Abstract
MicroRNAs (miRNAs) are a large family of post-transcriptional regulators of gene expression that control cellular and developmental processes by targeting messenger RNAs (mRNA). These small non-coding RNAs (ncRNAs) are aberrantly expressed in cancer, and are known to contribute to tumorigenesis and disease progression. Therapeutic strategies based on modulating miRNAs activity are emerging due to the ability of these ncRNAs to influence cellular behavior. MiRNA levels predict disease prognosis and overall patient survival, and reconstituting their basal levels has been proven to inhibit tumor growth and metastasis. Different delivery mechanisms have been tested in vivo, however many challenges need to be overcome before their utilization in the clinic. Moreover, it has been found that circulating miRNAs in body fluids have the potential to reshape cancer diagnosis and prognosis by functioning as biomarkers and indicators of progression and metastasis. These miRNAs as biofluids-based biomarkers provide an alternative strategy for early diagnosis and treatment of cancer patients.
Keywords: microRNA, Epigenetics, Cancer, Diagnosis, Therapy, Biomarkers, Pathobiology
INTRODUCTION
MicroRNAs are small non-coding RNAs of ~22 nt which post-transcriptionally regulate gene expression. The majority of these small RNA molecules are transcribed by RNA polymerase II as independent genes, or as introns of protein-coding genes [1]. MiRNAs target messenger RNAs (mRNAs) causing critical changes and instability that consequently inhibit translation. A percentage of approximately 60% of genes in the human genome are predicted to be targeted by miRNAs, as they conserve pairing homology [2].
Mechanisms of action
MiRNAs are initially transcribed by RNA polymerase II enzyme as long primary transcripts (pri-miRNAs). This transcript folds on itself forming a double stranded hairpin structure. An RNase endonuclease III Drosha, along with DGCR8 (molecular anchor part of a microprocessor complex), cleaves this secondary structure resulting in ~70 nt precursor miRNAs (pre-miRNAs). Exportin 5 and RAN-GTP promote nuclear translocation of the pre-miRNAs to the cytoplasm. Dicer, another double stranded RNA-specific nuclease located in the cytoplasm, defines a cleavage site that results in ~22 nt long double stranded dsRNA containing a mature miRNA guide strand and a passenger strand [3]. The guide strand promotes the association of a protein complex termed RNA-induced silencing complex (RISC) with the 3′unstranslated region of the targeted mRNAs. The primary component of the RISC complex is an Argonaute (Ago) protein, which cause miRNA-mediated mRNA repression. The Ago protein is crucial for the miRNA to form a complex with RISC, moreover it allows miRNA maturation even in the absence of Dicer by acquiring the ability to cleave pre-miRs. This evidence was identified by Cheloufi and collegues who observed that miR-451 could enter RISC through an alternative biogenesis pathway. After Drosha-mediated cleavage, the pre-miR was loaded directly into Ago omitting the catalytic activity of Dicer and forming a complex with RISC [4].
Roles and genomic regulation
MiRNAs regulate the expression of specific targets. However, since the nucleotide pairing by complementarity of these sequences is imperfect, it allows key miRNAs to regulate the expression levels of hundreds of genes simultaneously [5]. MiRNA profiles in different tissues are subject to regulating mechanisms that increase or decrease their downstream effect(s). This mechanism of action implicates them in various important processes such as tissue morphogenesis, apoptosis and signal transduction simultaneously [6, 7]. This, along with other evidences has proved that miRNA expression signatures are associated with human pathologies such as cancer.
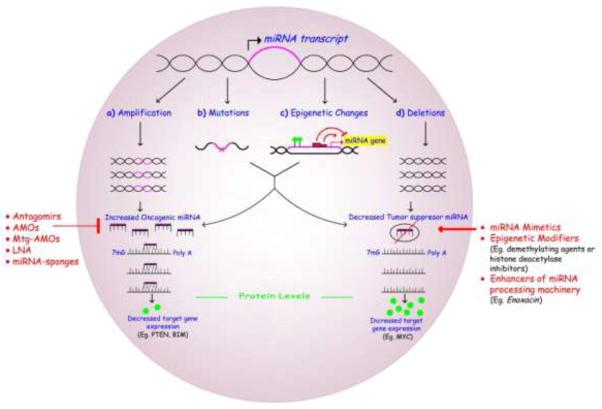
MiRNAs are subject to genomic regulation, which can be segregated in four groups: amplification, deletions, mutations/translocations and epigenetic alterations [8]. Allelic amplification of miRNAs results in decreased expression of the target gene, proportional to miRNA levels in the cell. Genomic deletion decreases the expression of miRNAs, increasing the genetic expression of the targeted genes. Translocations and sequence variations such as mutations or single nucleotide polymorphisms (SNPs) can alter the transcribed miRNAs causing changes in the binding sites and inhibiting or promoting repression. Finally, epigenetic alterations such as methylation of the promoter region, also cause changes in the miRNAs that mature in a cell, decreasing their basal levels [8] (Figure 1).
