Abstract
Haloarchaea are the predominant microflora of hypersaline econiches such as solar salterns, soda lakes, and estuaries where the salinity ranges from 35 to 400 ppt. Econiches like estuaries and solar crystallizer ponds may contain high concentrations of metals since they serve as ecological sinks for metal pollution and also as effective traps for river borne metals. The availability of metals in these econiches is determined by the type of metal complexes formed and the solubility of the metal species at such high salinity. Haloarchaea have developed specialized mechanisms for the uptake of metals required for various key physiological processes and are not readily available at high salinity, beside evolving resistance mechanisms for metals with high solubility. The present paper seeks to give an overview of the main molecular mechanisms involved in metal tolerance in haloarchaea and focuses on factors such as salinity and metal speciation that affect the bioavailability of metals to haloarchaea. Global transcriptomic analysis during metal stress in these organisms will help in determining the various factors differentially regulated and essential for metal physiology.
1. Introduction
Many metal ions have a key role in the physiology of cells. Metals such as calcium, cobalt, chromium, copper, iron, potassium, magnesium, manganese, sodium, nickel, and zinc are essential and serve as micronutrients. These metals act as the redox centers for metalloproteins such as cytochromes, blue copper proteins, and iron-sulfur proteins which play a vital role in electron transport [1]. As the transition metals exist in numerous oxidation states, they efficiently act as electron carriers during redox reactions of electron transport chains to generate chemical energy [2, 3]. Metal ions also function as cofactors and confer catalytic potential and stability to proteins [4]. Other metals like silver, mercury, lead, aluminum, cadmium, gold, and arsenic have no biological roles and are potentially toxic to microbes [5]. The toxicity is exerted by the displacement of essential metals from native binding sites or through ligand interactions [6]. Both essential and nonessential metals at high concentrations disrupt cell membrane, alter enzymatic specificity, hinder cellular functions, and damage DNA [5]. Thus, as any disturbance in metal ion homeostasis could produce toxic effects on cell viability, the concentrations of metals within cells are stringently controlled. An increase in the ambient metal concentration leads to activation of metal resistance mechanisms to overcome metal stress. Metal homeostasis has been well studied in bacteria and eukaryotes and is attributed to differential regulation of transporters like P1B-type ATPases, ABC transporters, cation diffusion facilitators (CDFs), and metallochaperones in response to metals [7–9]. Among the Archaea, thermophiles and hyperthermophiles of the Crenarchaeota and the methanogens and thermophiles of Euryarchaeota utilize P1B-type ATPases and ABC transporters for metal transport and homeostasis [10, 11]. However, metal homeostasis in haloarchaea from the phylum Euryarchaeota has not been extensively studied [11].
Haloarchaea are members of the third domain of life, the Archaea, within which a total of 36 genera and 129 species have been identified to date [12]. These organisms require between 10% and 35% salt for optimum growth and are the predominant microflora of hypersaline environments such as solar salterns, salt lakes, soda lakes, salt deposits, and so forth [13]. However, some low salt tolerant haloarchaea can be found in estuarine environments [14]. Estuaries serve as interfacial mixing zones between rivers and seawaters which determine the flux of chemical species into the ocean [15]. Econiches like estuaries [16] and solar crystallizer ponds [17] may contain high concentrations of metals, since they serve as ecological sinks for metals and as effective traps for river borne metals [18]. Anthropogenic activities like urbanization and industrialization, including mining, agriculture, and waste disposal, further contribute towards metal pollution at these sites [19, 20]. Haloarchaea have developed various mechanisms of resistance in order to thrive under metal stress [21–23]. However, studies on metal resistance in haloarchaea are still in their infancy.
Most of the reports until now are limited to MIC (minimum inhibitory concentration) studies [24–26]. However, in a study on comparative gene analysis of the role of P1B-type ATPases in maintaining metal homeostasis in bacteria and archaea, Coombs and Barkay (2005) [10] have shown that P1B-type ATPases containing N-terminal metal-binding motifs are distributed across bacteria and archaea, including haloarchaea. These ATPases along with the ABC transporters, transcriptional regulators, and certain metallochaperones were found to be involved in metal resistance and homeostasis in the haloarchaeon Halobacterium sp. strain NRC-1 [22]. Haloarchaea exhibit a high degree of variation in the concentration of metals that they can tolerate [22, 24–26]. Interestingly, at low concentrations, certain metal ions like Mn(II), Fe(II), Co(II), Ni(II), and Zn(II) were found to enhance growth [22, 24]. An in-depth study at molecular level may help in better understanding of this variation. This paper seeks to give an overview of the main molecular mechanisms involved in metal tolerance in haloarchaea and to outline the factors such as salinity and metal speciation that affect the bio-availability of the metals to haloarchaea. The need for further studies on metal homeostasis and resistance in haloarchaea is highlighted. The elucidation of complete pathways of metal resistance from uptake to transformation/detoxification and efflux will help to determine the final fate of metals. The final metal species could be either volatile or chelated intracellularly and thereby rendered nontoxic to the organism.
2. Bioavailability of Metals to Haloarchaea
For a metal to act either as a micronutrient or as a toxicant, it has to be available for uptake by the organism [27]. The metal species determines the solubility, bio-availability, and membrane transport, besides influencing the phenomenon of adsorption, oxidation/reduction, and oceanic residence times [28]. Metal speciation is governed by alkalinity, pH, hardness (presence of Ca/Mg), natural dissolved organic matter, redox potential, and salinity [29]. Strongly complexed and thus nonlabile and particulate metal species are less available to organisms for uptake [30]. As haloarchaea inhabit hypersaline environments with salinity in the range of 2%–35%, salinity is proposed to be the most important factor affecting bio-availability. The salt content in hypersaline econiches of solar salterns is about 10-fold of its concentration in seawater [31] due to evaporation. This process also concentrates other anions and cations present in the sea water including the metal salts fed through contaminated estuaries [32]. The high chloride ion (Cl−) content in these environments results in the formation of metal chlorocomplexes.
The type of complex formed depends upon the chelating ligand, that is, organic or inorganic ligands, and the kind of heavy metal present in the system [33]. Metals like Zn and Cu that have small ionic radii preferentially complex with hard donors containing oxygen like OH−, CO2 3−, HCO3 −, and SO4 2− to form inorganic complexes [34]. Soft acceptors like Hg, Cd, and Ag are easily ionized and are thus, more likely to form chlorocomplexes. Although inorganic species exist in natural waters, organic metal species predominate [35]. The complexation of metals with organic ligands reduces bioavailability as organic-metal complexes are not readily transported across cell membranes [30, 36]. Inorganic species, on the other hand, are readily available to the biota as the complexes are weak and dissociate rapidly to form free ions which bind to the transporters or are chelated by biotic ligands secreted by the organisms [37–40]. Table 1 shows the type of inorganic species formed at different salinities for five major metals.
Table 1.
Various inorganic complexes formed in natural waters, seawater, estuarine waters (variable salinity), and hypersaline waters. As haloarchaea inhabit hypersaline environments where inorganic ligands predominate, inorganic metal speciation is described. The availability of metal depends upon the kind of inorganic complex formed. Lipophilic soluble chlorocomplexes of Hg and Ag are easily available in hypersaline waters. Insoluble (precipitated) ZnCl2 and CuCl2 are unavailable to the organism. Fe (II), Co (II), Ni (II), and Mn (II) form weak complexes with Cl− that easily dissociate and can be taken up by organisms [28, 41–46].
| Metal | Hypersaline (5–35% salinity) |
Sea water (3.5% salinity) |
Estuarine (variable salinity) |
River water/natural water |
|---|---|---|---|---|
| Cd | CdCl2, CdCl+ | CdCl+ | CdCl2, CdCl+ | Cd2+, CdCO3 |
| Ag | AgCl0, AgCl2−, AgCl3 2−, AgCl4 3− |
AgCl0, AgHS0 | AgCl0, AgHS0, AgCl2−, AgCl3 2−, AgCl4 3− |
Ag+, AgCl0 |
| Hg | HgCl0, HgCl−, HgCl4 2− |
HgCl− | HgCl0, HgCl−, HgCl4 2− |
Mixture of Hg- chloro and hydroxy complex |
| Zn | ZnCl2 | Zn2+, ZnCl2, ZnCO3, Zn(HCO3)2, Zn(OH)2, ZnSO4 |
Zn2+, ZnCl2, ZnCO3, Zn(HCO3)2, Zn(OH)2, ZnSO4 |
Hydrated Zn2+ |
| Cu | CuCl2 | Carbonato and hydroxy complexes | CuCl2, Carbonato and hydroxy complexes | Cu2+, CuCO3 |
While metal bio-availability, uptake, and toxicity decrease in presence of natural dissolved organic ligands, metals differ in their behavior at high salinities. For example, in case of cadmium, speciation is highly dependent on the complexing ligands. In river water it exists either as CO3 − complex or free cation, and in oceanic waters it exists as highly soluble CdCl2, whereas in estuarine waters, it forms a strong CdCl+ complex which is biologically unavailable [41–43]. In case of silver, insoluble AgCl0 is formed in estuarine and oceanic waters, while under hypersaline conditions, soluble AgCl2-, AgCl3 2−, and AgCl4 3− complexes are formed [44]. Soluble HgCl− and sparingly soluble HgCl2 are the predominant complexes of Hg at high salinities [45]. HgCl2 and the soluble silver-chloro complexes are lipophilic and can easily diffuse through cellular membranes [46]. Zn and Cu exist as ZnCl+ and CuCl+ which coprecipitate at higher salinities, due to decrease in the net negative charge on macromolecular suspended particles and therefore are not available for uptake. Unlike Zn(II) and Cu(II), Fe(II), Co(II), Ni(II), and Mn(II) form weak complexes with Cl−, that easily dissociate and can be taken up by organisms [28]. Table 2 summarizes the bioavailability of metal-chloro complexes.
