Abstract
For several well-documented reasons, it has been challenging to develop artificial small molecule inhibitors of protein/protein complexes. Such reagents are of particular interest for transcription factor complexes given links between their misregulation and disease. Here we report parallel approaches to identify regulators of a hypoxia signaling transcription factor complex, involving the ARNT subunit of the HIF (Hypoxia Inducible Factor) activator and the TACC3 (Transforming Acidic Coiled Coil Containing Protein 3) coactivator. In one route, we used in vitro NMR and biochemical screening to identify small molecules that selectively bind within the ARNT PAS (Per-ARNT-Sim) domain that recruits TACC3, identifying KG-548 as an ARNT/TACC3 disruptor. A parallel, cell-based screening approach previously implicated the small molecule KHS101 as an inhibitor of TACC3 signaling. Here, we show that KHS101 works indirectly on HIF complex formation by destabilizing both TACC3 and the HIF component HIF-1α. Overall, our data identify small molecule regulators for this important complex and highlight the utility of pursuing parallel strategies to develop protein/protein inhibitors.
Introduction
While many small molecule enzyme inhibitors and receptor ligands that modulate cellular signaling pathways have been discovered for research and therapeutic use, comparable reagents that target non-enzymatic protein/protein interactions are relatively rare. While such compounds are available for several systems, technical issues – from the suitability of compounds in screening libraries to the difficulty of predicting “druggable” sites1, 2 – complicate the development of specific inhibitors of targeted protein/protein interactions.
Such inhibitors have been particularly sought for transcription factors and their associated regulatory proteins1, 3, given well-validated links between misregulation of these proteins and disease. Here we focus on one such complex as a model: hypoxia inducible factor (HIF), the central regulator of the mammalian hypoxia response.4 HIF is a heterodimer of two bHLH-PAS (basic Helix Loop Helix - Per-ARNT-Sim) subunits, including a HIF-α paralog (HIF-1α,-2α,-3α) and aryl hydrocarbon receptor nuclear translocator (ARNT, also known as HIF-β) (Fig. 1a). While O2-dependent post-translational hydroxylation normally lowers both of HIF-α abundance and activity, these modifications are reduced under hypoxia and allow HIF-α to accumulate in the nucleus.5 Subsequently, HIF complexes form and control the expression of several hundred genes, including potent angiogenic and growth factors.6 As such, abnormally high levels of HIF correlate with several forms of cancer, suggesting that HIF inhibitors could potentially block tumor formation and progression.5 Such inhibition might be achieved by blocking the HIF-α and ARNT interaction, which uses interchain contacts between bHLH and PAS (Per-ARNT-Sim) domains.7–11 While we have successfully found inhibitors that use this approach by exploiting a ligand-binding cavity within one of the HIF-2 PAS domains9, 10, differences among HIF-α sequences suggest that this route is paralog-specific.
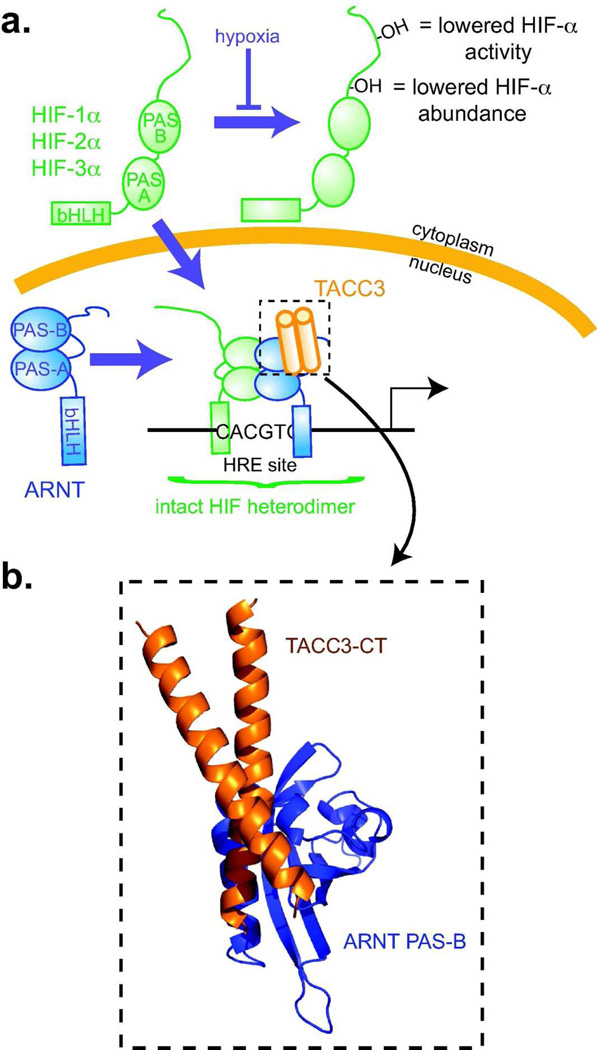
Figure 1. Overview of the ARNT/TACC3 complex.
a. Schematic of HIF complexes, which are bHLH-PAS heterodimers that include an O2-sensitive HIF-α subunit and a constitutive ARNT subunit. Under normoxia, O2-dependent hydroxylation of HIF-α decreases its abundance and activity.5 Hypoxia stops these modifications, allowing HIF-α to accumulate in the nucleus and dimerize with ARNT. This heterodimer binds to hypoxia responsive enhancer (HRE) sites, controlling target gene transcription. In addition to binding HIF-α, ARNT PAS-B directly recruits CCC proteins.12, 14 b. Structural model of an ARNT/CCC complex14 showing how the TACC3 coiled coil interacts with the helical surface of ARNT PAS-B, opposite from where HIF-α PAS-B binds.
To simultaneously inhibit all HIF complexes, we considered targeting interactions between the ARNT subunit, shared among these complexes, with transcriptional coactivators. This strategy is predicated on the ARNT PAS-B domain (Fig. 1) directly recruiting coiled coil coactivators (CCCs) to HIF for proper transcriptional regulation.12–14 By depleting endogenous proteins or overexpressing mutants, we found that HIF complexes differentially utilize several CCC proteins including thyroid hormone receptor interacting protein 230 (TRIP23015), Coiled-Coil Coactivator (CoCoA16) and transforming acidic coiled-coil 3 (TACC317) at different promoters.14 Combining biophysical and mutagenesis data, we generated a structural model of the ARNT/TACC3 complex, showing that CCC proteins use a coiled coil to bind a helical surface on ARNT PAS-B (Fig. 1b).12, 14 Notably, the CCC-binding surface on ARNT is near where other PAS domains bind cofactors that modulate their protein/protein interactions18, leading us to hypothesize that artificial ARNT-binding compounds might similarly control ARNT PAS-B/CCC interactions to regulate HIF activity.