Figure 1. MICRORNA ALTERATIONS IN HUMAN CANCERS AND THERAPEUTIC STRATEGIES.
MiRNA levels are altered in cancer. a | Amplification of oncogenic miRNAs results in decreased expression of target genes. b | Mutations can decrease the expression of tumor suppressive miRNAs or could potentiate the expression of oncogenic ones. c | Epigenetic changes such as promoter methylation, or trans-acting factors change the expression profile of miRNAs in a cell either decreasing or increasing their basal levels. d | Deletions eliminate the expression of tumor suppressive miRNAs increasing the expression of target genes.
THERAPEUTIC OPPORTUNITIES IN CANCER
MicroRNAs negatively regulate genes involved in eukaryotic survival and proliferation, and can cause a disturbance in protein expression, translation and stability. The first associations observed between miRNAs expression and cancer development was the miR-15 and miR-16 dysregulation in B cell chronic lymphocytic leukemia, as a result of chromosome 13q14 deletion deletion [9]. Many miRNA genes are located in fragile genomic sites (regions more susceptible to mutations, rearrangements and loss of heterozygosity), which are frequently found in tumor development [10]. The alteration of proteins involved in miRNA regulation and the targeting of important messenger RNAs (mRNAs) are involved in tumor initiation and progression, as these coding genes behave as oncogenes or tumor suppressor genes [11]. Tumor suppressive miRNAs function by reducing the expression of proto-oncogenes, delaying carcinogenesis, and tumor maintenance. The miR-200 family is an example of a tumor suppressive miRNA, proven to influence the epithelial phenotype of cells by downregulation of E-cadherin (promoting epithelial-to-mesenchymal transition) [12]. On the other hand, oncogenic miRNAs reduce expression of tumor suppressor coding genes and are over-expressed or amplified in tumor cells, contributing to tumor development. Some oncogenic miRNAs inhibit repressors, causing an increase in transcription factors or growth factors (eg. R-Smads, ER-α, p53) and auxiliary proteins (eg. hnRNPA1). In this way they influence the mRNA processing machinery and cause alterations in their basal levels [13]. The dysregulation of miRNAs in cells, and the subsequent gain/loss of functions dictate treatment efficacy, therapeutic response and patient prognosis.
Two important strategies have guided the use of these molecules as novel therapies. First, miRNA molecules have been modified to achieve a prolonged in vivo half-life and efficiency (e.g. anti-miRNA and locked nucleic acid-modified oligonucleotides, and antagomirs). Second, in vivo development of miRNA transgenic mice (such as the miR-155, miR-21, miR17~92) and knockouts (such as miR-15, miR-16, miR-146 and miR-29) have offered valuable information that has guided therapeutic opportunities for cancer patients [8]. The strategies are based on two principles: targeting oncogenic miRNAs (to decrease their levels), or restoring tumor suppressive miRNA (to rescue their basal levels).
Targeting oncogenic miRNAs
Cancer cells contain many genetic and epigenetic abnormalities, but despite their complexity, their growth and survival can often be impaired by inactivating a single oncogene. This phenomenon, called “oncogene addiction,” provides a rationale for molecular targeted therapy [14]. Correlations between regulatory miRNAs and cancer have revealed that this concept applies to miRNA dysregulation in patients.
Therapies against oncogenic miRNA focus on decreasing miRNA levels by inhibiting them through complementary base pairing. Although inhibiting an individual miRNA role was the initial strategy, studies have highlighted the importance of targeting the “collaborative role” of miRNA groups as these cooperate in the acquisition of a pleiotropic biology in cancer cells. Both of these approaches have been pointed out in miR-17~92, an oncogenic polycistronically expressed cluster composed of six miRNAs: miR-17, -18a, -19a, -20a, -19b-1 and -92a-1 on chromosome 13. Overexpression of the miR-19 family members (relative to their cluster), proved to increase the latency of lymphomas, while their inactivation promoted MYC-induced lymphomagenesis [15]. Both of these results suggest an important individual role of the miR-19 family. Furthermore, multiple members of the cluster are capable of individually promoting NOTCH1-induced T cell acute lymphoblastic leukemia in a mouse model [16]. These findings on the other hand emphasize the collaborative role of three miRNAs of the cluster (miR-19b-1, -20a and -92a-1) in reducing disease latency by decreasing the expression of the tumor suppressors PTEN and BIM (typically downregulated in T-ALL) [16]. Because of the diversity of mechanisms by which miRNA levels contribute to tumor initiation and progression, several therapeutic models have been developed to target these processes.