Table 2.
Bioavailability of metal-ligand complexes in hypersaline conditions depending upon the nature of the complex formed.
| Availability | Type of complex |
|---|---|
| Biologically unavailable | (i) Strong insoluble inorganic metal-chloro complexes (ZnCl2, CuCl2) |
| (ii) Soluble not easily dissociable metal-chloro complexes (CdCl2) | |
| (iii) Biosorbed metal complexes (i.e., metals sorbed on a biotic ligand) | |
|
| |
| Biologically available | (i) Strong soluble lipophilic inorganic metal-chloro complexes (AgCl2−, AgCl3 2−, AgCl4 3−, and HgCl2) |
| (ii) Weak metal-chloro complexes (Fe, Co, Ni, and Mn) | |
| (iii) Metal complexes sorbed to abiotic ligands | |
The bioavailability of chlorocomplexes also depends upon the type of biotic ligands present. Biotic ligands are the receptors on an organism where metal binding takes place which results in the manifestation of its toxic effects [47]. Metal receptors and ion transporters such as Na(II) and Ca(II) transporters present on fish gill surfaces, algal membranes, phytoplankton membranes, and so forth act as biotic ligands [47]. Binding of metals to biotic ligands is unaffected by changes in salinity. However, metal complexes adsorbed to abiotic ligands such as sediments are desorbed with increase in salinity. Thus, biotic ligands render the metals unavailable to other organisms for uptake. For example, in case of silver, with increase in salinity, desorption of Ag(I) complexed with suspended sediments and formation of soluble chlorocomplexes, which are bioavailable have been observed. However, biosorbed Ag(I) is not influenced by the increase in salinity, and desorption of Ag(I) does not occur [48].
The toxicity of a metal to microorganisms does not have a linear relationship with its concentration, and it depends strongly upon chemical speciation [49, 50]. For certain metals such as Zn(II) and Cu(II), complexation with chloride ions may result in precipitation at high salinities. Therefore, these complexes are not available to micro-organisms for uptake. However, metals such as Hg(II), Ag(II), Fe(II), Co(II), Ni(II), and Mn(II) either form lipophilic soluble chlorocomplexes or weak chlorocomplexes that dissociate easily and are thus available to organisms for uptake. Therefore, while studying metal resistance in haloarchaea, metal speciation and the bio-availability of metals should be taken into consideration.
3. Metal Resistance
Organisms inhabiting the metal polluted environments develop resistance mechanisms that enable efficient detoxification and transformation of toxic forms to nontoxic forms. The majority of bacteria and eukarya tolerate metals by a reduced influx/enhanced efflux [51, 52] or enzymatic detoxification sometimes followed by volatilization [6, 53]. Intracellular compartmentalization is observed only in eukaryotes [51]. Figure 1 shows the various mechanisms of metal resistance exhibited by all the three domains of life.
Figure 1.
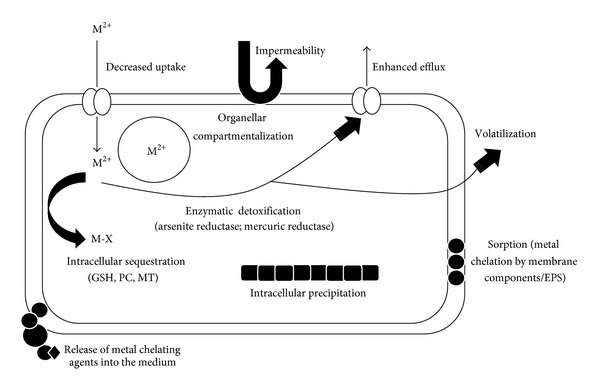
General mechanisms adapted by bacteria, eukaryotes, and archaea for metal resistance. All the three domains exhibit sorption of metals, volatilization, release of metal chelating compounds in the medium, enhanced efflux, impermeability, decreased uptake, enzymatic detoxification, and intracellular chelation as mechanisms for metal resistance. Organellar compartmentalization is observed only in eukaryotes, with the exception of magnetosomes in magnetotactic bacteria.
Intracellular chelation (Figure 1) by a variety of cysteine- (Cys-) rich metal-binding peptides like glutathione (GSH) and proteins like metallothioneins (MTs) and phytochelatins (PCs) also confers resistance to metals in many microbes [54]. MTs are genetically coded small molecular weight polypeptides that are classified based upon the number of Cys residues [55]. They typically have two Cys-rich domains that bind heavy metals through mercaptide bonds, giving these proteins a dumbbell-shaped conformation comprising an N-terminal β-domain that usually binds 3 metal ions and a C-terminal α-domain that binds 4 metal ions [56, 57]. PCs comprise (γ-GluCys)n-Gly where n is usually in the range of 2 to 5. They are enzymatically synthesized by PC synthase using GSH as the substrate [58, 59]. The thiol group of the cysteine residue in PCs sequesters heavy metals. Apart from these cysteine-rich peptides, cells may secrete other metal sequestering proteins like siderophores and DNA-binding protein from nutrient starved cells (Dps) (Figure 1). Siderophores are a class of low molecular weight iron chelating compounds which store iron and are overexpressed during conditions of stress or iron deficiency [60]. They are chemically diverse and generally possess oxygen-donor-type chelating functional groups [61]. Once chelated, the Fe-siderophore complex is transported to the periplasm through the energy-coupled transport involving TonB dependent transporters (TBDT) and the inner membrane TonB-complex, composed of TonB, ExbB, and ExbD. The Fe-siderophore can then be transported to the cytoplasm through ABC transporters like ferrichrome or permeases [62]. TonB protein is responsible for transducing cytoplasmic membrane energy to the outer membrane which results in TonB associating with or changing its affinity towards the outer membrane, while ExbB/D components antagonize this association or affinity of TonB to cytoplasmic membrane [63].Siderophores have also been shown to chelate metals other than Fe [62]. Dps are structurally homologous to ferritins, the primary iron storage/detoxification proteins, are usually expressed in response to excess of iron, and are found in all three domains of life [64]. These proteins have pores that are lined with acidic residues that bind cations like Fe(II) [65]. The binding of Fe(II) to Dps protects cells from oxidative stress by inhibiting the Fe-catalyzed production of hydroxyl radicals [66].
Haloarchaea have γ-glutamylcysteine (γ-GC) [67, 68] which is analogous to GSH and is involved in maintaining a reducing environment within the cell, overcoming oxidative and disulfide stress and detoxification of xenobiotics [69]. The thiol group of cysteine in γ-GC can chelate the toxic metal ions thereby conferring resistance. A unique phenomenon observed in archaea is the heavy metal-induced multimerization of metal chelating proteins such as CutA- and DpsA-like proteins that result in the precipitation of the protein-metal ion complex [23]. This precipitate resolubilizes and the multimers disintegrate when the metal stress decreases [70, 71]. Although these proteins are known to be involved in divalent metal tolerance in bacteria and eukaryotes, the multimerization of these proteins has been observed only in archaea. The aspartate residue in position 48 has been found to be critical for metal-induced multimerization and metal ion binding of CutA protein in Pyrococcus horikoshii. Substitution of Asp48 with alanine decreases the amount of aggregate formation [70]. Similarly, the multimeric non-haem ferritin DpsA-like protein of Halobacterium salinarum ensued from an assembly of 12 units and was found to sequester iron in response to the oxidative stress exerted by excess iron [72]. This protein was downregulated under iron-deficient conditions unlike the other dps that are upregulated under these conditions. It exhibits the features of non-haem bacterial ferritins that are expressed to sequester the excess iron. Their expression is repressed under conditions of iron starvation [73]. Overexpression of siderophores in haloarchaea may increase chelation in case of iron deficiency. On the other hand, repression of these siderophores in presence of excess iron may avoid uptake [68, 74]. MTs are absent in archaea [23].