Here we characterize the mechanisms of action of two small molecule inhibitors of ARNT/TACC3 signaling, identified from independent in vitro target-based and cell-based phenotypic screens. The first approach took advantage of our NMR studies of ARNT PAS-B19, letting us use this method to screen over 760 compounds20, 21 for protein binding. One ARNT-binding compound, KG-548, binds in a cavity adjacent to the TACC3 binding site and displaces CCCs from ARNT in vitro. In parallel, we investigated KHS101, a thiazole derivative originally identified in a cell-based phenotypic screen22 and that specifically induces neuronal differentiation.23 Based on KHS101 affecting aspects of the neuronally-expressed ARNT homolog, ARNT2, in neural progenitor cells (NPCs) and crosslinking to TACC3 in NPC lysates, the question arises as to whether KHS101 directly disrupts the ARNT2/TACC3 interaction. We show that KHS101 does not directly bind to the minimal ARNT or TACC3 interacting domains, but instead promotes TACC3 protein turnover within cells, indicating an indirect mode of function. Notably, KHS101 also promotes the turnover of the HIF-1α subunit itself and interferes with HIF-driven transcription in living cells under hypoxia, as anticipated by these effects on the levels of both activator (HIF-1α) and coactivator (TACC3) components. Taken together, KG-548 and KHS101 provide useful molecules for studies of HIF signaling, and more broadly, valuable examples of different ways to control protein complex formation with small molecules.
Results
Identifying direct inhibitors of ARNT/TACC3 interactions by NMR screening
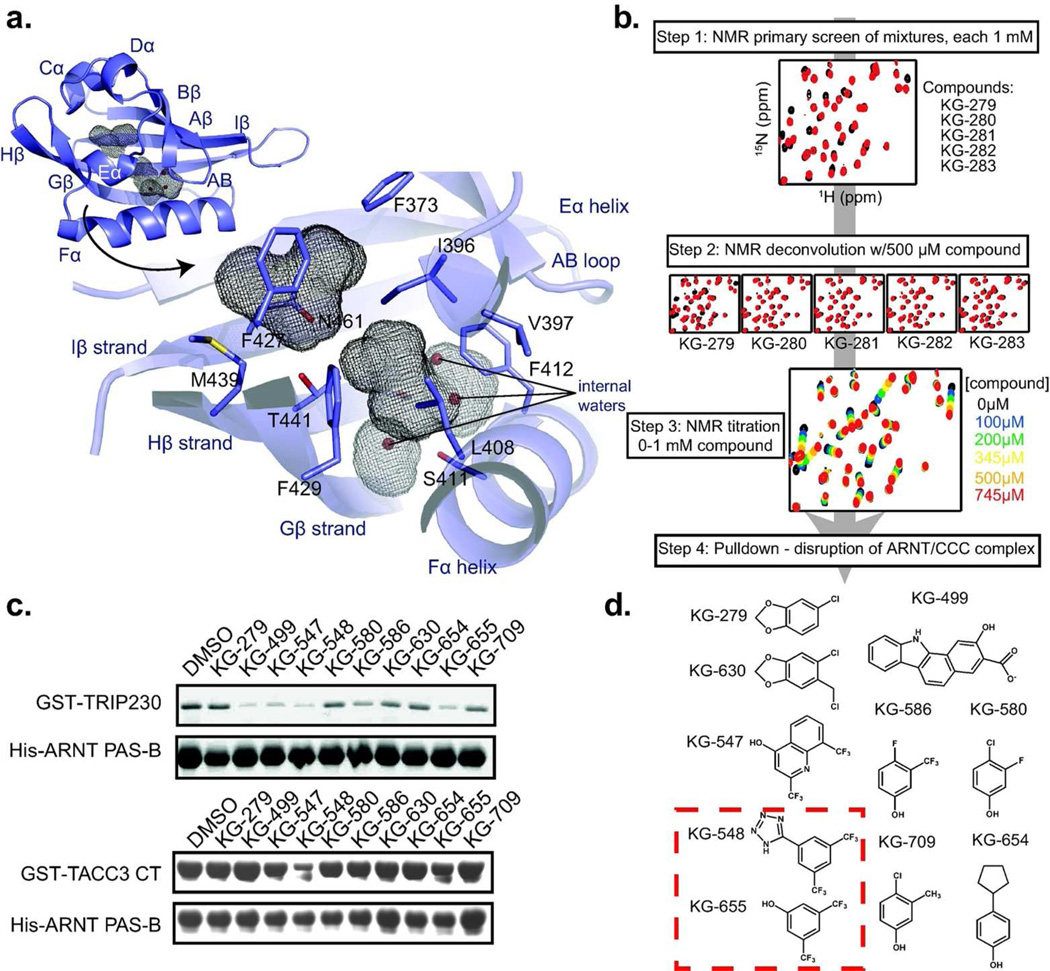
We have solved a new 1.6 Å resolution X-ray diffraction structure of ARNT PAS-B (Table 1; Fig. 2a), revealing two cavities near the regulatory cofactor binding sites in other PAS domains (e.g. flavins in photosensors; heme in oxygen sensors24, 25). The larger 65 Å3 cavity is flanked by the E.α/ Fα helices, Gβ / Hβ / Iβ strands and the AB loop, and includes several polar residues (e.g. S411, T441 and S443) that facilitate the binding of three waters at typical cofactor sites. A hydrophobic side of the cavity, involving V397, L408, F412 and F427, is shared with the CCC-binding surface in our ARNT/TACC3 model.14 The second cavity is slightly smaller (40 A3) and comparable to the chromophore binding site in photoactive yellow protein.26 While these cavities are relatively small, their locations (and potential for merge into a single larger cavity) raised the potential for them to bind artificial compounds and modulate CCC binding, similarly to other PAS domains.
Table 1.
X-ray Crystallography Data Processing and Refinement Statistics
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 93.3, 61.7, 55.5 |
| β (°) | 124.6 |
| Resolution (Å) | 20.7 to 1.6 |
| (1.63 to 1.60) | |
| Rsym or Rmerge | 4.7 (43.9) |
| I / σI | 38.8 (2.2) |
| Observed reflections | 155,415 |
| Unique reflections | 34402 |
| Completeness % (highest shell) | 99.9 (100) |
| Refinement | |
| Resolution (Å) | 22.5 to 1.6 |
| (1.63 to 1.60) | |
| Rwork / Rfree | 20.15 / 22.65 |
| No. atoms | |
| Protein | 1832 |
| Water | 206 |
| Avg. B-factor | |
| Protein | 20.3 |
| Water | 35.4 |
| R.M.S. deviations | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.45 |
Figure 2. Screening ARNT PAS-B for small molecule effectors of the ARNT/TACC3 interaction.
a. Diagram of the ARNT PAS-B crystal structure, shown inset with secondary structure designations and internal cavities (grey mesh). The larger cavity is adjacent to the TACC3 binding site and contains three waters; the.smaller cavity is primarily hydrophobic. b. Schematic of the NMR-based screen for compounds that bind ARNT PAS-B. Initial 15N/1H HSQC spectra were acquired with 250 µM 15N-labeled ARNT PAS-B with a mixture of five compounds (1 mM each); mixtures producing large chemical shift perturbations (compared to DMSO) were deconvoluted as shown. c. Lead compounds (500 µM each) from NMR-based screen were tested for their ability to disrupt complexes of ARNT PAS-B with CCC fragments of TRIP230 (1583–1716) and TACC3 (561–631 = TACC3-CT). d. Summary of ARNT PAS-B binding and ARNT/CCC disruption of tested compounds. All ten compounds generated chemical shift perturbations when titrated into ARNT PAS-B; KG-548 and KG-655 (red box) also disrupted TRIP230 and TACC3 binding to ARNT PAS-B in vitro.