Therapies to decrease the effect of a specific miRNA have been proposed by using antisense oligonucleotides (ASOs) that inhibit targets by adhering to the miRNA forming miRNA-anti-miRNA binding complexes. Three types have been described: antagomirs, locked nucleic acids (LNAs) and ASOs with chemical alterations to optimize efficacy (AMOs) [17–19] (Table 1). With the use of ASOs, Fontana et al., demonstrated that tumor growth was inhibited when injecting the antagomir-17-p in therapy resistant neuroblastoma cell lines [20]. In reference to the chemically modified, Ma et al., used AMOs to intravenously inhibit miR-10b in a mammary mouse tumor model, and observed that metastasis was inhibited [21]. Park and collegues tested another chemically modified antagomir, chol-anti-miR-221. In their orthotopic mouse model of hepatocellular carcinoma, they proved that chol-anti-miR-221 significantly reduced miR-221 levels in the liver, and that this, correlated with a reduction in tumor cell proliferation, an increase in apoptosis markers and cell cycle arrest [22]. LNA’s, another type of ASO have also proven to be efficient, as a liver regeneration mouse model tested by Sapra et al., demonstrated that using a locked nucleic acid antisense oligonucleotide against survivin (an apoptosis inhibitor), reduced its mRNA levels in 80% [23].
Table 1.
INHIBITORY MIRNA DRUGS
| Agent | What are these agents? | Mechanism of action | Example |
|---|---|---|---|
| Antagomirs | Single-stranded 23 nt RNA molecules complementary to the targeted microRNA that have been modified to increase the stability of the RNA and protect it from degradation. The modifications included a partial phosphorothioate backbone in addition to 2′-O-methoxyethyl. | The mechanism is not completely known, however it is believed to act by forming a duplex: (miRNA/antagomir) that induces the degradation of the targeted miRNA and recycling of the antagomir. |
Antagomir-17-5p: Shown to abolish the growth of MYCN-amplified and therapy-resistant neuroblastoma leading to cell cycling blockade and apoptosis [20]. |
| AMOs anti-miRs | Single-stranded antisense oligonucleotides (ASO), chemically modified to enhance binding affinity to a complementary microRNA. It is usually shorter than their target miRNA ~17–20 nt in length and inhibits in a specific manner the genetic expression of its target gene. | They produce a ASO-miRNA double stranded complex through complementary base binding, leading to non-specific endonuclease cleavage of the targeted miRNA. |
Anti-miR-10b: Chemically modified anti-miRNA oligonucleotide shown to suppress breast cancer metastasis in mouse mammary tumor model [21]. |
| LNA anti-miRs | A nucleic acid analogue containing one or more LNA nucleotide monomers with a bicyclic furanose unit locked in an RNA mimicking sugar conformation. | Same as the AMO. |
EZN-3042: A LNA-ASO proven to downregulate surviving (apoptosis inhibitor), and thereby inhibit tumor growth in vivo [23]. |
| MTg-AMOs | Modified AMO, in which multiple antisense units are engineered into a single unit. | Same as the AMO, except that the multiplicity of units enables it to simultaneously silence multiple target miRNAs. |
MTg-AMO against miR-21, miR-155 and miR-17-5p : Produced a greater inhibitory effect on cancer cell growth, compared with the regular single-target AMOs [24]. |
| microRNA sponges | Transcripts that contain multiple tandem-binding sites to a miRNA of interest and are transcribed from expression vectors. | By competing with the native targets of miRNAs, these highly expressed transcripts result in increased expression of the miRNA’s native targets. |
miRNA sponge to increase miR-9 targeting: Metastasis formation was inhibited in highly malignant breast cancer cells [25]. |
Researchers have recently engineered a single subunit termed “multiple-target anti-miRNA antisense oligodeoxyribonucleotide” (MTg-AMO), through which simultaneous silencing has been achieved [24]. MTg-AMO’s have been proven to allow the restoration of dysregulated miRNA levels by targeting several key aspects of the biology of cancer cells in tumor tissue at once. Lu et al., demonstrated that the MTg-AMO targeting miR-21, miR-155 and miR-17-5p produced a greater inhibitory effect on cancer cell growth, compared with the regular single-target AMOs [24].
Another therapeutic approach to decrease miRNA levels involves expressing competitive inhibitors of their function. A type of these are “microRNA sponges” which are vectors containing multiple artificial miRNA binding sites that are placed under the control of strong promoters to produce large quantities of transcript [25]. These transcripts express multiple tandem sites to a miRNA of interest [26]. MiRNA sponges were used in metastatic breast cancer mouse model, where in vivo experiments demonstrated that the downregulation of an over-expressed breast cancer miRNA (miR-19) could be achieved, along with metastasic inhibition [27]. Approaches have already been developed to rapidly generate single or combined miRNA sponges that can be used for long-term miRNA loss-of-function studies [28].
Restoring expression of tumor suppressive miRNAs
Although specific miRNAs are overexpressed in cancer tissue, several miRNAs are downregulated in tumors [29]. Global repression of miRNA expression has been proven to increase tumorigenesis in both in vitro and in vivo models [30]. The downregulation of elements of the biogenesis pathway (processing machinery), is one of the mechanisms by which mature miRNA levels are reduced in some types of cancer. An example of this was seen by Merrit and collegues when they studied Dicer and Drosha levels in patients with ovarian cancer. They found that patients with a lower expression were significantly associated with low miRNA levels, as well as advanced tumor stage and suboptimal surgical cytoreduction [31].