Biosorption of metals by the organisms at the surface or by the exopolysaccharides (EPS) secreted to form the biofilms enables organisms to tolerate metals [75, 76]. Biofilm forming organisms exhibit an altered phenotype with respect to growth rate and gene transcription [77]. Haloarchaea synthesize EPS as a protective mechanism for survival under adverse conditions such as nutrient starvation, temperature fluctuation, and presence of toxic compounds [78]. Similarly the hyperthermophilic archaeon, Archaeoglobus fulgidus, was found to form a biofilm in response to toxic concentrations of metals, where the toxic metal was proposed to be trapped within the EPS matrix [78]. Thus, it is probable that under metal stress, haloarchaea may secrete EPS to make the cell impermeable to metals. In a study by Kawakami et al. (2007) [79], it has been found that Halobacterium salinarum CCM 2090 has a Ca(II)-dependent aggregation system, where the Ca(II) binds to certain aggregation factors present on the cell surface and induces ionic crossbridging between the EPS resulting in aggregation of the haloarchaeal cells. The presence of certain receptor proteins on the cell surface that interact with Ca(II) to form cell aggregates/flocs has also been demonstrated [79]. Four haloarchaeal genomes, Haloquadratum walsbyi, Haloarcula marismortui, Haloterrigena turkmenica, and Halobacterium sp. strain NRC-1, have been annotated with cbp encoding the cell surface calcium-binding acidic-repeat protein [80–83] that has been proposed to be the factor involved in Ca(II)-dependent aggregation, although its role in this process remains to be demonstrated. A similar dependence on Ca(II) and/or Fe(II) for biofilm formation is observed in Vibrio cholerae [84] and Pseudomonas aeruginosa [85]. Ca(II) is the twentieth element found in the fourth row of the periodic table, which could be replaced by other transition metal ions such as Mn(II), Cr(II), Fe(II), Co(II), Ni(II), Cu(II), and Zn(II), belonging to the same row. The distinctive electronic configuration of these metals, characterized by preferential filling of the 4s subshell before the 3d subshell, may be responsible for these metals substituting Ca(II) during aggregate formation [79]. Thus, tolerance to these metals may be mediated through binding with EPS. This view is supported by the observation that there is no aggregation in presence of certain other metals lacking this electronic configuration, such as Mg(II) and Sr(II) (alkali earth metals), Mo(II), Cd(II) and Sn(II) (fifth period), and Hg(II) and Pb(II) (sixth period) [79]. Aggregation in haloarchaeal cells results in formation of nonadherent floating multicellular aggregates which is different from biofilm formation, where the adherent multicellular structures are attached to diverse surfaces [86]. Recently, biofilm formation involving EPS (glycoconjugates and extracellular DNA) matrix was demonstrated in five haloarchaeal genera, Halobacterium, Haloferax, haloalkaliphilic Halorubrum, psychrotolerant Halorubrum, and a new genus of psychrotolerant haloarchaea isolated from Deep Lake, Antarctica [87]. Here Ca(II) ion did not have an effect on surface adhesion, suggesting the involvement of flagellar-twitching-motility-induced cellular aggregation and adhesion to substratum. Biofilm formation by other archaea like Pyrococcus furiosus, Sulfolobus solfataricus, and Methanococcus maripaludis involves cellular appendages such as archaeal type IV pili [88–90]. These are similar in structure and function to the type IV pili present in bacteria that facilitate cell-cell interactions, surface adhesion, and motility [91]. Archaeal type IV pili have been shown to be involved in biofilm formation by Haloferax, Halobacterium, and Halorubrum [87, 92, 93]. The biofilm formed may trap the metals within the EPS matrix and prevent the diffusion of metals inside the cell, thus conferring resistance to haloarchaea.
Most bacteria carry the resistance determinants for metals as operons, on their plasmids [94]. The metal resistance operons usually include genes for transporters and an enzyme for detoxification. Haloarchaea exhibit resistance mechanisms similar to those of bacteria. The ars operon conferring arsenite and arsenate resistance in Halobacterium sp. strain NRC-1 is present on one of its two plasmids [83]. A comparative genome analysis of bacteria and archaea revealed some common elements responsible for maintenance of metal homeostasis and resistance. P1B-type ATPases involved in cation transport with a high diversity in the N-terminal metal-binding motifs were found to be distributed throughout the bacterial and archaeal lineages [10]. Genome of Halobacterium salinarum NRC-1 was found to carry two distinct phylogenetic clusters, CopA1 (Cu(II) influx) and CopA2 (Cu(II) influx and efflux). These clusters were also found to span the entire diversity of the bacterial domain. Coombs and Barkay (2005) [10] have proposed that variation in N-terminal metal-binding motifs does not affect the metal translocation function of P1B-type ATPases and therefore concluded that divergence in consensus sequence of the N-terminal metal-binding motif might have been tolerated during evolution [10, 83]. But this is just one of the few studies on metal homeostasis in archaea. Similar studies understanding the phylogenetic variation within the family Halobacteriaceae will enable a better understanding of metal homeostasis, by giving a snapshot of substrate specificity, variation in active sites, and so forth [23].
3.1. Operons in Metal Resistance
Many metal resistance determinants have been characterized in the bacterial system [95–105], of which mer operon for mercury resistance [95], ars for arsenic resistance [97, 98], and cad operon for cadmium resistance [99] have been extensively studied. All archaea except haloarchaea have been shown to carry such metal resistance determinants [106, 107]. The most comprehensively studied among these are the mer operon of the thermoacidophilic archaeon Sulfolobus solfataricus [108, 109], ars operon of acidophilic archaeon Ferroplasma acidarmanus Fer1 [110] and Thermoplasma acidophilum [111], and the cop operon for copper resistance of Sulfolobus solfataricus P2 [112–114]. Although many heavy metal transporters like CbiNOQ, HemUV, NosFY, and so forth are present in haloarchaea, their arrangement in an operon has not been shown, except for the ArsA ATPase transporter as a part of ars operon for arsenic resistance in Halobacterium sp. strain NRC-1 [21].
Most haloarchaea have large plasmids in addition to their genomes (chromosomes) known as minichromosomes/megaplasmids. These minichromosomes harbor genes for antibiotic resistance or metal resistance that may be essential for haloarchaeal survival [115]. The pNRC100, one of the two minichromosomes of model organism Halobacterium sp. strain NRC-1, harbors the arsADRC gene cluster, responsible for conferring arsenate (As(V)) and arsenite (As(III))/antimonite (Sb(III)) resistance [83]. A fifth gene for arsenic resistance, arsB, is present on the main chromosome. The arsADRC operon was annotated for As(III) transport due to its homology to previously characterized genes [116], but later, by gene knockout studies, it was shown to confer resistance to As(III) and Sb(III) [21]. As(V) can be taken up by the cells through phosphate transporters (pit/pst) and As(III) by aquaglycerophorins (glycerophorin membrane transport proteins) [117] or hexose transporters [118]. As(V) is then converted to As(III) by arsenate reductase encoded by arsC [119]. arsA codes for P 1B-type ATPase transporters that help in extrusion of As(III)/Sb(III) from the cell. arsR and arsD encode trans-acting repressors of the operon. ArsR and ArsD bind to As(III)/Sb(III) resulting in expression of the arsA and arsC. Arsenate reductase encoded by arsC is expressed weakly in Halobacterium sp. strain NRC-1, and therefore deletion of arsC and arsADRC was found to be ineffective in conferring arsenate sensitivity [21]. The operon arsADRC was found to be inducible by arsenite and antimonite [21].
In bacteria, ArsB, an inner membrane protein, along with ArsA, the membrane-bound anion-transporting ATPase, forms the anion-conducting channel for arsenite extrusion [120]. Halobacterium sp. strain NRC-1 also harbors both arsA in ars operon on the megaplasmid pNRC100 and arsB on the main chromosome. However, arsB was found to play no role in arsenic resistance in this organism. Thus, it has been proposed that Halobacterium sp. strain NRC-1 harbors a novel transporter unrelated to ArsB but with a similar function [21]. Arsenic resistance in the Gram-negative acidophilic bacterium Acidthiobacillus ferrooxidans is determined by the chromosomally located arsCRBH operon comprising four genes [121]. The unique and common feature between the arsADRC and arsCRBH operons is the bidirectional nature of translation; that is, the arsAD and arsCR genes are translated in an opposite direction to arsRC and arsBH, respectively [21, 121] (Figure 2).
Figure 2.
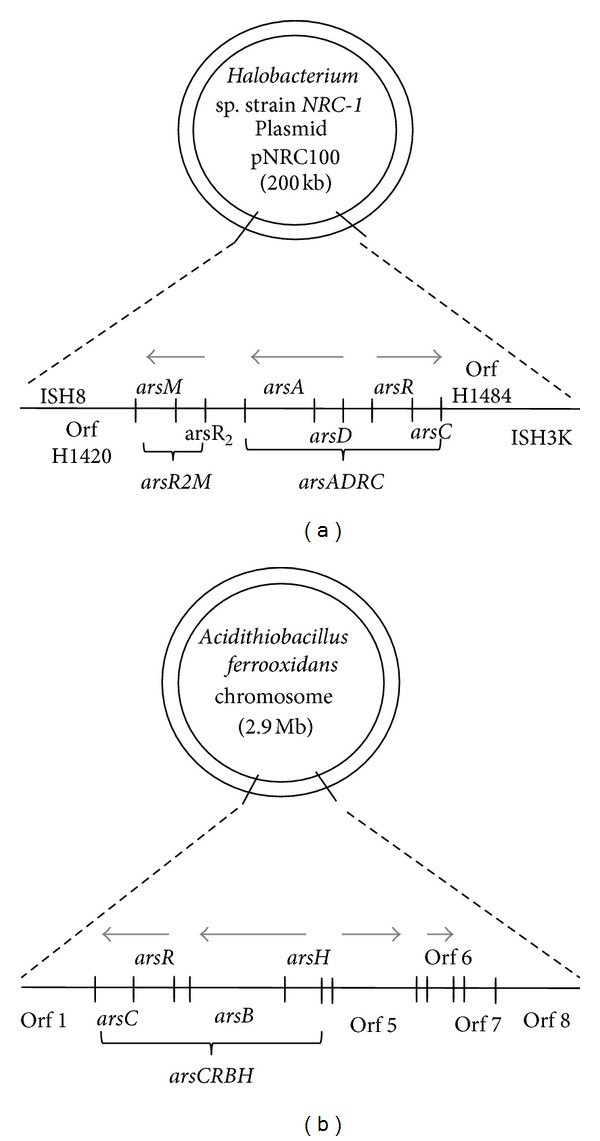
Arsenic resistance is determined by the presence of ars operon, which codes for an arsenite P1-type ATPases transporter ArsA/ArsB, an arsenate reductase ArsC, and arsenite responsive repressors ArsD and ArsR. The arsADRC and arsR2M operons are present on the plasmid in haloarchaeon Halobacterium sp. strain NRC-1 (a). The acidophilic bacterium Acidithiobacillus ferrooxidans has chromosomally encoded arsCRBH (b). The unique feature of these operons is the bidirectional nature transcription.