To identify such compounds, we used solution NMR to search over 760 small molecules9, 20, 21 for ARNT PAS-B binding. This library consists of low molecular weight fragments (average MW: 203 ± 73 Da, Table S1) containing “privileged” moieties enriched in protein-binding compounds.27 Further, this collection has previously provided us several compounds (or analogs) which bind to other PAS domains9, 10, 20 and disruptors of protein/protein interactions in other transcriptional regulators.9, 21
We quickly evaluated these compounds for ARNT PAS-B binding using protein-detected NMR (Fig. 2b). In the primary screen, 15N/1H HSQC spectra were acquired on 15N-labeled ARNT PAS-B samples mixed with five candidate compounds (250 µM protein, 1 mM each compound) or DMSO. Compound mixtures that produced changes in peak locations or intensities indicated that one or more compounds bound the protein target; sixteen such mixtures were subsequently deconvoluted by acquiring spectra on ARNT PAS-B with individual compounds. Eighteen hits from these steps were tested in titrations at concentrations up to 1 mM to establish binding potency and location; ten of these (Fig. 2c) were soluble throughout this concentration range and further studied.
To examine whether these ARNT-binding compounds affected the stability of ARNT PAS-B/CCC complexes, we initially checked their disruption of ARNT-mediated pulldowns of fragments of the TRIP230 and TACC3 coactivators (Fig. 2c). The fragments of both CCC proteins contained the coiled coil that binds ARNT PAS-B12, 14, facilitating robust interaction without compounds present. However, several chemicals markedly interfered with ARNT binding to one or both coactivators. Among these, KG-548 exhibited the greatest reduction of ARNT/CCC complex formation for both coactivators, with a smaller effect seen for the structurally related fragment KG-655 (Fig. 2d).
KG-548 binds to the cavity of ARNT PAS-B
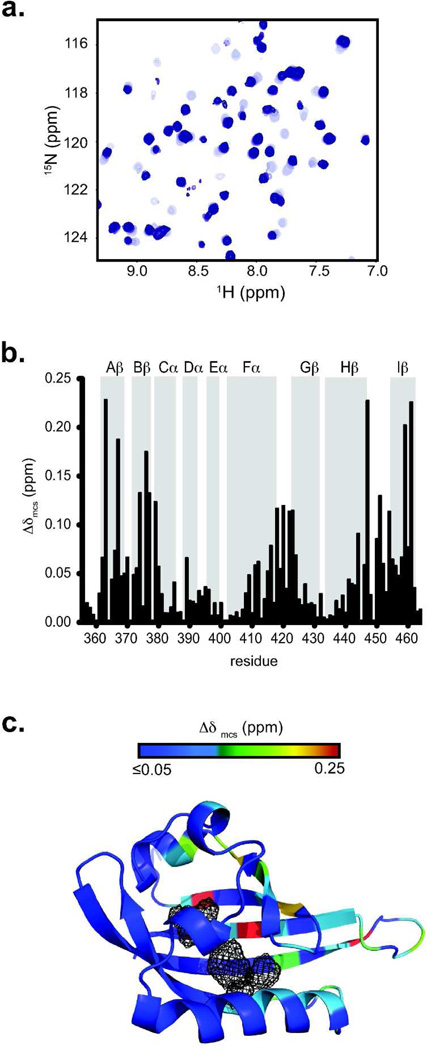
Next, we further characterized compound-mediated ARNT/CCC disruption with solution NMR to identify ligand-binding sites within ARNT PAS-B. Using KG-548 as the most effective disruptor of this complex, we observed slow exchange behavior in compound titrations monitored by ARNT PAS-B 15N/1H HSQC spectra (Fig. 3a), suggesting micromolar (or tighter) dissociation constants. We used our prior chemical shift assignments19 to map ligand-induced changes onto the ARNT PAS-B structure using minimum chemical shift difference analyses, assuming correlations between the nearest pairs of peaks in apo- and KG-548 saturated spectra (Fig. 3b). Residues that were most strongly affected by KG-548 addition were located close to the internal ARNT PAS-B cavities (Fig. 3b,c) and analogous to where other PAS domains often bind ligands28. Notably, these ligand-mediated structural and functional effects were sensitive to minor changes in compound structure. Two KG-548 variants with small changes – CF3 to Cl substitutions on the phenyl ring, and addition of methyl or ethyl moieties on the tetrazole – bound ARNT more weakly and were unable to disrupt ARNT/TACC3 complexes (Fig. S1a–e). Coupled with the limited number of leads from the library screen, these data demonstrate specificity of the ARNT/ligand interaction.
Figure 3. KG-548 appears to bind within the ARNT PAS-B cavities.
a. 15N/1H HSQC spectra of a KG-548 titration (0–1 mM from light to dark crosspeaks) into 320 µM 15N ARNT PAS-B. Slow exchange behavior was observed, indicated by the disappearance of apo-crosspeaks and concomitant appearance of new peaks. b. Minimum chemical shift analysis45 of KG-548 titration into ARNT PAS-B, mapped onto the sequence and secondary structure. c. Chemical shift mapping suggests that KG-548 binds the ARNT PAS-B cavities, as shown by a heat map of KG-548-induced chemical shift changes on the ARNT PAS-B crystal structure (Δδmcs colored from low (blue) to high (red)) with the largest changes near the internal cavities (mesh).
KG-548 binding shows selectivity among PAS domain targets
Having demonstrated specificity from the ligand perspective, we next explored the potential for other bHLH-PAS PAS-B domains to bind KG-548. We started by examining ARNT2, a closely-related ARNT homolog that is chiefly expressed in neuronal and kidney tissue29, 30 (Fig. S2). Using solution NMR methods, we verified that the ARNT and ARNT2 PAS-B domains adopt comparable structures, as expected from the 80% sequence identity between them. Similarities in chemical shifts and TALOS+-derived secondary structures31 (Fig. S3a,b) strongly indicate ARNT2 adopts the same overall fold as seen in our crystal (Fig. 2 and ref. 9) and solution19 structures of ARNT PAS-B, giving us confidence in a ARNT2 PAS-B homology model to suggest the placement of residues involved in ARNT/CCC interactions14 (Fig. S3c).
To functionally test these structural similarities, we examined the ability of ARNT2 PAS-B to bind coactivator fragments and KG-548. Pulldown experiments demonstrated that ARNT2 PAS-B directly interacts with GST-tagged TACC3-CT (=C-terminal 70 aa of TACC3, including residues 561–631) in vitro (Fig. S3d), as seen for ARNT (Fig. 2c). Mutations to ARNT2 PAS-B residues E372 and K391 weakened this interaction (Fig. S3d), chosen for their similarity to ARNT E398 and K417 (Fig. S3c).14 15N/1H HSQC spectra of both ARNT2 E372A and K391A PAS-B mutants were similar to wildtype protein (Fig. S3e), suggesting that both point mutations had minimal structural effects. Next, we asked if KG-548 could also bind to ARNT2 PAS-B and compete away TACC3. Ligand titrations monitored with 15N/1H HSQC spectra showed the same slow exchange (Fig. S4a) observed with ARNT PAS-B, with the largest effects (Fig. S4b,c) clustered in the same internal sites (Fig. S2). Finally, as expected from the similar CCC and ligand binding modes, KG-548 also disrupts ARNT2 PAS-B/TACC3 interactions in pulldown assays (Fig. S4d), underscoring the high degree of similarity between ARNT and ARNT2.