For tumors with reduced expression of miRNAs, restoration of their basal levels is the key strategy, which can be achieved through miRNA mimetics or by regenerating “miRNAome” (full spectrum of expressed miRNAs in a cell at a specific time) functionality (Table 2). The most widely used approach is using miRNA mimetics which are synthetic small RNAs that contain the exact sequence of the endogenous ones. To achieve the delivery of a stable molecule, miRNA’s are delivered as perfectly complementary duplexes, similar in architecture to siRNAs [32].
Table 2.
DRUGS THAT RESTORE MIRNA
| Agent | What are these agents? | Mechanism of action | Examples |
|---|---|---|---|
| miRNA mimetics | Synthetic small RNAs that contain the exact sequence of endogenous miRNA. They can be modified to have enhanced efficiency by increasing the affinity for a specific target and reducing other unwanted miRNA effects. | They are delivered as perfect complementary duplexes and they act by improving the RISC loading of miRNA. |
miR-26a: In a hepatocellular carcinoma mouse model this mimetic inhibited cell growth and apoptosis [34]. |
| Epigenetic modifiers | Molecules that reverse DNA methylation and posttranslational histone modifications | They either inhibit methylation, or interact with enzymes that remove acetyl groups. |
5-aza-2′-deoxycytidine and 4-phenylbutyric acid: Restored tumor suppressive miR-127 levels and subsequently inactivated BCL6 proto-oncogene [36]. |
| Enhancers of miRNA processing machinery | Small molecule/drug that enhances RNAi and promotes miRNA processing. | Increase the production of tumor suppressive miRNA y interacting with the binding products of the proteins that regulate miRNA maturation. |
Enoxacin: A fluoroquinolone that has been proven to increase the production of tumor suppressive miRNA by binding to the transactivation-responsive RNA-binding protein 2 (TRBP) [38]. |
The majority of the therapies that have tried to restore tumor suppressive miRNA with mimetics have achieved their goal by administering them locally. However, nowadays the challenge of developing a systemic delivery in a tissue/cell-type specific manner has been proven to be achievable through different delivery systems. Ibrahim et al., used both systemic and local application of low molecular weight polyethylenimine/miR-145 and miR-33a complexes, which were delivered into mouse xenograft tumors of colon carcinoma, obtaining antitumor effects such as reduced proliferation and increased apoptosis [33]. In another model, the systemic administration of miR-26a through an adenoviral vector was given to mice with hepatocellular carcinoma, resulting in the inhibition of cancer cell proliferation, induction of tumor cell apoptosis and protection from disease progression [34]. A very recent orthotopic model for hepatocellular carcinoma achieved a higher therapeutic efficacy than previous ones; consistently decreased levels of miR-34a where targeted with liposomal delivery of the miRNA in a relevant NOV340/miR-34a formulation. The effectiveness of decreasing tumorigenesis in this model has led the researchers to anticipate the initiation of clinical trials in 2013, with miR-34a being one of the first miRNA mimics to reach the clinic [35].
Along with mimetics, other therapies have aimed at restoring miRNA levels in a “global manner”. Two of these therapies are epigenetic modifiers and enhancers of the miRNA processing machinery.
Epigenetic modifiers address changes such as hypermethylation and histone modification patterns, which have been linked to miRNA dysregulation. Saito and colleagues aimed at targeting the hypermethylation that silenced the tumor suppressor miR-127 in human cancer cells. For this, they used chromatin-modifying drugs such as 5-aza-2′-deoxycytidine and 4-phenylbutyric acid in human cancer cells. After the treatment they observed a restoration in miR-127 levels and subsequent inactivation of BCL6 proto-oncogene [36]. Other research groups have furthermore combined strategies using DNA demethylating agents and histone deacetylase inhibitors. Huang and colleagues tested both therapies to regenerate basal levels of tumor suppressor miR-129-2. Their treatment resulted in the reactivation of miR-129-2 in cancer cells, with the downstream effect of decreasing SOX4 (transcription factor) levels and reducing proliferation in endometrial cancer cells [37].
The disruption of the miRNA processing machinery has also been known to contribute to the development of human tumors as previously mentioned [31]. Scientists have recently come up with a small molecule/drug that enhances RNAi and promotes miRNA processing: Enoxacin [38]. This is a fluoroquinolone that increases the production of tumor suppressive miRNA by binding to the transactivation-responsive RNA-binding protein 2 (TRBP). In human cultures and a metastatic mouse model, the use of this drug had a cancer-specific growth-inhibitory effect [38]. These results suggest that novel strategies aiming to restore the miRNAome might be promising.