In Halobacterium sp. strain NRC-1, a second arsenite resistance operon, arsR2M, is present upstream of arsADRC on pNRC100 (Figure 2), where arsR2 is constitutively expressed while As(III)/Sb(III) induce the expression of arsM [21]. The arsR2 is analogous to arsR and arsM encodes a putative As(III)-methyltransferase very similar to human methyltransferases and S-adenosyl methionine-dependent methyltransferases of Magnetospirillum magnetotacticum. ArsM is involved in converting As(III) to methylated species like dimethylarsinate (DMA), trimethylarsine oxide (TMAO), or trimethylarsine (TMA) gas [122]. Deletion of arsM exhibited as increased sensitivity to arsenite but not towards arsenate or antimonite [21]. Thus, two possible mechanisms of As(III) resistance have been proposed to be conferred by arsM. First, the generation of a concentration gradient results in the movement of methylated arsenite (negatively charged/uncharged) out of the cell. Second, the volatile trimethylarsine formed diffuses out of the cell thus eliminating As(III) [123, 124]. Although arsenite methylation as a resistance mechanism is present in bacteria, arsM gene functions independently and is not a part of ars operon [123]. However, in Halobacterium sp. strain NRC-1, the arsM gene is present as a part of the arsR2M operon involved in arsenite resistance [21].
Mercury resistance in archaea and bacteria is conferred by the mer operon involved in detection, regulation, transport, and reduction of Hg(II) [125, 126]. One of the best studied mercury resistance operons in Archaea is the merRHAI operon of thermoacidophilic archaeon Sulfolobus solfataricus [108, 109]. The operon is under the control of the regulator MerR, which represses the operon in absence of Hg(II) and enhances transcription in its presence. MerH is the metallochaperone with a TRASH (trafficking, resistance, and sensing of heavy metals) domain that binds Hg(II), and MerA is the mercuric reductase for reduction and detoxification to volatile Hg(0) [108, 109]. Some mer operons carry additional mer genes, notably merB, an organomercurial lyase, that cleaves the C-Hg bonds of organomercurials, and the released Hg(II) is reduced to Hg(0) by MerA [126]. Among all haloarchaeal genomes sequenced to date, only Halobacterium sp. strain NRC-1 and Haloterrigena turkmenica have been annotated with merA and merB genes, respectively [83, 127].
3.2. Transporters in Metal Resistance
Membrane transporters may act as the first line of defense against toxic or heavy metals. In order to exert their toxicity, metals need to gain entry within the cell. Thus, the organism may downregulate the transporters responsible for influx or induce the expression of efflux pumps to enable faster removal of toxic metals from within the cell [76]. The use of these membrane transporters and efflux pumps is one of the most common mechanisms of resistance to inorganic ions in microbes.
Both influx and efflux types of transporters for various metals have been annotated in all haloarchaeal genomes sequenced to date (Table 3). The following membrane transporters have been implicated in haloarchaeal metal resistance.
Table 3.
Annotated transporters for various metals in haloarchaeal genomes. Ten haloarchaeal genomes have been completely sequenced while others are partially sequenced. All these organisms have been annotated with transporters belonging to the following type of transporters-cation efflux type, P1B-type ATPases, cation diffusion facilitator (CDF) family, and ATP-binding cassette (ABC) family. The most abundant transporters were for iron followed by copper. Only one haloarchaeon, Halogeometricum borinquense, was annotated with silver transporters [135].
| Transporters for metals | H.s. | H.m. | H.v. | H.w. | H.l. | H.mu. | H.u. | H.b. | H.t. | H.j. | N.p. | N.m. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Copper | + | + |
+ |
+ |
+ |
+ |
+ |
+ |
· | + |
+ |
+ |
| Iron | + |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| Manganese | · | + |
+ |
+ |
+ |
· | · | · | + |
· | + |
+ |
| Zinc | + |
+ |
+ |
+ |
+ |
+ |
+ |
· | + |
+ |
+ |
+ |
| Cobalt | + |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| Nickel | + |
· | · | + |
· | · | · | + |
· | + |
+ |
+ |
| Molybdenum | · | + |
+ |
· | · | · | · | · | · | · | · | · |
| Arsenic | + |
+ |
+ |
+ |
+ |
+ |
+ |
· | + |
+ |
· | · |
| Cadmium | + |
+ |
· | + |
· | · | · | · | · | · | + |
+ |
| Magnesium | · | + |
+ |
· | · | · | + |
+ |
· | + |
· | · |
| Silver | · | · | · | · | · | · | · | + |
· | · | · | · |
(+) present; (·) not annotated yet; H.s., Halobacterium sp. strain NRC-1; H.m., Haloarcula marismortui; H.v., Haloferax volcanii; H.w., Haloquadratum walsbyi; H.l., Halorubrum lacusprofundi; H.mu., Halomicrobium mukohataei; H.u., Halorhabdus utahensis; H.b., Halogeometricum borinquense; H.t., Haloterrigena turkmenica; H.j., Halalkalicoccus jeotgali; N.p., Natronomonas pharaonis; N.m., Natrialba magadii.
3.2.1. P1B-Type ATPases
The P1B-type ATPases are a large family of integral membrane proteins driven by ATP hydrolysis [128]. Members of this family are of vital importance to all kingdoms of life, as they generate and maintain electrochemical gradients across membranes by transporting cations and heavy metals [129]. A wide variety of heavy metal ions like Mg(II), Ca(II), Cu(II), Ag(II), Zn(II), and Cd(II) act as substrates to these ATPases [130]. These transporters serve the purpose of uptake (import) of essential elements and efflux (export) of toxic elements, thus conferring resistance to the expelled metal ion [131, 132]. All haloarchaeal genomes have been annotated with metal transporting ATPases.
A putative Cd(II)-efflux ATPase was annotated on Halobacterium sp. strain NRC-1 genome [83]. In a system level analysis of Halobacterium sp. strain NRC-1, the functionality and role of such transporters in metal resistance was shown [22]. They exhibited upregulation of yvgX, a P 1B-type ATPase, in response to Cu(II) and Zn(II) metal stress. In bacteria, the yvgX family is known to encode two kinds of CopA proteins, CopA1 and CopA2 [133]. CopA1 is essential for copper influx and tolerance, while CopA2 is involved in the influx/efflux of Cu and its transport to Cu-containing enzyme cytochrome oxidase c [133, 134]. A diverse range of organisms contain CopA2-like proteins, suggesting that coding genes appeared early in evolution via gene duplication or horizontal transfer but were kept only in some organisms for a specific biological function [134]. A comparative genome analysis for ATPases in bacteria and archaea showed the preference for CopA2 over CopA1 [10]. It has been proposed that CopA2 may represent the ancestral form of CopA1 protein that may have coevolved with the other metal influx proteins [10]. The yvgX of Halobacterium sp. strain NRC-1 was found to be more specific for Cu(II) efflux family as the ΔyvgX strain was susceptible to Cu(II) and not to Zn(II) or Co(II) and therefore belongs to the CopA2 family of proteins. CopA2 is also found in other haloarchaea like Haloarcula marismortui, Haloarcula hispanica, and Haloquadratum walsbyi [135].
The As(III)/Sb(III) transporting P1B-type ATPase, ArsA discussed in Section 3.1, is present in almost all haloarchaea sequenced to date, including Halobacterium sp. strain NRC-1, Halalkalicoccus jeotgali, Haloarcula hispanica, Natrialba magadii, Haloarcula marismortui, Haloquadratum walsbyi, and Natronomonas pharaonis [135]. ArsB was found to play no role in arsenite resistance in Halobacterium sp. strain NRC-1 [21].
Heavy metal cation-transporting CPx P1B-type ATPases are of two types, that is, Cu-CPx-type-ATPases involved in efflux of monovalent cations, Cu(I) and Ag(I), and Zn-CPx-type ATPases involved in the efflux of divalent cations of Zn, Cd and Pb [137–139]. However, Cu-CPx-type ATPases have also been shown to be involved in uptake of copper to meet cellular demands [140, 141]. The cpx gene that encodes CPx P1B-type ATPases was found to be downregulated by Fe(II), Cu(II), and Ni(II) to avoid influx in Halobacterium sp. strain NRC-1 [22]. This mechanism of resistance involving the downregulation of uptake systems avoids toxic metal buildup within the cell.
3.2.2. Cation Diffusion Facilitators (CDF Family) Metal Transporters
The CDF family of transport proteins is ubiquitously present in all three domains of life [142]. Although CDFs are primarily Zn(II) efflux pumps, bacterial CDFs have been shown to transport Hg(II), Pb(II), Zn(II), Co(II), Fe(II), and Cd(II) from the cytoplasm to the outside of the cell or into subcellular compartments [132, 143]. Based upon their substrate specificity, CDFs have been classified as Zn(II)-CDF, Fe/Zn-CDF, and Mn-CDF [144]. They usually possess six transmembrane domains (TMDs) with a cytoplasmic N- and C-terminal and a histidine loop of variable length between TMD IV and V [145, 146]. The amphipathic domains TMD I, II, V, and VI are involved in metal transfer and are the most conserved, while the hydrophobic TMD III and IV are critical for zinc specificity and mutations within these domains alter substrate specificity [144]. All the proteins of this family of transporters possess a characteristic cation efflux C-terminal domain [147]. These kinds of transporters serve as secondary cation filters in bacteria [132]. The genome of Halobacterium sp. strain NRC-1 was annotated with putative CDF Cd (II) transporter ZntX, which confers resistance against Ni(II), Cu(II), and Zn(II) besides Cd(II) [22]. The role of Znt family of CDFs in Cu(II) and/or Zn(II) homeostasis and resistance has been discussed in detail by Haney et al. (2005) [145]. The upregulation of ZntA in response to heavy metals (Cu and/or Zn) and poor growth of ΔzntA strain in presence of Ni(II), Cu(II), Zn(II), and Cd(II) have confirmed the role of this transporter in metal resistance [22]. The broad specificity of this transporter to various metals has been putatively attributed to the preference of metals by zntA based on charge and species rather than size [148]. Haloarcula hispanica and Haloarcula marismortui have also been annotated with ZntA for Zn(II) transport. A putative CDF family protein has also been found on the chromosome of Natrialba magadii for inorganic metal ion transport [135].