In contrast, similar titrations of KG-548 into two more widely diverged PAS-B domains from the BMAL-1 and HIF-2α bHLH-PAS proteins (38% and 34% identity to ARNT PAS-B) showed specificity in the protein/ligand interaction. We observed very few chemical shift changes caused by KG-548 addition to BMAL-1, and virtually none with HIF-2α (Fig. S5) despite large cavities within both PAS domains.9, 10, 32 We interpret these data to indicate that these two PAS-B domains have (much) lower affinities for KG-548 than ARNT PAS-B, showing little to no interaction at the tested concentrations and thus establishing that observed ligand-binding specificity is not solely based on simple accessibility. More broadly, we have not observed KG-548 binding to other PAS and non-PAS targets20, 21, consistent with the specificity that can be observed in small fragments from other libraries.33
KG-548 breaks up the ARNT/TACC3 complex in vitro and in cell lysate
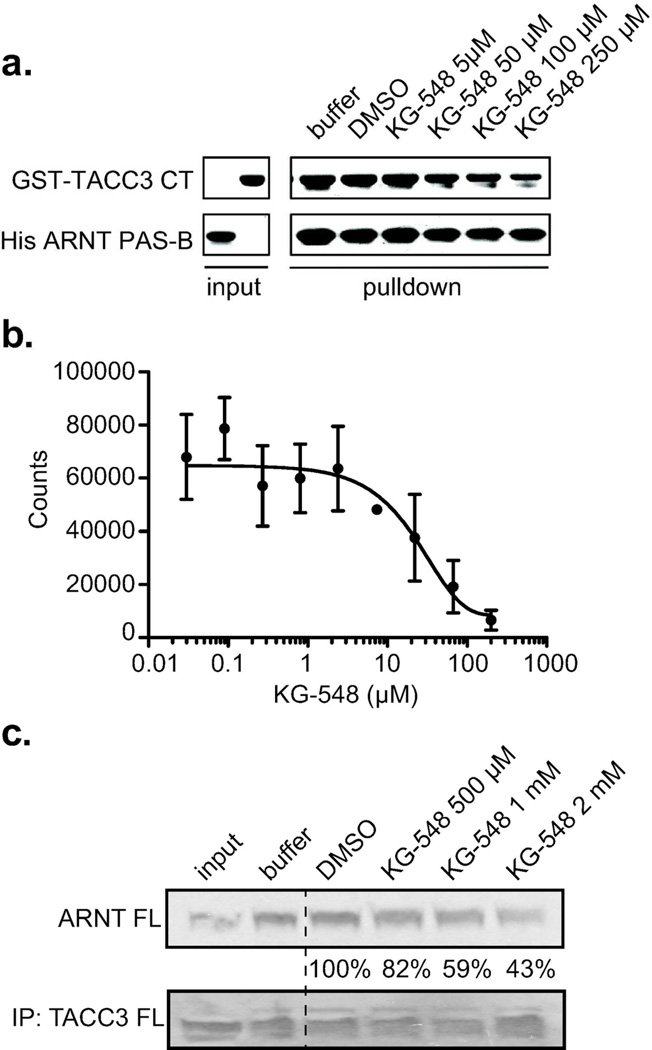
To further characterize KG-548 induced disruption of ARNT/TACC3, we examined the dose dependence of this effect in two ways. Pulldown assays using His-ARNT PAS-B and GST-TACC3-CT, tagged versions of the minimal interacting components of both proteins14, showed that levels of GST-TACC3-CT pulled down by His-ARNT PAS-B decreased in a KG-548 dose-dependent manner (Fig. 4a) at concentrations that perturbed ARNT PAS-B NMR spectra. We quantified the potency of KG-548 using AlphaScreen, a luminescence proximity assay of complex formation between the two tagged fragments (Fig. S6a). The assay was validated by showing that untagged ARNT PAS-B or TACC3 competed against complex formation between tagged proteins with IC50~2–3 µM (Fig. S6b). Similar AlphaScreen assays with KG-548 showed a 25 µM IC50 inhibition (Fig. 4b), consistent with the apparent ARNT PAS-B/KG-548 affinity seen in NMR-based titrations.
Figure 4. KG-548 disrupts in vitro ARNT/TACC3 interactions.
a. Titration of KG-548 into an in vitro pulldown assay of minimal ARNT PAS-B and TACC3-CT interacting fragments shows a dose-dependent reduction in ARNT/TACC3 complex formation. b. Quantification of KG-548 potency for disrupting the ARNT PAS-B/TACC3-CT interaction as provided by AlphaScreen, showing an apparent IC50 of 25 µM. c. Co-immunoprecipitation assays of full length ARNT and TACC3 proteins in HEK293T cell lysates show that KG-548 weakens the ARNT/TACC3 interaction as demonstrated by the dose-dependent decrease in the intensity of the ARNT protein band (quantitated with % remaining compared to DMSO control) associated with immunoprecipitated TACC3 protein.
To examine how well these results from isolated domains translate to full length proteins, we surveyed the ability of KG-548 to disrupt the complex between full length human ARNT and mouse TACC3 proteins. While cell-based qPCR and HRE reporter assays of this effect were hampered by issues with cell toxicity at mid-micromolar KG-548 concentrations, an alternative was provided by immunoprecipitation experiments in lysates of HEK 293T cells transfected with expression vectors for both proteins. Within this system, the ARNT/TACC3 interaction can clearly be observed by co-IP without ligand present; addition of increasing concentrations of KG-548 leads to a progressive decrease in the amount of TACC3-associated ARNT protein (Fig. 4c). While the apparent potency of KG-548 is lower in this more complex setting, our data clearly demonstrate that this ARNT-binding compound can inhibit ARNT/TACC3 complex formation in truncated or full length proteins. Notably, two negative control compounds KG2-006 and KG2-007 did not show such inhibition at comparable concentrations, consistent with the isolated domain results (Fig. S1f).
Discovery of KHS101 as a TACC3 inhibitor
Complementing our targeted in vitro screening, we examined a presumed disruptor of ARNT/CCC interactions provided by the HTS-derived compound KHS101 (Fig. S7a) that accelerates NPC differentiation in the adult rat.23 Crosslinking data initially associated TACC3 with KHS101 by showing that a benzophenone derivative bound TACC3 at an uncharacterized location. This linkage between KHS101 and TACC3 was underscored by the observation of similar cellular effects with either KHS101 treatment or anti-TACC3 siRNAs in several settings 23, 34, suggesting that this compound is a general TACC3 inhibitor. However, this study did not characterize the mechanism of KHS101 action on TACC3, leading us to ask if KHS101 1). directly interferes with TACC3 binding to its partners ARNT or ARNT2 PAS-B in vitro; 2). alters TACC3 protein levels in cells; or 3). affects transcription from ARNT/CCC-reliant promoters, such as HIF-driven genes.14
KHS101 is not a direct regulator of the ARNT/TACC3 complex
To address how KHS101 regulates ARNT/TACC3, we performed in vitro pulldown experiments in the presence of KHS101 using the minimal constructs of both proteins. We did not observe any substantial KHS101-dependent effect on TACC3 pulldowns with either ARNT or ARNT2 (Fig. S7b,c), suggesting that KHS101, in contrast to KG-548, works indirectly to disrupt ARNT/TACC3 function. To test this hypothesis, we used solution NMR spectroscopy to examine KHS101 binding to ARNT PAS-B and TACC3. 15N/1H HSQC spectra of ARNT PAS-B titrated with KHS101 showed only minor changes (Fig. S7d; compare to Fig. 3a for KG-548), indicating no binding. Analogous spectra using a 15N-labeled minimal version of TACC3 with both the ARNT-interacting C-terminal 21 residues of TACC3 and a stabilizing GCN4 coiled coil (GCN4-TACC3-min) also showed no differences between DMSO and KHS101-treated samples (Fig. S7e) at concentrations that trigger biological responses (vide infra and ref. 23). These negative data strongly suggested that KHS101 perturbs the ARNT/TACC3 complex in a different, likely indirect, mechanism than KG-548.