DELIVERY MECHANISMS
In vivo delivery is the main focus of miRNA based therapies, and either local or systemic techniques have been developed. Local strategies, such as direct site injection, have several advantages as they evade nuclease degradation, decrease miRNA modulator uptake by non-targeted tissues and increase bioavailability in tumor sites. Nevertheless they are merely applicable to a very limited list of target tissues such as ocular, brain, sarcomas, mesotheliomas [39]; and even in these tissues not all targeted tumor cells can be reached by the miRNA modulators [40]. The second approach, a systemic delivery, consists of a bloodstream injection that - in theory - should have a much more efficient dissemination to the target tissues. However, these mechanisms need to overcome in vivo barriers such as nuclease degradation and non-specific targeting of all the tissues that are reached by blood vessels, specifically the liver, jejunum, lung and kidney [40]. Chemical modifications, encapsulations and conjugations have aimed to protect these therapeutic miRNAs, because once in the bloodstream they are required to maintain intact. Conserving their structure allows them to traverse cellular membranes and more importantly reach the cytosol once released from their endosome vesicle [41].
Carriers
Synthesizing a stable, biodegradable and biocompatible miRNA modulator is very important, but ensuring an adequate cellular penetration of the delivery molecule is also crucial. Carriers can be divided in two groups: viral and non-viral, and both types aim to target tumor systems while simultaneously evading immune toxicity.
Viral strategies have been used with vectors such as lentiviruses, adenoviruses and adeno-associated viruses (AAVs), and genetic manipulation can target these entities to the cells of choice [42, 43]. Although preclinical mouse models using virus as vector mediated delivery have shown promising results, they present with some flaws that still need to be addressed before advancing into the clinical practice. Some of the strategies to overcome these flaws could focus in: obtaining efficient nuclear localization, evading the activation of oncogenes or inactivation of tumor suppressor genes, and eliminating undesired immunogenic responses in patients [41].
Non-viral strategies, such as liposomal delivery have been widely used and they have been demonstrated to induce tumor suppression in several mice models [44–46]. Their success has been attributed to the protection of oligonucleotides from nuclease mediated degradation, along with their ability to increase the circulating half-life when systemically delivered [39]. However, in vivo systems have had adverse effects related to the positive charge of the lipid component [47, 39].
One strategy to overcome these toxic effects is development of neutral nano-liposomes based on 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) [48]. These nano-liposomes can deliver microRNA in vivo into tumor cells 10- and 30-fold more effectively than cationic liposomes [49], and have been proven in mouse models. For example, in a mouse model for lung cancer, Trang et al., systemically delivered the tumor suppressor miR-34a in a neutral lipid emulsion and obtained a 60% reduction in tumor area compared to controls [50]. In another model, mice with ovarian cancer tumors were treated with siRNA incorporated into the neutral-DOPC liposomes to target the oncoprotein EphA2. In these experiments Landen et al., observed reduced tumor growth when compared with a nonsilencing siRNA [49].
Other alternatives include coating cationic bilayers with hydrophilic polyethylene glycol (PEG), which also decreases immunogenic responses [51]. Finally the addition of biodegradable synthetic or natural polymers could also represent an effective alternative to facilitate sustained delivery in vivo (e.g. polyethyleneimines), nevertheless toxicity remains a challenge.
BODY FLUID EXPRESSED miRNAS: BIOMARKERS AND TREATMENT-RESPONSE PREDICTORS
Different types of cancers have specific miRNA profiles that allow tumors to be classified based on their miRNA expression levels. The lack of complex transcriptional and translational modifications compared to mRNAs and proteins suggest that the use of miRNAs as biomarkers for cancer has great potential. Since there must be some method of communication between cancer cells and their microenvironment, research has suggested that microRNAs should be considered molecules of secretion along with cytokines, growth factors and other proteins [52]. Serum and plasma can both be used to measure specific miRNA levels as there comparison in several studies has led to equal distribution of miRNAs. Mitchell et al., found a strong correlation when comparing both of these in the blood of mice in prostate cancer xenograft model [53], moreover a profile study done in serum and plasma of lung cancer patients led to the same conclusions [54].
Studies have already demonstrated correlation between circulating miRNA-expression levels and response to anticancer treatment [55], giving miRNA a crucial role in therapeutic management of cancer patients. Research groups have focused on segregating circulating miRNAs in specific patterns based on the different cancer types [56] (for a more detailed review see Allegra et al., 2012). In this review I will refer to four of the cancer types with the highest incidence (lung, breast, colon and hematologic malignancies), and current research findings on characteristic fluid expressed miRNAs.
Lung Cancer
More people die from lung cancer than from any other type, and in the US over 200,000 cases were diagnosed in 2012, and approximately 160,000 died from the disease [57]. This has increased research including studies in the miRNA field. In lung cancer circulating miRNAs have been linked to pre-disease patterns, disease staging, tumor activity and metastasis development. Research has recently generated a high-throughput study were for the first time it was demonstrated that pre-disease signatures of miRNA expression in plasma samples can predict development of lung cancer before acquiring a diagnosis from conventional techniques [58]. MiR-21 was recently described as a miRNA that could differentiate early stage lung cancer patients from healthy individuals; furthermore, its plasma levels not only serve as a circulating tumor biomarker, but could also determine sensitivity to platinum based chemotherapy [59, 60]. High circulating miR-155, miR-197 and miR-182 were also correlated with disease stage I, as plasma samples from patients demonstrated significantly higher levels than their respective controls [61]. Finally, miR-486-5p has been also studied in plasma of lung cancer patients, and a tumor suppressive role has been associated with its downregulation [62, 59]. These findings suggest that specific miRNAs serve as markers of disease stage and a prognosis indicator [61].