3.2.3. ATP-Binding Cassette (ABC) Transporters
The multisubunit ABC transporters are one of the largest protein families with a variety of physiological functions. These transporters are ubiquitously present in all living forms from bacteria to eukaryotes including archaea. They are involved in various functions such as nutrient uptake [149], oligopeptide and protein transport [150], metal extrusion [151, 152], and drug efflux [153, 154].
Although many ABC transporter genes for a variety of substrates have been annotated in all the 10 haloarchaeal genomes sequenced to date, experimentally, very few have been shown to be functional. ABC transporters for sugar and polypeptide have been found in Haloferax volcanii [155], Haloarcula marismortui [81], Halobacterium sp. NRC-1 [83], Natronomonas pharaonis [156], and Haloquadratum walsbyi [80]. All haloarchaea possess at least one copy of metal ion ABC transporter. Some of the ABC transporters in Halobacterium sp. NRC-1 with their functions are listed in Table 4. Many of the ABC transporters are metal ion transporters such as cbiNOQ for Co(II) transport [157], hemUV for iron uptake [158, 159], nosFY for copper [160], and zurMA for zinc transport (Figure 3). Although most of the ABC transporter proteins exhibit stringent specificity towards their substrate, a few, such as phosphate transporters, oligopeptide transporters, and dipeptide transporters, have been shown to have multiple specificities and were found to be differentially regulated by more than one metal ion. This has been proposed to facilitate transport of metal ions in addition to their usual function [150]. Kaur et al. (2006) [22] have shown that deletion of transporters like phoX (phosphate transport), appA (peptide transport), and ycdH (Mn(II) transport) along with two putative subunits of Fe(II) transport system does not prove deleterious for Halobacterium sp. NRC-1. Due to the large repertoire of ABC transport proteins, they concluded that deletion/mutation of a single ABC transporter is easily managed by the organism by substituting the deleted/mutated ABC transporter product with functional ABC transporter product of similar role.
Table 4.
ABC transporters with various functions present in some model haloarchaea. ABC transporters have three components that together help in uptake of nutrients or for the efflux of extracellular proteins, enzymes, and toxicants. Permease is the transmembrane component and is responsible for the uptake of ions or macromolecules, while the ATP-binding component is the water soluble domain that binds ATP. Substrate binding at the substrate binding site brings about a conformational change in the ATP-binding component resulting in ATP hydrolysis. The presence or absence of the three components of ABC transporters for sugar, peptide, amino acids, phosphate, and iron transport is shown in the following table [135, 136].
| ABC transporters | H.s. | H.v. | H.m. | H.w. | H.l. | N.p. | N.m. |
|---|---|---|---|---|---|---|---|
| Sugar transport system components | |||||||
| Permease | + | + | + | + | − | − | − |
| ATP binding | + | + | + | + | − | − | − |
| Substrate binding | − | − | + | + | + | − | − |
| Phosphate transport system components | |||||||
| Permease | + | + | + | + | + | + | + |
| ATP binding | − | + | + | + | + | + | + |
| Susbtrate binding | + | + | + | + | + | + | + |
| Dipeptide/oligopeptide transport system components | |||||||
| Permease | + | + | + | + | − | + | − |
| ATP binding | + | + | + | + | + | + | + |
| Susbtrate binding | − | − | + | + | − | + | + |
| Amino acid transport system components | |||||||
| Permease | − | + | + | + | − | + | − |
| ATP binding | + | + | + | + | − | + | − |
| Susbtrate binding | − | + | − | + | + | + | + |
| Fe(III) transport system components | |||||||
| Permease | + | + | + | + | − | + | + |
| ATP binding | + | + | − | − | − | + | − |
| Susbtrate binding | − | − | − | + | + | + | − |
H.s., Halobacterium sp. strain NRC-1; H.v., Haloferax volcanii; H.m., Haloarcula marismortui; H.w. Haloquadratum walsbyi; H.l., Halorubrum lacusprofundi; N.p, Natronomonas pharaonis; Natrialba magadii; (+) present; (−) absent.
Figure 3.
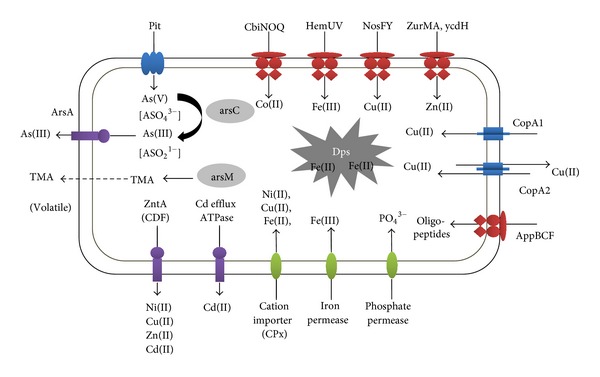
Various transporters playing a role in metal transport, homeostasis maintenance, and resistance in Halobacterium sp. strain NRC-1, reported to date. The efflux pumps of ATPases and CDF family involved in Cd, Ni, Cu, Zn, Cd, and arsenite transport are represented in purple. ABC transporters (represented in red) involved in metal uptake are many. Certain toxic metals that do not have a dedicated uptake system may gain entry into the cell through other ABC transporters like oligopeptide and phosphate transporters. For example, the arsenate oxyanion gains entry into the cell through the pit/pst phosphate transporters due to its structural similarity to phosphate. The metal ions upon uptake can be detoxified either by enzymatic detoxification (ArsC and ArsM) or by chelation by peptides like Dps (DNA-binding protein of nutrient starved cells).
The differential regulation of all three classes of metal transporters discussed above is in congruence with the general norm of micro-organisms utilizing enhanced efflux or decreased influx to resist metals. However, the P1B-type ATPases and CDF family have a greater role in maintaining metal homeostasis than the ABC transporters in haloarchaea [22].
3.3. Transcriptional Changes in Response to Metal Stress
Under unfavourable conditions of growth, all organisms make adjustments at the system level to overcome the stress imposed by the stressor. Presence of heavy metals in their environment triggers global transcriptional regulations either to prevent their entry into the cell or to transform the metal to nontoxic form. This response can be transitory, with perturbations of a few genes within minutes of metal exposure, but once the cell acclimatizes to the new environment, the transcript levels of some early response genes return to preperturbation levels. The early response to a stressor usually results in the upregulation of transcription and translation. As a consequence, the transcripts damaged due to the stressor are replaced and new proteins are synthesized [161, 162].
In haloarchaea, only one study on transcriptional changes in response to heavy metal stress (Fe(II), Cu(II), Co(II), Ni(II), Zn(II), and Mn(II)) in Halobacterium sp. strain NRC-1 has been reported [22]. Besides studying the transcriptional changes by microarray analysis and mutant constructions, Kaur et al. (2006) [22] elucidated a systemic overview to metal stress response, thus providing a snapshot of various mechanisms involved in stress management. A total of 623 genes were found to be differentially regulated in presence of any of the six transition metals used for the study. Around 69% of these genes were early response genes; that is, they exhibited deviation from normal transcript levels within 0–25 minutes of metal exposure. However, 91% of these early response genes transcript levels reverted to preperturbation levels within 25–40 minutes. These included transcriptional regulator genes, transporter genes for phosphate, metals, and peptides, ribosomal protein genes, and protein export genes. Therefore, once the various damaged transcripts and proteins were replaced with the new proteins for managing metal stress and acclimatizing the cells to the new environment, the early response genes were found to revert to preperturbation levels [22].
One of the major toxic effects elicited by heavy metals is the rapid generation of reactive oxygen species (ROS) that damages the cellular machinery [136, 163]. The ROS are usually scavenged by specific enzymatic detoxification systems like superoxide dismutases (SOD), peroxidases, dehydrogenases, and antioxidants like glutathione (GSH) [164]. Therefore, it follows that the genes involved in oxidative stress management are differentially regulated early in the stress management. Model haloarchaeal genomes, including Halobacterium salinarum and Haloferax volcanii, have been annotated with SOD [165] and catalase-peroxidase (KatG) genes [166]. Metal-induced ROS results in an early increase in the levels of transcripts of genes related to oxidative stress management like dehydrogenases and peroxidases in Halobacterium sp. strain NRC-1 [22].