KHS101 reduces intracellular TACC3 stability
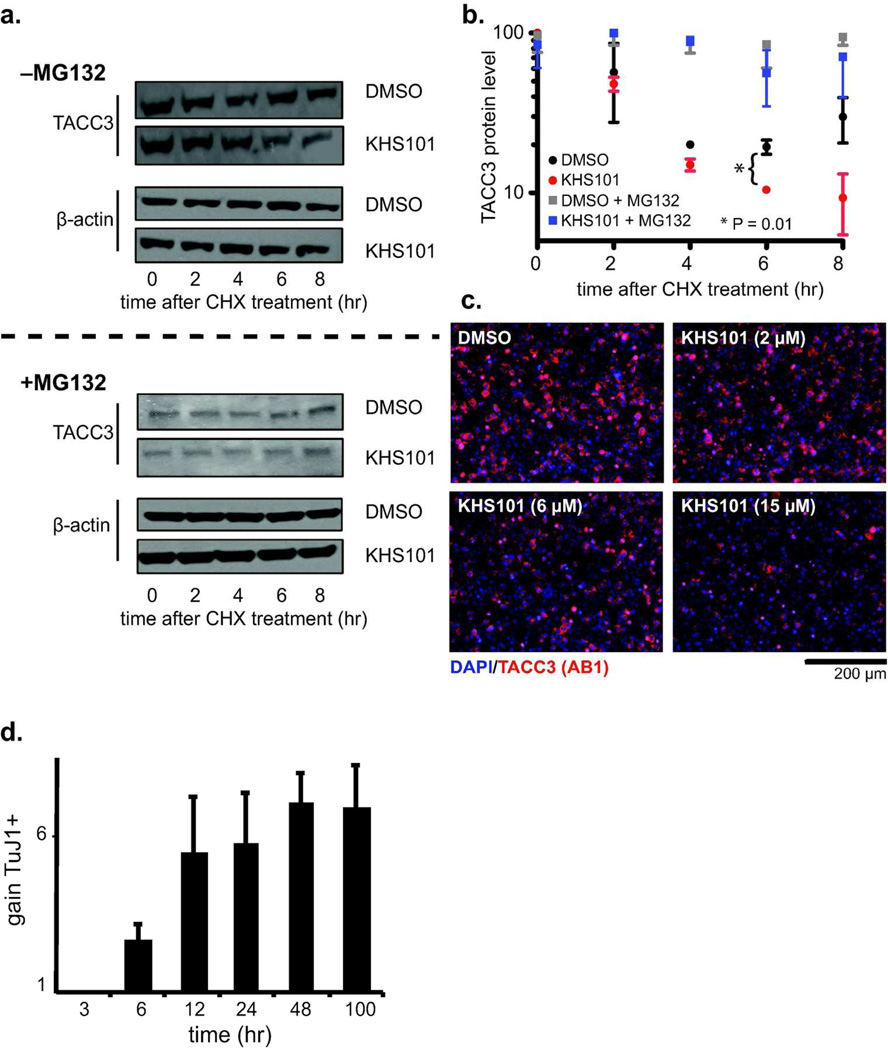
Coupling prior data implicating KHS101 interference with ARNT/TACC323 with our observation that KHS101 does not directly block the minimal ARNT PAS-B/TACC3-CT interaction, we hypothesized that KHS101 might function indirectly by modulating TACC3 stability. To test this, we examined TACC3 protein stability in HEK293T cells using pulse chase experiments conducted with the translation inhibitor cycloheximide (CHX) and either 5 µM KHS101 or DMSO control. Cells were harvested at different times post-CHX treatment and TACC3 protein levels were monitored by immunoblot (Fig. 5a). KHS101 treatment decreased the stability of TACC3 compared to DMSO controls (Fig. 5a,b), with substantial differences after 6 hr of treatment. We observe a statistically-significant difference at 6 hr and a net three-fold drop in TACC3 protein levels at 8 hr (relative levels of 9.3 [KHS101] and 29.9 [DMSO], Fig. 5b), whereas ARNT levels were barely affected (Fig. S8a,b). In contrast, cells treated with KG-548 or an inactive KHS101 analog, KHS9123, showed no change in TACC3 level compared with DMSO, indicating specificity in the ligand-induced degradation (Fig. S8c–g). Parallel experiments including the proteasome inhibitor MG132 muted this loss of TACC3 protein, without any marked difference between KHS101 and control groups (Fig. 5a,b). Finally, we verified that KHS101 affects steady state TACC3 levels in the absence of CHX, showing a dose-dependent and saturable drop in TACC3 protein levels with increasing concentrations of KHS101 (Fig. 5c, S9a–c). While this drop was smaller than we observed with CHX treatment, the consistent observation of a 10% drop in TACC3 levels in independent experiments with different sample types (using either intact cells or cell lysates) and different primary antibodies (Fig. S9) gives us confidence in this trend. Taken together, our results suggest that KHS101 treatment leads to increased proteasome-mediated degradation of TACC3 protein in cells, conceivably mediated by interactions outside of the ARNT-binding motif.
Figure 5. KHS101 decreases TACC3 levels in cells and regulates HIF gene expression.
a. KHS101 facilitates TACC3 degradation in a proteasome-dependent manner. HEK293T cells were treated with 5 µM KHS101, 100 µg/ml cycloheximide (CHX) (upper panel); additional cells were similarly treated with KHS101 and CHX plus 20 µM MG132 (lower panel). Cells were harvested 0µ8 hr post-treatment and prepared for TACC3 immunoblot analyses. b. Quantification of TACC3 protein levels from data shown in panel a. Without MG132, TACC3 protein levels decreased, with greater drops observed in KHS101-treated cells (compared to DMSO) after 6 hr incubation. A statistically-significant difference was observed 6 hr post-treatment (p < 0.01 by Student’s t-test); the 8 hr timepoint also shows a substantial decrease in TACC3 levels, but this is not statistically significant due to large variations in data values. In the presence of MG132, we observed little decrease in TACC3 levels with no KHS101-dependent effects, implicating a proteasomal-dependent degradation pathway. c. Steady state treatment with KHS101 reduces TACC3 levels. HEK293T cells were treated with KHS101 (0 – 15 µM) and immunostained with TACC3 AB1 after 14 hr. TACC3 intensity was negatively affected by KHS101. d. KHS101 induces cell differentiation with a minimum of 6 hr exposure. Adult rat NPCs were exposed to 5 µM KHS101 (or DMSO) for the indicated times, after which media were replaced with compound-free versions and incubated for a total of 100 hr. Neuronal differentiation was assessed by expression of the TuJ1 marker, showing upregulation after times consistent with TACC3 levels falling in CHX-treated cells (panels a, b).