Breast Cancer
Breast cancer is the most common cancer among American women (aside from skin), and miRNAs have also acquired significance in this disease. In the United States in 2008, 210,203 women were diagnosed with the disease, and 40,589 women died from it [63]. Altered miRNAs have been studied by several groups: Roth and colleagues reported findings of altered levels in the serum of breast cancer patients [64]. Moreover, Mitchell et al., and Heneghan et al., demonstrated tumorigenic roles of miR-195 and let-7a in breast cancer, as they proved that levels were increased with the presence of the tumor, and decreased in serum 2 weeks after surgical resection [53, 65].
Colorectal Cancer (CRC)
Of cancers affecting both men and women, colorectal cancer is the second leading cancer killer in the United States [66]. CRC has also been studied to find alternative biomarkers of disease progression. For staging techniques, Cheng et al., found that circulating miR-141 was significantly associated with stage IV colon cancer in a cohort of plasma samples. They also observed that combining the detection of miR-141 and carcinoembryonic antigen (CRC marker) further improved the accuracy of detection and patients that have both of them were associated with a poor prognosis [67]. Other potential biomarkers studied have been the circulating levels of miRs-17-3p and -92. These were shown to decrease after surgery, a finding that served to differentiate CRC patients in recovery from other gastric cancer patients and from normal individuals [68].
Hematological Malignancies
In the US in 2009, there were approximately 271, 880 men and women alive who had a history of leukemia, and over 10% of these are diagnosed before the age of 20 [69]. In the reach of discovering new therapeutic modalities for these patients, the role of miRNAs is trying to be elucidated [69]. In hematological malignancies miRNAs have an important role, as they participate in leukemogenesis by coordinating effects with key transcription factors required for hematopoiesis. Moussay et al., studied 14 miRNAs associated with CLL and they found that three of them strongly separated controls vs. CLL patients, and these were miR-195, miR-29-a and miR-222. Moreover increased levels of miRNA-150 increased with disease severity, suggesting the potential of this biomarker in disease staging [70]. In other malignancies, such as B cell lymphoma, miRNAs such as miR-21, miR-155 and miR-210 were found to correlate with favorable clinical outcome, as they were associated with relapse-free survival [71]. Although not many studies have been done of circulating miRNA in acute leukemias, recent evidence have pointed out that decreased plasma levels of miRNA-92 is a significant biomarker for patients presenting acute myeloid leukemia [72].
Conclusion
The general idea of reconstituting the miRNAome as “a whole” has resulted in improving strategies to achieve a reduction in tumor growth and metastasis inhibition. The development of miRNA-based therapies (such as molecular drugs, mimetics and AMOs) has continued to progress, as the in vivo models and delivery mechanisms being developed aim to be less toxic and more efficient in reaching their targets. The synthesis of nanoparticles composed of polymers, neutral lipids, chimeras or conjugates, will continue improving local as well as systemic delivery. Nevertheless, several immunogenic/nonspecific effects, point to the necessity of validating the efficiency of therapeutic responses in a more personalized, patient-specific manner.
Circulating miRNAs not only contribute to disease staging, but they can improve prognosis by emphasizing disease progression or regression, and by predicting metastatic patterns individually in patients. Combining the use of known therapeutic mechanisms along with fluid miRNA profiles could result in an improved strategy against cancer. However, it should be taken into consideration that miRNA levels in tumor microenvironment might be a contribution from cells that are not cancer related. Moreover other types of pathologies could also be generating similar miRNA profiles which could overlap the analysis done in cancer patients. Finally, the fact that RNA-extracting and quantification methods, as well as normalizers vary between published studies, make conclusions less precise and uniform.
Biological fluid-based miRNAs profiles, along with miRNA-based therapies represent and attractive entity of the current cancer research because they could potentially revert cancer cells from their transformed phenotype back to their normal one. In this way, pharmaceutical therapies are headed to be individualized, and the efficiency of treatments could potentially increase.
Acknowledgments
Dr. Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. He is supported also as a Fellow at The University of Texas MD Anderson Research Trust, as a University of Texas System Regents Research Scholar, and by the CLL Global Research Foundation. Work in Dr. Calin’s laboratory is supported in part by the NIH/NCI (CA135444), a Department of Defense Breast Cancer Idea Award, Developmental Research Awards in Breast Cancer, Ovarian Cancer, Brain Cancer, Prostate Cancer, Multiple Myeloma, Leukemia (P50 CA100632) and Head and Neck (P50 CA097007) SPOREs, a SINF MDACC_DKFZ grant in CLL, a SINF grant in colorectal cancer, the Laura and John Arnold Foundation, the RGK Foundation, and the Estate of C. G. Johnson, Jr.