Few of the early response genes found to be differentially regulated were transcriptional regulators like tfbB and SirR. TfbB is the transcription initiation factor IIB. Its upregulation indicates a global response towards stress by increasing the rate of transcription to increase protein turnover. Similarly, the upregulation of SirR (silent information regulator) repressed the active uptake of Mn(II), thus providing the organism the ability to overcome the stress. Similar upregulation has also been observed in certain bacteria and yeast [167, 168]. Staphylococcus aureus and Staphylococcus epidermidis were shown to carry several copies of sirR genes that act as divalent metal cation-dependent transcriptional repressor [169]. cbiN, cbiM, and cbiQ involved in cobalt transport and zurM, zurA, and ycdH that encode Mn/Fe ABC-transporters were predicted to be putatively regulated by sirR in Halobacterium sp. strain NRC-1 [170]. This was found to be consistent with the observation that sirR is essential for survival during metal-induced stress. This was evident from the upregulation of Mn(II) uptake genes zurM, zurA, and ycdH in ΔsirR strain as compared to parent strain in Halobacterium sp. strain NRC-1. Thus, in the haloarchaeon Halobacterium sp. strain NRC-1, sirR acts as a Mn(II)-dependent autorepressor [22].
A putative Lrp (leucine-responsive regulatory protein) family protein VNG1197C was reported to upregulate the Cu(II)-P1B type ATPases gene yvgX in Halobacterium sp. strain NRC-1 [22]. VNG1197C was found to be a Cu(II)-dependent transcriptional activator carrying a metal-binding TRASH (trafficking, resistance, and sensing of heavy metals) domain. Kaur et al. (2006) [22] proposed that putative metallochaperones VNG0702H and/or VNG2581H deliver Cu(II)/Zn(II) to the TRASH domain of VNG1197C. This binding activates the transcription of yvgX as well as the metallochaperones, thus providing Cu(II) resistance to Halobacterium sp. strain NRC-1. A similar pattern involving a metallochaperone (CopM), a transcriptional regulator with C-terminal TRASH domain (CopT), and a P-type Cu(II) exporting ATPase (CopA) forming the cop gene cluster for Cu(II) resistance has been described in Sulfolobus solfataricus, a thermoacidophilic crenarchaeon [113, 114].
Thus, organisms have the ability to differentiate between metal ions and therefore elicit responses that enable better survival. These responses could be local or global but in effect would be to efficiently handle the stress. The transcriptional regulation exhibited by Halobacterium sp. strain NRC-1 is an example of how metal homeostasis is maintained. Transient changes in transcripts to resist metals may play a major role in haloarchaea.
4. Conclusion
Haloarchaea encounter metals in their natural environment and utilize some of these metals for various key physiological functions. However, at higher concentrations, these metals can be toxic, and thus haloarchaea exhibit metal resistance mechanisms. Knowledge about metal physiology in haloarchaea is cursory, and therefore global studies for gaining insights into the metabolic regulations in response to metal stress are required. Metal physiology studies in model haloarchaeon Halobacterium sp. strain NRC-1 show that they have the ability to elicit a tailor-made response to metal stress. Other genera of the halophilic Euryarchaeota have not yet been subjected to such detailed studies with regard to metal homeostasis and resistance. The development of standard molecular and genetic tools for haloarchaea may facilitate better understanding of the various components involved in metal resistance including detoxifying enzymes, metallochaperones, and metal chelators and transporters. While assessing metal resistance in haloarchaea, the metal speciation should be given importance, as the metal might be unavailable to the cell for uptake, thus giving a higher MIC value. Therefore, beside understanding the molecular mechanisms underlying the resistance to metals, metal speciation and bio-availability studies should be carried out to obtain a complete picture. Further, this could facilitate the use of haloarchaea for bioremediation of metal-polluted hypersaline environments.
Acknowledgments
The authors thank the Ministry of Earth Science (MoES and the Government of India for their funding of the project MoES/11-MRDF/1/38/P/10-PC-III.
References
- 1.Gray HB, Ellis WR., Jr. Electron transfer. In: Bertini I, Gray HB, Lippard SJ, Valentine JS, editors. Bioinorganic Chemistry. Mill Valley, Calif, USA: University Science Books; 2006. pp. 315–364. [Google Scholar]
- 2.Bott AW. Redox properties of electron transfer metalloproteins. Current Separations. 1999;18(2):47–54. [Google Scholar]
- 3.Messerschmidt A, Huber R, Wieghart K, Poulos T. Handbook of Metalloproteins. 1–3. New York, NY, USA: Wiley; 2005. [Google Scholar]
- 4.Gray HB, Stiefel EI, Valentine JS, Bertini I. Biological Inorganic Chemistry: Structure and Reactivity. Herndon, Va, USA: University Science Books; 2007. [Google Scholar]
- 5.Bruins MR, Kapil S, Oehme FW. Microbial resistance to metals in the environment. Ecotoxicology and Environmental Safety. 2000;45(3):198–207. doi: 10.1006/eesa.1999.1860. [DOI] [PubMed] [Google Scholar]
- 6.Nies DH. Microbial heavy-metal resistance. Applied Microbiology and Biotechnology. 1999;51(6):730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 7.Hanikenne M, Krämer U, Demoulin V, Baurain D. A comparative inventory of metal transporters in the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschizon merolae . Plant Physiology. 2005;137(2):428–446. doi: 10.1104/pp.104.054189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chemical Reviews. 2009;109(10):4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolchi A, Ruotolo R, Marchini G, et al. Genome-wide inventory of metal homeostasis-related gene products including a functional phytochelatin synthase in the hypogeous mycorrhizal fungus Tuber melanosporum. Fungal Genetics and Biology. 2011;48(6):573–584. doi: 10.1016/j.fgb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Coombs JM, Barkay T. New findings on evolution of metal homeostasis genes: evidence from comparative genome analysis of bacteria and archaea. Applied and Environmental Microbiology. 2005;71(11):7083–7091. doi: 10.1128/AEM.71.11.7083-7091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartolucci S, Contursi P, Fiorentin G, Limauro D, Pedone E. Responding to toxic compounds: a genomic and functional overview of Archaea. Frontiers in Bioscience. 2013;18:165–189. doi: 10.2741/4094. [DOI] [PubMed] [Google Scholar]
- 12.Oren A. Taxonomy of the family Halobacteriaceae: a paradigm for changing concepts in prokaryote systematics. International Journal of Systematic and Evolutionary Microbiology. 2012;62(2):263–271. doi: 10.1099/ijs.0.038653-0. [DOI] [PubMed] [Google Scholar]
- 13.Zafrilla B, Martínez-Espinosa RM, Alonso MA, Bonete MJ. Biodiversity of Archaea and floral of two inland saltern ecosystems in the Alto Vinalopó Valley, Spain. Saline Systems. 2010;6(1):p. 10. doi: 10.1186/1746-1448-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bochiwal C. Adaptation of Low-Salt Tolerant Haloarchaea in Estuarine Environments. Essex, UK: University of Essex; 2009. [Google Scholar]
- 15.Sholkovitz ER. Flocculation of dissolved organic and inorganic matter during the mixing of river water and seawater. Geochimica et Cosmochimica Acta. 1976;40(7):831–845. [Google Scholar]
- 16.Kumar CSR, Joseph MM, Kumar TRG, Renjith KR, Manju MN, Chandramohanakumar N. Spatial variability and contamination of heavy metals in the inter-tidal systems of a tropical environment. International Journal of Environmental Research. 2010;4(4):691–700. [Google Scholar]
- 17.Pereira F, Kerkar S, Krishnan KP. Bacterial response to dynamic metal concentrations in the surface sediments of a solar saltern (Goa, India) Environmental Monitoring and Assessment. 2012 doi: 10.1007/s10661-012-2814-7. [DOI] [PubMed] [Google Scholar]
- 18.Chapman PM, Wang F. Assessing sediment contamination in estuaries. Environmental Toxicology and Chemistry. 2001;20(1):3–22. [PubMed] [Google Scholar]
- 19.Tabak HH, Lens P, Van Hullebusch ED, Dejonghe W. Developments in bioremediation of soils and sediments polluted with metals and radionuclides-1. Microbial processes and mechanisms affecting bioremediation of metal contamination and influencing metal toxicity and transport. Reviews in Environmental Science and Biotechnology. 2005;4(3):115–156. [Google Scholar]
- 20.Ross SM. Retention, transformation and mobility of toxic metals in soils. In: Ross SM, editor. Toxic Metals in Soil-Plant Systems. Chichester, UK: John Wiley & Sons; 1994. pp. 63–152. [Google Scholar]
- 21.Wang G, Kennedy SP, Fasiludeen S, Rensing C, DasSarma S. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. Journal of Bacteriology. 2004;186(10):3187–3194. doi: 10.1128/JB.186.10.3187-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur A, Pan M, Meislin M, Facciotti MT, El-Gewely R, Baliga NS. A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Research. 2006;16(7):841–854. doi: 10.1101/gr.5189606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bini E. Archaeal transformation of metals in the environment. FEMS Microbiology Ecology. 2010;73(1):1–16. doi: 10.1111/j.1574-6941.2010.00876.x. [DOI] [PubMed] [Google Scholar]