KHS101 treatment affects two independent TACC3-containing pathways in cells
To evaluate the functional implications of the KHS101-triggered drop in TACC3 levels, we looked for correlations between KHS101 effects on TACC3 levels and the activities of two TACC3-dependent pathways. We initially examined the kinetics of compound-induced NPC differentiation, which involves both TACC3 and ARNT2.23 Here we used KHS101 washout experiments, exposing rat NPCs to KHS101 for various times before switching to compound-free media for the remainder of the 100 hr incubation. Afterwards, differentiation was assessed using immunofluorescence-based detection of the pan-neuronal TuJ1 marker. We saw a KHS101-dependent increase in TuJ1 expression only after 6 hr of treatment, maximizing after 12–24 hr of exposure (Fig. 5d). A reasonable hypothesis for this delay in KHS101 efficacy is that its proneurogenic effects are directly related to the time required to alter TACC3 protein levels (4–8 hr by pulse chase; 16 hr under steady state conditions).
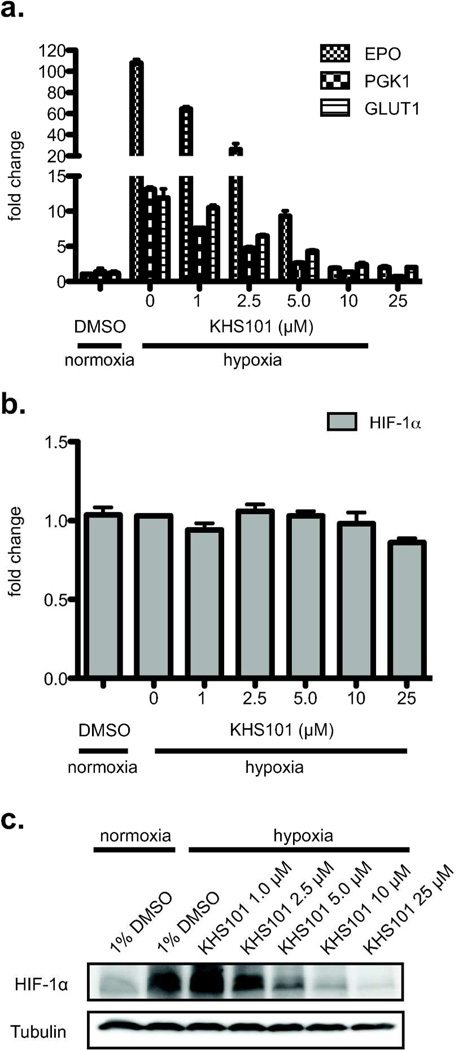
To independently assess KHS101 effects in another TACC3-dependent activity, we examined the dose dependence of KHS101 on the transcription of endogenous HIF target genes using qPCR.14 Since KHS101 decreases steady state TACC3 levels, we suspected that compound treatment would lower TACC3 participation in hypoxia-induced HIF complexes, analogously to TACC3 knockdown and ARNT point mutations which weaken TACC3 binding.14 To test this possibility, we measured mRNA levels for three HIF-responsive genes in Hep3B cells, which utilize ARNT in both HIF-1 and HIF-2 signaling. Though some genes are regulated by both HIF paralogs, others are controlled only by either HIF-1 (e.g. PGK-1) or HIF-2 (e.g. Epo).35, 36 As expected, levels of these HIF-regulated transcripts increase (from 5- to 130-fold) upon exposure to hypoxia (Fig. 6a). Concomitant treatment with KHS101 and hypoxia led to dose-dependent reductions in the levels of all three transcripts, with apparent IC50 values below 5 µM (Fig. 6a), correlating the dose dependencies of KHS101 on TACC3 levels (Fig. 5c). To further evaluate the mechanism of this effect, we quantitated HIF-1α mRNA and protein levels by qPCR and Western blot. While HIF-1α mRNA levels were unaffected by KHS101 treatment (Fig. 6b), HIF-1α protein levels were drastically decreased in a KHS101 dose-dependent manner (Fig. 6c). We suspect HIF-2α is similarly destabilized in KHS101-treated hypoxic cells, but quantitation of this effect is complicated by technical issues with available anti-HIF-2α antibodies; compound treatment has no effect on HIF-2α mRNA levels (data not shown). While further studies are needed to fully characterize the breadth of KHS101-induced protein destabilization with respect to several parameters (protein target, cell type, growth conditions), our functional data provide a mode of action for KHS101 and demonstrate its efficacy in two distinct pathways depending on cellular context.
Figure 6. KHS101 inhibits HIF target gene expression and decreases HIF-1α protein levels.
a. KHS101 treatment potently reduces HIF target gene expression. Hep3B cells were treated with KHS101 (0–25 µM) and incubated under hypoxia (1% O2) for 16 hr. The expression of three HIF-driven genes (EPO, PGK1, GLUT1) were assessed by qPCR, demonstrating KHS101 inhibiting transcription with IC50 < 5 µM. b. HIF-1α mRNA level was measured by qPCR, showing no significant change with increasing KHS101 concentration. c. HIF-1α protein levels are reduced in a KHS101 dose-dependent manner, using anti HIF-1α Western blot.
Discussion
Protein/protein interactions are often difficult to modulate with chemical reagents. Here we describe two compounds that affect the ARNT/TACC3 complex, an important component of HIF. These chemicals work by two different mechanisms: KG-548 directly interferes with ARNT/TACC3 complex formation by competing with TACC3 for binding to the ARNT PAS-B domain while KHS101 modulates the abundance of both TACC3 and the HIF component, HIF-1α. The different origins of these compounds underscores the merits of parallel in vitro target-based and cell-based phenotypic screens, each having strengths and weaknesses in drug discovery In vitro target-based methods are appealing in their use of mechanism-driven hypotheses and focused searches for inhibitors of disease-associated targets, such as HIF or TACC3.37, 38 However, this route often cannot address issues of potency, specificity and metabolism that are essential for cellular applications, and which can be problematic to subsequently incorporate into lead compounds. In contrast, cell-based phenotypic methods have tremendous power to identify new inhibitors of biological activities, but require downstream mechanistic studies to clarify modes of action. In our case, the linkage between ARNT2/TACC3 interactions and neuronal differentiation might have remained cryptic without the phenotypic screen and initial characterization of KHS101.22, 23 While KHS101 demonstrates the potential of TACC3 destabilization to alter HIF transcriptional activation, we suggest that a target-based approach embodied by KG-548 can provide compounds that work more specifically than by simply destabilizing TACC3 and HIF-1α, which may have potential secondary effects. Retrospective analyses of drug discovery successes underscore the utility of combining phenotypic and targeted approaches: the former still generate the majority of first-in-class new molecular entities but are often followed by target-based screens which identify many more candidates using the foundation established by the phenotypic efforts.39
From the standpoint of small molecule HIF inhibitors, blocking ARNT/TACC3 interactions may hold several advantages over previously described strategies.9, 40–42 Most notably, this strategy targets a mechanistically-defined interaction12, 14 common among all three HIF paralogs, allowing a single compound to simultaneously block transcription from multiple HIFs given their shared use of ARNT. While our approach shares the general concept of disrupting activator/coactivator interactions with the HIF inhibitor chetomin41, it is worth emphasizing that chetomin targets a completely different complex (HIF-α C-terminal transcriptional activation domain with the CH1 domain of the p300/CBP coactivator) that lacks the small-molecule binding pockets that confer specificity to ARNT PAS-B targeting compounds.