Footnotes
Disclosure No potential conflicts of interest relevant to this article were reported.
Contributor Information
Paloma del C. Monroig, Email: Paloma.D.Monroig@uth.tmc.edu, Department of Experimental Therapeutics, University of Texas M. D. Anderson Cancer Center, Houston TX 77030, USA.
George A. Calin, Email: gcalin@mdanderson.org, Department of Experimental Therapeutics, University of Texas M. D. Anderson Cancer Center, Houston TX 77030, USA.
References
Papers of particular interest have been highlighted as:
• Of importance
•• Of major importance
- 1.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis function and decay. Nat Rev. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denli AM, Tops BB, Plasterk RH, et al. Processing of primary microRNAs by the Microprocessor complex. Nat Rev. 2004:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 4.Cheloufi S, Dos Santos CO, Chong MM, et al. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11(4):252–63. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Lee CG. MicroRNA and cancer--focus on apoptosis. J Cell Mol Med. 2009;13(1):12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev. 2011 Dec;11(2011):849–64. doi: 10.1038/nrc3166. This study discusses the progress using mouse models to understand the roles of miRNAs in cancer and the potential for manipulating miRNAs for cancer therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. This report presents evidence for the involvement of miRNA genes in human tumors, and provides further evidence of the significance of this growing family of regulatory genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer--new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21(3):470–9. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Davalos V, Moutinho C, Villanueva A, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncog. 2012;31(16):2062–74. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis-Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. The J of Biochemistry. 2010;148(4):381–92. doi: 10.1093/jb/mvq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng CJ, Slack FJ. The duality of oncomiR addiction in the maintenance and treatment of cancer. Cancer J. 2012;18(3):232–7. doi: 10.1097/PPO.0b013e318258b75b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23(24):2839–49. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806–11. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elmén J, Lindow M, Schütz S, et al. LNA-mediated microRNA silencing in non-human primates. Nat. 2008;452(7189):896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 18•.Esau C. Inhibition of microRNA with antisense oligonucleotides. Methods in Molecular Biology. 2008;41(1):55–60. doi: 10.1016/j.ymeth.2007.11.001. The focus of this review was the use of antisense oligonucleotides (antimiRs) in miRNA inhibition for loss-of-function studies. It summarizes the employed antisense chemistries and their utility in designing antimiR oligonucleotides, and describes in vivo delivery strategies and approaches for assessment of miRNA inhibition and potential off-target effects. [DOI] [PubMed] [Google Scholar]
- 19••.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nat. 2005;438(7068):685–9. doi: 10.1038/nature04303. The findings of this article show that antagomirs are powerful tools to silence specific miRNAs in vivo and may represent a therapeutic strategy for silencing miRNAs in disease. [DOI] [PubMed] [Google Scholar]
- 20.Fontana L, Fiori ME, Albini S, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3(5) doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28(4):341–7. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Park JK, Kogure T, Nuovo GJ, et al. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 2011;71(24):7608–16. doi: 10.1158/0008-5472.CAN-11-1144. These findings proved the preclinical efficacy of chol-anti-miR-221 in a valid orthotopic mouse model of hepatocellular carcinoma. Results suggest that targeting this miRNA serves a therapeutic benefit for patients with advanced disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sapra P, Wang M, Bandaru R, et al. Down-modulation of survivin expression and inhibition of tumor growth in vivo by EZN-3042, a locked nucleic acid antisense oligonucleotide. Nucleosides Nucleotides Nucleic Acids. 2010;29(2):97–112. doi: 10.1080/15257771003597733. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Xiao J, Lin H, et al. A single anti-microRNA antisense oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an improved approach for microRNA interference. Nucleic Acids Res. 2009;37(3) doi: 10.1093/nar/gkn1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 26.Stenvang J, Kauppinen S. MicroRNAs as targets for antisense-based therapeutics. Expert Opin Biol Ther. 2008;8(1):59–81. doi: 10.1517/14712598.8.1.59. [DOI] [PubMed] [Google Scholar]
- 27.Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12(3):247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kluiver J, Slezak-Prochazka I, Smigielska-Czepiel K, et al. Generation of miRNA sponge constructs. Methods in Molecular Biology. 2012;58(2):113–7. doi: 10.1016/j.ymeth.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–68. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 30.Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 31.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359(25):2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love TM, Moffett HF, Novina CD, Love TMMH, Novina CD. Not miR-ly small RNAs: big potential for microRNAs in therapy. J Allergy Clin Immunol. 2008;121(2):309–19. doi: 10.1016/j.jaci.2007.12.1167. [DOI] [PubMed] [Google Scholar]