- 24.Sehgal SN, Gibbons NE. Effect of some metal ions on the growth of Halobacterium cutirubrum . Canadian Journal of Microbiology. 1960;6:165–169. doi: 10.1139/m60-018. [DOI] [PubMed] [Google Scholar]
- 25.Nieto JJ, Ventosa A, Ruiz-Berraquero F. Susceptibility of halobacteria to heavy metals. Applied and Environmental Microbiology. 1987;53(5):1199–1202. doi: 10.1128/aem.53.5.1199-1202.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popescu G, Dumitru L. Biosorption of some heavy metals from media with high salt concentrations by halophilic archaea. Biotechnology and Biotechnology Equipment. 2009;23(2):791–795. [Google Scholar]
- 27.Worms I, Simon DF, Hassler CS, Wilkinson KJ. Bioavailability of trace metals to aquatic microorganisms: importance of chemical, biological and physical processes on biouptake. Biochimie. 2006;88(11):1721–1731. doi: 10.1016/j.biochi.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Byrne RH. Inorganic speciation of dissolved elements in seawater: the influence of pH on concentration ratios. Geochemical Transactions. 2002;3:11–16. doi: 10.1186/1467-4866-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markich SJ, Brown PL, Batley GE, Apte SC, Stauber JL. Incorporating metal speciation and bioavailability into water quality guidelines for protecting aquatic ecosystems. Australian Journal of Ecotoxicology. 2001;7(2):109–122. [Google Scholar]
- 30.Campbell PGC. Interactions between trace metals and aquatic organisms: a critique of the free-ion activity model. In: Tessier A, Turner DR, editors. Metal Speciation and Bioavailability in Aquatic Systems. Chichester, UK: Wiley; 1995. pp. 45–102. [Google Scholar]
- 31.DasSarma S, Arora P. Encyclopedia of Life Sciences. Vol. 8. London, UK: Nature Publishing Group; 2002. Halophiles; pp. 458–466. [Google Scholar]
- 32.Pereira F, Krishnan KP, Sinha RK, Kerkar S. Insights on metal-microbe interactions in Bacillus sp. and Chromohalobacter sp. from a solar saltern. Journal of Ecobiotechnology. 2012;4(1):14–24. [Google Scholar]
- 33.Florence TM. Development of physico-chemical speciation procedures to investigate the toxicity of copper, lead, cadmium and zinc towards aquatic biota. Analytica Chimica Acta. 1982;141:73–94. [Google Scholar]
- 34.Tipping E, Lofts S, Lawlor AJ. Modelling the chemical speciation of trace metals in the surface waters of the Humber system. Science of the Total Environment. 1998;210-211:63–77. doi: 10.1016/s0048-9697(98)00045-x. [DOI] [PubMed] [Google Scholar]
- 35.Han Bin Xue, Sigg L. Zinc speciation in lake waters and its determination by ligand exchange with EDTA and differential pulse anodic stripping voltammetry. Analytica Chimica Acta. 1994;284(3):505–515. [Google Scholar]
- 36.Zamuda CD, Wright DA, Smucker RA. The importance of dissolved organic compounds in the accumulation of copper by the American Oyster, Crassostrea virginica. Marine Environmental Research. 1985;16(1):1–12. [Google Scholar]
- 37.Nurnberg HW. Investigation on heavy metal speciation in natural waters by voltammetric procedures. Fresenius’ Zeitschrift für Analytische Chemie. 1983;316(6):557–565. [Google Scholar]
- 38.L’her Roux LL, Le Roux SL, Appriou P. Behaviour and speciation of metallic species Cu, Cd, Mn and Fe during estuarine mixing. Marine Pollution Bulletin. 1998;36(1):56–64. [Google Scholar]
- 39.Scoullos MJ, Pavlidou AS. Speciation studies of trace metals in the gulf of Elefsis, Greece. Croatica Chemica Acta. 1997;70(1):289–307. [Google Scholar]
- 40.Sunda WG, Gillespie PA. The response of a marine bacterium to cupric ion and its use to estimate cupric ion activity in seawater. Journal of Marine Research. 1979;37:761–772. [Google Scholar]
- 41.Turner DR. Speciation and cycling of arsenic, cadmium, lead and mercury in natural waters. In: Hutchinson TC, Meema KM, editors. Lead, Mercury, Cadmium and Arsenic in the Environment. New York, NY, USA: John Wiley and Sons Ltd.; 1987. pp. 175–186. [Google Scholar]
- 42.Peakall D, Burger J. Methodologies for assessing exposure to metals: speciation, bioavailability of metals, and ecological host factors. Ecotoxicology and Environmental Safety. 2003;56(1):110–121. doi: 10.1016/s0147-6513(03)00055-1. [DOI] [PubMed] [Google Scholar]
- 43.Jensen A, Bro-Rasmussen F. Environmental cadmium in Europe. Reviews of Environmental Contamination and Toxicology. 1992;125:101–181. doi: 10.1007/978-1-4612-2890-5_3. [DOI] [PubMed] [Google Scholar]
- 44.Luoma S. Fate, bioavailability and toxicity of silver in estuarine environments. In: Andren A, Bober T, editors. Proceedings of the 2nd International Conference: Transport, Fate and Effects of Silver in the Environment; September 1994; Madison, Wis, USA. University of Wisconsin Sea Grant Institute; pp. 11151–14155. [Google Scholar]
- 45.Noegrohati S. Sorption-desorption characteristics of heavy metals and their availability from the sediment of Segara anakan estuary. Indonesian Journal of Chemistry. 2005;5(3):236–244. [Google Scholar]
- 46.Engel DW, Sunda W, Fowler BA. Factors affecting trace metal uptake and toxicity to estuarine organisms. I. Environmental parameters. In: Vernberg FJ, Calabrese A, Thurberg FT, Vernberg WB, editors. Biological Monitoring of Marine Pollutants. New York, NY, USA: Academic Press; 1981. [Google Scholar]
- 47.Paquin PR, Zoltay V, Winfield RP, et al. Extension of the biotic ligand model of acute toxicity to a physiologically-based model of the survival time of rainbow trout (Oncorhynchus mykiss) exposed to silver. Comparative Biochemistry and Physiology C. 2002;133(1-2):305–343. doi: 10.1016/s1532-0456(02)00105-9. [DOI] [PubMed] [Google Scholar]
- 48.Sanders JG, Abbe GR. The role of suspended sediments and phytoplankton in the partitioning and transport of silver in estuaries. Continental Shelf Research. 1987;7(11-12):1357–1361. [Google Scholar]
- 49.Blust R, Baillieul M, Decleir W. Effect of total cadmium and organic complexing on the uptake of cadmium by the brine shrimp, Artemia franciscana . Marine Biology. 1995;123(1):65–73. [Google Scholar]
- 50.Satish Nair P, Robinson WE. Histidine-rich glycoprotein in the blood of the bivalve Mytilus edulis: role in cadmium speciation and cadmium transfer to the kidney. Aquatic Toxicology. 2001;52(2):133–142. doi: 10.1016/s0166-445x(00)00138-7. [DOI] [PubMed] [Google Scholar]
- 51.Gadd GM. Interactions of fungi with toxic metals. New Phytologist. 1993;124(1):25–60. [Google Scholar]
- 52.Blaudez D, Jacob C, Turnau K, et al. Differential responses of ectomycorrhizal fungi to heavy metals in vitro. Mycological Research. 2000;104(11):1366–1371. [Google Scholar]
- 53.Silver S, Phung LT. Heavy metals, bacterial resistance. In: Schaechter M, editor. Encyclopedia of Microbiology. Oxford, UK: Elsevier; 2009. pp. 220–227. [Google Scholar]
- 54.Avery SV. Metal toxicity in yeasts and the role of oxidative stress. Advances in Applied Microbiology. 2001;49:111–142. doi: 10.1016/s0065-2164(01)49011-3. [DOI] [PubMed] [Google Scholar]
- 55.Cobbett C, Goldsbrough P. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annual Review of Plant Biology. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- 56.Cherian GM, Chan HM. Biological functions of metallothioneins- a review. In: Suzuki KT, Imura N, Kimura M, editors. Metallothionein III: Biological Roles and Medical Implications. Basel, The Switzerland: Birkhauser Verlag; 1993. pp. 87–109. [Google Scholar]
- 57.Wang H, Zhang Q, Cai B, et al. Solution structure and dynamics of human metallothionein-3 (MT-3) FEBS Letters. 2006;580(3):795–800. doi: 10.1016/j.febslet.2005.12.099. [DOI] [PubMed] [Google Scholar]
- 58.Glaeser H, Coblenz A, Kruczek R, Ruttke I, Ebert-Jung A, Wolf K. Glutathione metabolism and heavy metal detoxification in Schizosaccharomyces pombe. Isolation and characterization of glutathione-deficient, cadmium-sensitive mutants. Current Genetics. 1991;19(3):207–213. [Google Scholar]
- 59.Rauser WE. Phytochelatins and related peptides. Structure, biosynthesis, and function. Plant Physiology. 1995;109(4):1141–1149. doi: 10.1104/pp.109.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neilands JB. Siderophores: structure and function of microbial iron transport compounds. Journal of Biological Chemistry. 1995;270(45):26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 61.Cotton FA, Wilkinson G. Advanced Inorganic Chemistry. 4th edition. New York, NY, USA: John Wiley and sons; 1980. [Google Scholar]
- 62.Schalk IJ, Hannauer M, Braud A. New roles for bacterial siderophores in metal transport and tolerance. Environmental Microbiology. 2011;13(11):2844–2854. doi: 10.1111/j.1462-2920.2011.02556.x. [DOI] [PubMed] [Google Scholar]
- 63.Letain TE, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli . Molecular Microbiology. 1997;24(2):271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 64.Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochimica et Biophysica Acta. 1996;1275(3):161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 65.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nature Structural Biology. 1998;5(4):294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 66.Nair S, Finkel SE. Dps protects cells against multiple stresses during stationary phase. Journal of Bacteriology. 2004;186(13):4192–4198. doi: 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sundquist AR, Fahey RC. The function of γ-glutamylcysteine and bis-γ-glutamylcystine reductase in Halobacterium halobium . Journal of Biological Chemistry. 1989;264(2):719–725. [PubMed] [Google Scholar]