To close, integrating two screening strategies has provided us with small molecules that are useful tools for continuing studies of the critical HIF signaling pathway. This should also refine our understanding of the roles of CCC proteins in HIF-driven gene activation and could potentially lead to development of new therapeutic routes for HIF-dependent cancers. Finally, we hope that this parallel direct/indirect inhibitor approach provides another example in the relatively limited number of small molecule inhibitors of protein/protein interactions.
Methods
Plasmids
For bacterial expression, human ARNT (356–470) and ARNT2 PAS-B domain (330–444) constructs were cloned into pHis-Parallel43, while murine TACC3 constructs (TACC3-CT=561–631; GCN4-TACC3-min=GCN4 coiled coil + residues 610–631) were cloned into pGST- and pHis-Parallel, respectively.43 Full-length human ARNT and murine TACC3 were cloned into pcDNA4B vector (Invitrogen) with a C-terminal FLAG (ARNT) or myc-His6 (TACC3) tags for mammalian cell transfection.
Compound sources
KHS101 was kindly provided by Dr. Peter Schultz (The Scripps Research Institute), synthesized as described.23 KG-548 was purchased from Fluorochem and resupplied from Matrix Scientific; the composition of the entire fragment-based library is described in Supporting Table S1.
Immunoblot, pulldown and immunofluorescence assays
For immunoblots, proteins resolved from SDS-PAGE gels were transferred to PVDF membranes (GE Healthcare) and immunoblotted with these antibodies: anti-ARNT (A-3), anti-TACC3 (T-17), anti-goat IgG-HRP (Santa Cruz Biotechnology); anti-TACC3 (ab56595) (abcam); anti-Myc (9E10), anti-HIF1α (BD Transduction Laboratories); anti-mouse IgG-HRP antibodies (Sigma). For pulldown experiments, 5 µM purified His-ARNT/ARNT2 PAS-B and 8–15 µM TACC3-CT were incubated with 15 µl Ni-NTA beads overnight at 4°C. Eluted protein was resolved in SDS-PAGE and stained with Coomassie Blue stain. Immunofluorescence studies of steady state TACC3 levels used HEK293 cells stained with an anti-TACC3 rabbit polyclonal antibody (Sigma-Aldrich HPA005781; here called TACC3 AB1); more details are provided in Supporting Information.
Protein expression and purification
Proteins were expressed in BL21(DE3) cells and purified using standard FPLC-based methods (detailed in Supporting Information).
Structural studies
Structural studies of ARNT PAS-B were conducted using standard crystallization and X-ray diffraction methods, detailed in Supporting Information. Coordinates of the resulting structure have been deposited in the RCSB (accession code 4EQ1), with refinement information in Table 1. Solution NMR studies of ARNT2 PAS-B utilized U-15N,13C labeled proteins and triple resonance assignment methods, also detailed in Supporting Information.
AlphaScreen assay
400 nM His-ARNT PAS-B and 400 nM GST-TACC3-CT E629A were mixed in a 384-well plate with a total volume of 15 µl and incubated at 4°C for 1 hr. AlphaScreen glutathione donor beads and AlphaLISA Ni chelate acceptor beads (5 µg/ml each; Perkin Elmer) were added under dim green light in 50 mM Tris (pH 7.5), 100 mM NaCl, 0.02% Tween 20 and 1 mM DTT. After incubating in a dark and humidified chamber for 3.5 hr, the plate was read using an Envision plate reader (Perkin Elmer).
KHS101 washout assay
Adult rat hippocampal NPCs were differentiated upon treatment with 5 µM KHS101 or a DMSO negative control as previously described23. After incubation for times shown in Fig. 5d, the culture medium was replaced with fresh (compound-free) media. After a total 100 hr incubation period, cells were fixed and stained for the neuronal marker TuJ123. Neuronal differentiation of KHS101-treated rat hippocampal NPCs was analyzed by microscopy, the percentage of TuJ1-positive cells was determined, and data were normalized to the DMSO control values.
TACC3 turnover assay
HEK293T cells were treated with 5 µM KHS101 and 100 µg/ml cycloheximide (CHX) with or without 20 µM MG132. DMSO treatment served as a negative control. Cells were harvested at several time points from 0 to 8 hr post-treatment and prepared for TACC3, ARNT and β-actin immunoblot analyses.
qPCR
Cells were collected with Trizol (Invitrogen) and total RNA was extracted using the RNeasy Mini Kit (Qiagen). Following DNase treatment, cDNA was synthesized from 1 µg of RNA using SuperScript II Reverse Transcriptase (Invitrogen) in final volume of 50 µl. Real-time PCR was performed (with target gene primers listed in Supporting Information) on 1.25 µl of cDNA in triplicate using Power SYBR Green PCR Master Mix and the 7900HT Fast Real-Time PCR System (Applied Biosystems). Target gene expression levels were normalized to the expression level of cyclophilin B in the same sample. Data were analyzed using the comparative CT method (2−ΔΔCT)44 and normalized to normoxia/1% DMSO conditions.
Supplementary Material
Acknowledgements
This work was supported by grants from the NIH (R01 GM081875 to K.H.G.; P01 CA095471 and R21 NS067624 to K.H.G. and R.K.B.; F32 CA130441 to C.L.P.) and CPRIT (RP100846 to K.H.G. and R.K.B.) and the Burroughs Wellcome Fund (R.K.B.). KHS101 was kindly provided as a gift from P. Schultz (The Scripps Research Institute, La Jolla, USA). Portions of this work were conducted in facilities constructed with support from the Research Facilities Improvement Program (Grant # C06 RR 15437-01) from the NIH/National Center for Research Resources, or at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, supported by U.S. Department of Energy, Office of Biological and Environmental Research Contract DE-AC02-06CH11357.
Footnotes
Supporting Information
Detailed experimental procedures, supplementary figures and supplementary table. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
Y.G., C.L.P., J.K., P.B.C., V.P., R.K.B., H.W. and K.H.G. designed research; Y.G., C.L.P., J.K., P.B.C., V.P., A.P. and H.W. performed research; Y.G., C.L.P., J.K., P.B.C., V.P., R.K.B., H.W. and K.H.G. analyzed data; Y.G., C.L.P., J.K., P.B.C., R.K.B., H.W. and K.H.G. wrote the paper.
The authors declare no competing financial interests.