- 33.Ibrahim AF, Weirauch U, Thomas M, et al. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71(15):5214–24. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 34.Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Bader AG. miR-34 – a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3(120) doi: 10.3389/fgene.2012.00120. This review focuses on the molecular mechanisms of miR-34-mediated tumor suppression, pharmacologies in animal models of cancer, and a status update of a miR-34 therapy that may be among the first miRNA mimics to reach the clinic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 37.Huang YW, Liu JC, Deatherage DE, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69(23):9038–46. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melo S, Villanueva A, Moutinho C, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci U S A. 2011;108(11):4394–9. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garzon R, Marcucci, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature Reviews Drug Discovery. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bader AG, Brown D, Stoudemire J, et al. Developing therapeutic microRNAs for cancer. Gene Therapy. 2011;18:1121–6. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Pereira DM, Rodrigues PM, Borralho PM, et al. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2012 doi: 10.1016/j.drudis.2012.10.002. This review focuses on in vivo strategies of miRNA modulator delivery in cancer models. [DOI] [PubMed] [Google Scholar]
- 42•.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–7. doi: 10.1038/nm.1880. This article proposed that miR-15a and miR-16 act as tumor suppressor genes in prostate cancer through the control of cell survival, proliferation and invasion. Moreover, they suggest that the therapeutic implications found may be exploited for future treatment of prostate cancer. [DOI] [PubMed] [Google Scholar]
- 43.Nadiminty N, Tummala R, Lou W, et al. MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0032832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akao Y, Nakagawa Y, Hirata I, et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17(6):398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 45.Pramanik D, Campbell NR, Karikari C, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10(8):1470–80. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rai K, Takigawa N, Ito S, et al. Liposomal delivery of MicroRNA-7-expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol Cancer Ther. 2011;10(9):1720–7. doi: 10.1158/1535-7163.MCT-11-0220. [DOI] [PubMed] [Google Scholar]
- 47.Pecot CV, Calin GA, Coleman RL, et al. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11(1):59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutiérrez-Puente Y, Tari AM, Ford RJ, et al. Cellular pharmacology of P-ethoxy antisense oligonucleotides targeted to Bcl-2 in a follicular lymphoma cell line. Leuk Lymphoma. 2003;44(11):1979–85. doi: 10.1080/1042819031000099733. [DOI] [PubMed] [Google Scholar]
- 49.Landen CN, Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910–8. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 50.Trang P, Wiggins JF, Daige CL, et al. Systemic Delivery of Tumor Suppressor microRNA Mimics Using a Neutral Lipid Emulsion Inhibits Lung Tumors in Mice. Mol Ther. 2011;19(6):1116–22. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Antonellis P, Medaglia C, Cusanelli E, et al. MiR-34a Targeting of Notch Ligand Delta-Like 1 Impairs CD15+/CD133+ Tumor-Propagating Cells and Supports Neural Differentiation in Medulloblastoma. PLos One. 2011;6(9) doi: 10.1371/journal.pone.0024584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wentz-Hunter KK, Potashkin JA. The Role of miRNAs as Key Regulators in the Neoplastic Microenvironment. Mol Biol Int. 2011 doi: 10.4061/2011/839872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 55.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nat. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 56••.Allegra A, Alonci A, Campo S, et al. Circulating microRNAs: New biomarkers in diagnosis, prognosis and treatment of cancer (Review) Int J Oncol. 2012;41(6):1897–912. doi: 10.3892/ijo.2012.1647. This manuscript emphasizes that miRNA profiling could improve the diagnosis of cancer and could predict patient outcome. Alterations in circulating miRNA may signal a predisposition to cancer, exalting the importance of these profiles as therapeutic targets in cancer patients. [DOI] [PubMed] [Google Scholar]
- 57.National Cancer Institute. [Accessed December 2012];Cancer Statistics. Available at http://www.cancer.gov/statistics/find.
- 58.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108(9):3713–8. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen J, Liu Z, Todd NW, et al. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011 doi: 10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei J, Gao W, Zhu CJ, et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer. 2011;30(6):407–14. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng D, Haddadin S, Wang Y, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4(6):575–86. [PMC free article] [PubMed] [Google Scholar]
- 62.Huang X, Ding L, Bennewith KL, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35(6) doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Center for Disease Control and prevention. [Accessed December 2012];Breast Cancer. Available at http://www.cdc.gov/cancer/breast/
- 64.Roth C, Rack B, Müller V, et al. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12(6) doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heneghan HM, Miller N, Kerin MJ. Circulating microRNAs: promising breast cancer Biomarkers. Breast Cancer Res. 2011;13(1) doi: 10.1186/bcr2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Center for Disease Control and prevention. [Accessed December 2012];Colorectal Cancer. Available at http://www.cdc.gov/cancer/colorectal/
- 67.Cheng H, Zhang L, Cogdell DE, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–81. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 69.National Cancer Institute. [Accessed December 2012];SEER Stat Fact Sheets: Leukemia. Available at http://seer.cancer.gov/statfacts/html/leuks.html.
- 70.Moussay E, Wang K, Cho JH, et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2011;108(16):6573–8. doi: 10.1073/pnas.1019557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 72.Tanaka M, Oikawa K, Takanashi M, et al. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4(5) doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]