno F, Delepierre M. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J. Biol. Chem. 1995;270:23520–23532. doi: 10.1074/jbc.270.40.23520. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Pastor JE, San Millan JL, Moreno F. The smallest known gene. Nature. 1994;369:281. doi: 10.1038/369281a0. [DOI] [PubMed] [Google Scholar]
- 54.Gonzalez-Pastor JE, San Millan JL, Castilla MA, Moreno F. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J. Bacteriol. 1995;177:7131–7140. doi: 10.1128/jb.177.24.7131-7140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novikova M, Metlitskaya A, Datsenko K, Kazakov T, Kazakov A, Wanner B, Severinov K. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J. Bacteriol. 2007;189:8361–8365. doi: 10.1128/JB.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metlitskaya A, Kazakov T, Kommer A, Pavlova O, Praetorius-Ibba M, Ibba M, Krasheninnikov I, Kolb V, Khmel I, Severinov K. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic Microcin C. J. Biol. Chem. 2006;281:18033–18042. doi: 10.1074/jbc.M513174200. [DOI] [PubMed] [Google Scholar]
- 57.Roush RF, Nolan EM, Lohr F, Walsh CT. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. J. Am. Chem. Soc. 2008;130:3603–3609. doi: 10.1021/ja7101949. [DOI] [PubMed] [Google Scholar]
- 58.Regni CA, Roush RF, Miller DJ, Nourse A, Walsh CT, Schulman BA. How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J. 2009;28:1953–1964. doi: 10.1038/emboj.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koehnke J, Bent A, Houssen WE, Zollman D, Morawitz F, Shirran S, Vendome J, Nneoyiegbe AF, Trembleau L, Botting CH, Smith MC, Jaspars M, Naismith JH. The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Nat. Struct. Mol. Biol. 2012 doi: 10.1038/nsmb.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montalban-Lopez M, Zhou L, Buivydas A, van Heel AJ, Kuipers OP. Increasing the success rate of lantibiotic drug discovery by Synthetic Biology. Expert Opin. Drug Discov. 2012;7:695–709. doi: 10.1517/17460441.2012.693476. [DOI] [PubMed] [Google Scholar]
- 61.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 62.Meindl K, Schmiederer T, Schneider K, Reicke A, Butz D, Keller S, Guhring H, Vertesy L, Wink J, Hoffmann H, Bronstrup M, Sheldrick GM, Sussmuth RD. Labyrinthopeptins: a new class of carbacyclic lantibiotics. Angew. Chem. Int. Ed. Engl. 2010;49:1151–1154. doi: 10.1002/anie.200905773. [DOI] [PubMed] [Google Scholar]
- 63.Muller WM, Schmiederer T, Ensle P, Sussmuth RD. In vitro biosynthesis of the prepeptide of type-III lantibiotic labyrinthopeptin A2 including formation of a C-C bond as a post-translational modification. Angew. Chem. Int. Ed. Engl. 2010;49:2436–2440. doi: 10.1002/anie.200905909. [DOI] [PubMed] [Google Scholar]
- 64.Leikoski N, Fewer DP, Sivonen K. Widespread occurrence and lateral transfer of the cyanobactin biosynthesis gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2009;75:853–857. doi: 10.1128/AEM.02134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sivonen K, Leikoski N, Fewer DP, Jokela J. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl. Microbiol. Biotechnol. 2010;86:1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McIntosh JA, Donia MS, Schmidt EW. Insights into heterocyclization from two highly similar enzymes. J. Am. Chem. Soc. 2010;132:4089–4091. doi: 10.1021/ja9107116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McIntosh JA, Robertson CR, Agarwal V, Nair SK, Bulaj GW, Schmidt EW. Circular Logic: Nonribosomal Peptide-like Macrocyclization with a Ribosomal Peptide Catalyst. J. Am. Chem. Soc. 2010;132:15499–15501. doi: 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McIntosh JA, Schmidt EW. Marine Molecular Machines: Heterocyclization in Cyanobactin Biosynthesis. Chembiochem. 2010;11:1413–1421. doi: 10.1002/cbic.201000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem. Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 70.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 71.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bowers AA, Walsh CT, Acker MG. Genetic interception and structural characterization of thiopeptide cyclization precursors from Bacillus cereus. J. Am. Chem. Soc. 2010;132:12182–12184. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim HJ, Ruszczycky MW, Liu HW. Current developments and challenges in the search for a naturally selected Diels-Alderase. Curr. Opin. Chem. Biol. 2012;16:124–131. doi: 10.1016/j.cbpa.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schowen KB, Schowen RL. The Use of Isotope Effects to Elucidate Enzyme Mechanisms. Bioscience. 1981;31:826–831. [Google Scholar]