References
- 1.Koehler AN. A complex task? Direct modulation of transcription factors with small molecules. Current opinion in chemical biology. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 3.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nature reviews. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 4.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Molecular cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev. 2007;17:71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix--loop--helix-PAS transcription factor hypoxia-inducible factor. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Zhang L, Erbel PJA, Gardner KH, Ding KM, Garcia JA, Bruick RK. Functions of the Per/ARNT/Sim (PAS) domains of the hypoxia inducible factor (HIF) Journal of Biological Chemistry. 2005;280:36047–36054. doi: 10.1074/jbc.M501755200. [DOI] [PubMed] [Google Scholar]
- 9.Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B. Journal of the American Chemical Society. 2009;131:17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao N, Whitelaw ML, Shearwin KE, Dodd IB, Chapman-Smith A. Identification of residues in the N-terminal PAS domains important for dimerization of Arnt and AhR. Nucleic Acids Research. 2011;39:3695–3709. doi: 10.1093/nar/gkq1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partch CL, Card PB, Amezcua CA, Gardner KH. Molecular basis of coiled coil coactivator recruitment by the aryl hydrocarbon receptor nuclear translocator (ARNT) The Journal of biological chemistry. 2009;284:15184–15192. doi: 10.1074/jbc.M808479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Partch CL, Gardner KH. Coactivator recruitment: a new role for PAS domains in transcriptional regulation by the bHLH-PAS family. Journal of cellular physiology. 2010;223:553–557. doi: 10.1002/jcp.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Partch CL, Gardner KH. Coactivators necessary for transcriptional output of the hypoxia inducible factor, HIF, are directly recruited by ARNT PAS-B. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7739–7744. doi: 10.1073/pnas.1101357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beischlag TV, Taylor RT, Rose DW, Yoon D, Chen Y, Lee WH, Rosenfeld MG, Hankinson O. Recruitment of thyroid hormone receptor/retinoblastoma-interacting protein 230 by the aryl hydrocarbon receptor nuclear translocator is required for the transcriptional response to both dioxin and hypoxia. The Journal of biological chemistry. 2004;279:54620–54628. doi: 10.1074/jbc.M410456200. [DOI] [PubMed] [Google Scholar]
- 16.Kim JH, Stallcup MR. Role of the coiled-coil coactivator (CoCoA) in aryl hydrocarbon receptor-mediated transcription. The Journal of biological chemistry. 2004;279:49842–49848. doi: 10.1074/jbc.M408535200. [DOI] [PubMed] [Google Scholar]
- 17.Sadek CM, Jalaguier S, Feeney EP, Aitola M, Damdimopoulos AE, Pelto-Huikko M, Gustafsson JA. Isolation and characterization of AINT: a novel ARNT interacting protein expressed during murine embryonic development. Mech Dev. 2000;97:13–26. doi: 10.1016/s0925-4773(00)00415-9. [DOI] [PubMed] [Google Scholar]
- 18.Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annual review of microbiology. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Card PB, Erbel PJA, Gardner KH. Structural basis of ARNT PAS-B dimerization: Use of a common beta-sheet interface for hetero- and homodimerization. Journal of Molecular Biology. 2005;353:664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 20.Amezcua CA, Harper SM, Rutter J, Gardner KH. Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation. Structure. 2002;10:1349–1361. doi: 10.1016/s0969-2126(02)00857-2. [DOI] [PubMed] [Google Scholar]
- 21.Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warashina M, Min KH, Kuwabara T, Huynh A, Gage FH, Schultz PG, Ding S. A synthetic small molecule that induces neuronal differentiation of adult hippocampal neural progenitor cells. Angew Chem Int Ed Engl. 2006;45:591–593. doi: 10.1002/anie.200503089. [DOI] [PubMed] [Google Scholar]
- 23.Wurdak H, Zhu S, Min KH, Aimone L, Lairson LL, Watson J, Chopiuk G, Demas J, Charette B, Halder R, Weerapana E, Cravatt BF, Cline HT, Peters EC, Zhang J, Walker JR, Wu C, Chang J, Tuntland T, Cho CY, Schultz PG. A small molecule accelerates neuronal differentiation in the adult rat. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16542–16547. doi: 10.1073/pnas.1010300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: Insights into light-mediated signal transduction. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genick UK, Borgstahl GE, Ng K, Ren Z, Pradervand C, Burke PM, Srajer V, Teng TY, Schildkamp W, McRee DE, Moffat K, Getzoff ED. Structure of a protein photocycle intermediate by millisecond time- resolved crystallography. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- 27.Hajduk PJ, Bures M, Praestgaard J, Fesik SW. Priveleged molecules for protein binding identified from NMR-based screening. Journal of Medicinal Chemistry. 2000;43:3443–3447. doi: 10.1021/jm000164q. [DOI] [PubMed] [Google Scholar]
- 28.Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, Saijo Y, Gotoh O, Sogawa K, Fujii-Kuriyama Y. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS factor (Arnt2) with close sequence similarity to the aryl hydrocarbon receptor nuclear translocator (Arnt) Molecular and cellular biology. 1996;16:1706–1713. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekine H, Mimura J, Yamamoto M, Fujii-Kuriyama Y. Unique and overlapping transcriptional roles of arylhydrocarbon receptor nuclear translocator (Arnt) and Arnt2 in xenobiotic and hypoxic responses. The Journal of biological chemistry. 2006;281:37507–37516. doi: 10.1074/jbc.M606910200. [DOI] [PubMed] [Google Scholar]
- 31.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of biomolecular NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science. 2012;337:189–194. doi: 10.1126/science.1222804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barelier S, Krimm I. Ligand specificity, privileged substructures and protein druggability from fragment-based screening. Curr Opin Chem Biol. 2011;15:469–474. doi: 10.1016/j.cbpa.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 34.Cappell KM, Sinnott R, Taus P, Maxfield K, Scarbrough M, Whitehurst AW. Multiple cancer testis antigens function to support tumor cell mitotic fidelity. Molecular and cellular biology. 2012;32:4131–4140. doi: 10.1128/MCB.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 36.Scortegagna M, Ding K, Zhang Q, Oktay Y, Bennett MJ, Bennett M, Shelton JM, Richardson JA, Moe O, Garcia JA. HIF-2alpha regulates murine hematopoietic development in an erythropoietin-dependent manner. Blood. 2005;105:3133–3140. doi: 10.1182/blood-2004-05-1695. [DOI] [PubMed] [Google Scholar]
- 37.Qing G, Simon MC. Hypoxia inducible factor-2alpha: a critical mediator of aggressive tumor phenotypes. Curr Opin Genet Dev. 2009;19:60–66. doi: 10.1016/j.gde.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lauffart B, Vaughan MM, Eddy R, Chervinsky D, DiCioccio RA, Black JD, Still IH. Aberrations of TACC1 and TACC3 are associated with ovarian cancer. BMC Womens Health. 2005;5:8. doi: 10.1186/1472-6874-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swinney DC, Anthony J. How were new medicines discovered? Nature reviews. Drug discovery. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 40.Park EJ, Kong D, Fisher R, Cardellina J, Shoemaker RH, Melillo G. Targeting the PAS-A domain of HIF-1alpha for development of small molecule inhibitors of HIF-1. Cell Cycle. 2006;5:1847–1853. doi: 10.4161/cc.5.16.3019. [DOI] [PubMed] [Google Scholar]
- 41.Kung AL, Zabludoff SD, France DS, Freedman SJ, Tanner EA, Vieira A, Cornell-Kennon S, Lee J, Wang B, Wang J, Memmert K, Naegeli HU, Petersen F, Eck MJ, Bair KW, Wood AW, Livingston DM. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of "parallel" expression vectors. Protein Expression and Purification. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 44.Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nuclear receptor signaling. 2003;1:e012. doi: 10.1621/nrs.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farmer BT, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind MG, Yanchunas JJ, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nature Structural Biology. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.