- 75.Kim HJ, Ruszczycky MW, Choi SH, Liu YN, Liu HW. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature. 2011;473:109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frey PA, Hegeman AD, Ruzicka FJ. The Radical SAM Superfamily. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 77.Roach PL. Radicals from S-adenosylmethionine and their application to biosynthesis. Curr. Opin. Chem. Biol. 2011;15:267–275. doi: 10.1016/j.cbpa.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 78.Booker SJ. Anaerobic functionalization of unactivated C-H bonds. Curr. Opin. Chem. Biol. 2009;13:58–73. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marsh EN, Patterson DP, Li L. Adenosyl radical: reagent and catalyst in enzyme reactions. Chembiochem. 2010;11:604–621. doi: 10.1002/cbic.200900777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu Y, Duan L, Zhang Q, Liao R, Ding Y, Pan H, Wendt-Pienkowski E, Tang G, Shen B, Liu W. Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem. Biol. 2009;4:855–864. doi: 10.1021/cb900133x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J. Am. Chem. Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 83.Mocek U, Zeng ZP, Ohagan D, Zhou P, Fan LDG, Beale JM, Floss HG. Biosynthesis of the Modified Peptide Antibiotic Thiostrepton in Streptomyces-Azureus and Streptomyces-Laurentii. J. Am. Chem. Soc. 1993;115:7992–8001. [Google Scholar]
- 84.Mocek U, Knaggs AR, Tsuchiya R, Nguyen T, Beale JM, Floss HG. Biosynthesis of the Modified Peptide Antibiotic Nosiheptide in Streptomyces-Actuosus. J. Am. Chem. Soc. 1993;115:7557–7568. [Google Scholar]
- 85.Duan L, Wang S, Liao R, Liu W. Insights into quinaldic acid moiety formation in thiostrepton biosynthesis facilitating fluorinated thiopeptide generation. Chem. Biol. 2012;19:443–448. doi: 10.1016/j.chembiol.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Q, Chen D, Lin J, Liao R, Tong W, Xu Z, Liu W. Characterization of NocL involved in thiopeptide nocathiacin I biosynthesis: a [4Fe-4S] cluster and the catalysis of a radical S-adenosylmethionine enzyme. J. Biol. Chem. 2011;286:21287–21294. doi: 10.1074/jbc.M111.224832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Banerjee R. Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem. Rev. 2003;103:2083–2094. doi: 10.1021/cr0204395. [DOI] [PubMed] [Google Scholar]
- 88.Babasaki K, Takao T, Shimonishi Y, Kurahashi K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Biochem. 1985;98:585–603. doi: 10.1093/oxfordjournals.jbchem.a135315. [DOI] [PubMed] [Google Scholar]
- 89.Shelburne CE, An FY, Dholpe V, Ramamoorthy A, Lopatin DE, Lantz MS. The spectrum of antimicrobial activity of the bacteriocin subtilosin A. J. Antimicrob. Chemother. 2007;59:297–300. doi: 10.1093/jac/dkl495. [DOI] [PubMed] [Google Scholar]
- 90.Kawulka K, Sprules T, McKay RT, Mercier P, Diaper CM, Zuber P, Vederas JC. Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to alpha-carbons of phenylalanine and threonine. J. Am. Chem. Soc. 2003;125:4726–4727. doi: 10.1021/ja029654t. [DOI] [PubMed] [Google Scholar]
- 91.Sit CS, van Belkum MJ, McKay RT, Worobo RW, Vederas JC. The 3D solution structure of thurincin H, a bacteriocin with four sulfur to alpha-carbon crosslinks. Angew. Chem. Int. Ed. Engl. 2011;50:8718–8721. doi: 10.1002/anie.201102527. [DOI] [PubMed] [Google Scholar]
- 92.Liu WT, Yang YL, Xu Y, Lamsa A, Haste NM, Yang JY, Ng J, Gonzalez D, Ellermeier CD, Straight PD, Pevzner PA, Pogliano J, Nizet V, Pogliano K, Dorrestein PC. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sit CS, McKay RT, Hill C, Ross RP, Vederas JC. The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links. J. Am. Chem. Soc. 2011;133:7680–7683. doi: 10.1021/ja201802f. [DOI] [PubMed] [Google Scholar]
- 94.Murphy K, O'Sullivan O, Rea MC, Cotter PD, Ross RP, Hill C. Genome mining for radical SAM protein determinants reveals multiple sactibiotic-like gene clusters. PLoS ONE. 2011;6:e20852. doi: 10.1371/journal.pone.0020852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zheng G, Hehn R, Zuber P. Mutational analysis of the sbo-alb locus of Bacillus subtilis: identification of genes required for subtilosin production and immunity. J. Bacteriol. 2000;182:3266–3273. doi: 10.1128/jb.182.11.3266-3273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zheng G, Yan LZ, Vederas JC, Zuber P. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J. Bacteriol. 1999;181:7346–7355. doi: 10.1128/jb.181.23.7346-7355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fluhe L, Knappe TA, Gattner MJ, Schafer A, Burghaus O, Linne U, Marahiel MA. The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 2012;8:737. doi: 10.1038/nchembio.798. [DOI] [PubMed] [Google Scholar]
- 98.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grove TL, Ahlum JH, Sharma P, Krebs C, Booker SJ. A consensus mechanism for Radical SAM-dependent dehydrogenation? BtrN contains two [4Fe-4S] clusters. Biochemistry. 2010;49:3783–3785. doi: 10.1021/bi9022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lanz ND, Booker SJ. Identification and Function of Auxiliary Iron-Sulfur Clusters in Radical SAM Enzymes. Biochim. Biophys. Acta. 2012 doi: 10.1016/j.bbapap.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 101.Zhang Q, van der Donk WA, Liu W. Radical-mediated enzymatic methylation: a tale of two SAMS. Acc. Chem. Res. 2012;45:555–564. doi: 10.1021/ar200202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. Metagenome Mining Reveals Polytheonamides as Posttranslationally Modified Ribosomal Peptides. Science. 2012 doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- 103.Houck DR, Chen LC, Keller PJ, Beale JM, Floss HG. Biosynthesis of the Modified Peptide Antibiotic Nosiheptide in Streptomyces-Actuosus. J. Am. Chem. Soc. 1988;110:5800–5806. [Google Scholar]
- 104.Zhou P, Ohagan D, Mocek U, Zeng ZP, Yuen LD, Frenzel T, Unkefer CJ, Beale JM, Floss HG. Biosynthesis of the Antibiotic Thiostrepton - Methylation of Tryptophan in the Formation of the Quinaldic Acid Moiety by Transfer of the Methionine Methyl-Group with Net Retention of Configuration. J. Am. Chem. Soc. 1989;111:7274–7276. [Google Scholar]
- 105.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 106.Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. Structural basis for methyl transfer by a radical SAM enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pierre S, Guillot A, Benjdia A, Sandstrom C, Langella P, Berteau O. Thiostrepton tryptophan methyltransferase expands the chemistry of radical SAM enzymes. Nat. Chem. Biol. 2012;8:957–959. doi: 10.1038/nchembio.1091. [DOI] [PubMed] [Google Scholar]
- 108.Layer G, Kervio E, Morlock G, Heinz DW, Jahn D, Retey J, Schubert WD. Structural and functional comparison of HemN to other radical SAM enzymes. Biol. Chem. 2005;386:971–980. doi: 10.1515/BC.2005.113. [DOI] [PubMed] [Google Scholar]
- 109.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes. EMBO J. 2003;22:6214–6224. doi: 10.1093/emboj/cdg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saleh O, Haagen Y, Seeger K, Heide L. Prenyl transfer to aromatic substrates in the biosynthesis of aminocoumarins, meroterpenoids and phenazines: the ABBA prenyltransferase family. Phytochemistry. 2009;70:1728–1738. doi: 10.1016/j.phytochem.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 111.Pernin R, Muyard F, Bevalot F, Tillequin F, Vaquette J. Efficient synthesis of octandrenolone and related dipyranoacetophenones. J. Nat. Prod. 2000;63:245–247. doi: 10.1021/np9902845. [DOI] [PubMed] [Google Scholar]
- 112.Pettus TRR, Inoue M, Chen X, Danishefsky SJ. A fully synthetic route to the neurotrophic illicinones: synthesis of tricycloillicinone and bicycloillicinone aldehyde. J. Am. Chem. Soc. 2000;122:6160–6168. [Google Scholar]
- 113.Quillinan AJ, Scheinmann F. Application of the Claisen rearrangement. J. Chem. Soc. 1971;D:966–967. [Google Scholar]
- 114.Hemphill HE, Gage I, Zahler SA, Korman RZ. Prophage-mediated production of a bacteriocinlike substance by SP beta lysogens of Bacillus subtilis, Can. J. Microbiol. 1980;26:1328–1333. doi: 10.1139/m80-220. [DOI] [PubMed] [Google Scholar]
- 115.Paik SH, Chakicherla A, Hansen JN. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 1998;273:23134–23142. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- 116.Oman TJ, Boettcher JM, Wang H, Okalibe XN, van der Donk WA. Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat. Chem. Biol. 2011;7:78–80. doi: 10.1038/nchembio.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang H, van der Donk WA. Substrate selectivity of the sublancin S-glycosyltransferase. J. Am. Chem. Soc. 2011;133:16394–16397. doi: 10.1021/ja2075168. [DOI] [PMC free article] [PubMed] [Google Scholar]