- 68.Malki L, Yanku M, Borovok I, Cohen G, Mevarech M, Aharonowitz Y. Identification and characterization of gshA, a gene encoding the glutamate-cysteine ligase in the halophilic archaeon Haloferax volcanii . Journal of Bacteriology. 2009;191(16):5196–5204. doi: 10.1128/JB.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fahey RC. Novel thiols of prokaryotes. Annual Review of Microbiology. 2001;55:333–356. doi: 10.1146/annurev.micro.55.1.333. [DOI] [PubMed] [Google Scholar]
- 70.Tanaka Y, Tsumoto K, Nakanishi T, et al. Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: the case of Pyrococcus horikoshii CutA. FEBS Letters. 2004;556(1–3):167–174. doi: 10.1016/s0014-5793(03)01402-9. [DOI] [PubMed] [Google Scholar]
- 71.Reindel S, Anemüller S, Sawaryn A, Matzanke BF. The DpsA-homologue of the archaeon Halobacterium salinarum is a ferritin. Biochimica et Biophysica Acta. 2002;1598(1-2):140–146. doi: 10.1016/s0167-4838(02)00361-8. [DOI] [PubMed] [Google Scholar]
- 72.Reindel S, Schmidt CL, Anemüller S, Matzanke BF. Expression and regulation pattern of ferritin-like DpsA in the Archaeon Halobacterium salinarum . BioMetals. 2005;18(4):387–397. doi: 10.1007/s10534-005-3713-y. [DOI] [PubMed] [Google Scholar]
- 73.Bereswill S, Greiner S, Van Vliet AHM, et al. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori . Journal of Bacteriology. 2000;182(21):5948–5953. doi: 10.1128/jb.182.21.5948-5953.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dave BP, Anshuman K, Hajela P. Siderophores of halophilic archaea and their chemical characterization. Indian Journal of Experimental Biology. 2006;44(4):340–344. [PubMed] [Google Scholar]
- 75.Harrison JJ, Turner RJ, Ceri H. Metal tolerance in bacterial biofilms. Recent Research Developments in Microbiology. 2009;9(1):33–55. [Google Scholar]
- 76.Gadd GM. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology. 2010;156(3):609–643. doi: 10.1099/mic.0.037143-0. [DOI] [PubMed] [Google Scholar]
- 77.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical Microbiology Reviews. 2002;15(2):167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poli A, Di Donato P, Abbamondi GR, Nicolaus B. Synthesis, production, and biotechnological applications of exopolysaccharides and polyhydroxyalkanoates by archaea. Archaea. 2011;2011:13 pages. doi: 10.1155/2011/693253.693253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kawakami Y, Hayashi N, Ema M, Nakayama M. Effects of divalent cations on Halobacterium salinarum cell aggregation. Journal of Bioscience and Bioengineering. 2007;104(1):42–46. doi: 10.1263/jbb.104.42. [DOI] [PubMed] [Google Scholar]
- 80.Bolhuis H, Palm P, Wende A, et al. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics. 2006;7:p. 169. doi: 10.1186/1471-2164-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baliga NS, Bonneau R, Facciotti MT, et al. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the dead sea. Genome Research. 2004;14(11):2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. http://archaea.ucsc.edu/lists/haloTurk1/refSeq-list.html.
- 83.Ng WV, Kennedy SP, Mahairas GG, et al. Genome sequence of Halobacterium species NRC-1. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(22):12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kierek K, Watnick PI. The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca 2+-dependent biofilm development in sea water. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(2):14357–14362. doi: 10.1073/pnas.2334614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banin E, Brady KM, Greenberg EP. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Applied and Environmental Microbiology. 2006;72(3):2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kawakami Y, Ito T, Kamekura M, Nakayama M. Ca2+-dependent cell aggregation of halophilic archaeon, Halobacterium salinarum . Journal of Bioscience and Bioengineering. 2005;100(6):681–684. doi: 10.1263/jbb.100.681. [DOI] [PubMed] [Google Scholar]
- 87.Frols S, Dyall-Smith M, Pfeifer F. Biofilm formation by haloarchaea. Environmental Microbiology. 2012;14(12):3159–3174. doi: 10.1111/j.1462-2920.2012.02895.x. [DOI] [PubMed] [Google Scholar]
- 88.Jarrell KF, Stark M, Nair DB, Chong JPJ. Flagella and pili are both necessary for efficient attachment of Methanococcus maripaludis to surfaces. FEMS Microbiology Letters. 2011;319(1):44–50. doi: 10.1111/j.1574-6968.2011.02264.x. [DOI] [PubMed] [Google Scholar]
- 89.Näther DJ, Rachel R, Wanner G, Wirth R. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. Journal of Bacteriology. 2006;188(19):6915–6923. doi: 10.1128/JB.00527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zolghadr B, Kling A, Koerdt A, Driessen AJM, Rachel R, Albers SV. Appendage-mediated surface adherence of Sulfolobus solfataricus . Journal of Bacteriology. 2010;192(1):104–110. doi: 10.1128/JB.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Albers SV, Pohlschröder M. Diversity of archaeal type IV pilin-like structures. Extremophiles. 2009;13(3):403–410. doi: 10.1007/s00792-009-0241-7. [DOI] [PubMed] [Google Scholar]
- 92.Tripepi M, Imam S, Pohlschröder M. Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. Journal of Bacteriology. 2010;192(12):3093–3102. doi: 10.1128/JB.00133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Albers SV, Szabó Z, Driessen AJM. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. Journal of Bacteriology. 2003;185(13):3918–3925. doi: 10.1128/JB.185.13.3918-3925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Annual Review of Microbiology. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 95.Nascimento AMA, Chartone-Souza E. Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genetics and Molecular Research. 2003;2(1):92–101. [PubMed] [Google Scholar]
- 96.Gupta A, Phung LT, Taylor DE, Silver S. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology. 2001;147(12):3393–3402. doi: 10.1099/00221287-147-12-3393. [DOI] [PubMed] [Google Scholar]
- 97.Branco R, Chung AP, Morais PV. Sequencing and expression of two arsenic resistance operons with different functions in the highly arsenic-resistant strain Ochrobactrum tritici SCII24T. BMC Microbiology. 2008;8:p. 95. doi: 10.1186/1471-2180-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sri S, Prashant LSM, Chari PVB, Rao P, Balaravi NS, Kishor PVK. Molecular identification of arsenic-resistant estuarine bacteria and characterization of their ars genotype. Ecotoxicology. 2012;21(1):202–212. doi: 10.1007/s10646-011-0779-x. [DOI] [PubMed] [Google Scholar]
- 99.Endo G, Silver S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. Journal of Bacteriology. 1995;177(15):4437–4441. doi: 10.1128/jb.177.15.4437-4441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Teixeira EC, de Oliveira JCF, Novo MTM, Bertolini MC. The copper resistance operon copAB from Xanthomonas axonopodis pathovar citri: gene inactivation results in copper sensitivity. Microbiology. 2008;154(2):402–412. doi: 10.1099/mic.0.2007/013821-0. [DOI] [PubMed] [Google Scholar]
- 101.Nawapan S, Charoenlap N, Charoenwuttitam A, Saenkham P, Mongkolsuk S, Vattanaviboon P. Functional and expression analyses of the cop operon, required for copper resistance in Agrobacterium tumefaciens . Journal of Bacteriology. 2009;191(16):5159–5168. doi: 10.1128/JB.00384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stoppel RD, Schlegel HG. Nickel-resistant bacteria from anthropogenically nickel-polluted and naturally nickel-percolated ecosystems. Applied and Environmental Microbiology. 1995;61(6):2276–2285. doi: 10.1128/aem.61.6.2276-2285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siunova TV, Siunov AV, Kochetkov VV, Boronin AM. The cnr-like operon in strain Comamonas sp. encoding resistance to cobalt and nickel. Russian Journal of Genetics. 2009;45(3):292–297. [PubMed] [Google Scholar]
- 104.Borremans B, Hobman JL, Provoost A, Brown NL, Van Der Lelie D. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. Journal of Bacteriology. 2001;183(19):5651–5658. doi: 10.1128/JB.183.19.5651-5658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh VK, Xiong A, Usgaard TR, et al. ZntR is an autoregulatory protein and negatively regulates the chromosomal zinc resistance operon znt of Staphylococcus aureus. Molecular Microbiology. 1999;33(1):200–207. doi: 10.1046/j.1365-2958.1999.01466.x. [DOI] [PubMed] [Google Scholar]
- 106.Ehrlich HL. Microbes and metals. Applied Microbiology and Biotechnology. 1997;48(6):687–692. [Google Scholar]
- 107.Dopson M, Baker-Austin C, Koppineedi PR, Bond PL. Growth in sulfidic mineral environments: metal resistance mechanisms in acidophilic micro-organisms. Microbiology. 2003;149(8):1959–1970. doi: 10.1099/mic.0.26296-0. [DOI] [PubMed] [Google Scholar]
- 108.Schelert J, Dixit V, Hoang V, Simbahan J, Drozda M, Blum P. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. Journal of Bacteriology. 2004;186(2):427–437. doi: 10.1128/JB.186.2.427-437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schelert J, Drozda M, Dixit V, Dillman A, Blum P. Regulation of mercury resistance in the crenarchaeote Sulfolobus solfataricus . Journal of Bacteriology. 2006;188(20):7141–7150. doi: 10.1128/JB.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baker-Austin C, Dopson M, Wexler M, et al. Extreme arsenic resistance by the acidophilic archaeon Ferroplasma acidarmanus Fer1. Extremophiles. 2007;11(3):425–434. doi: 10.1007/s00792-006-0052-z. [DOI] [PubMed] [Google Scholar]
- 111.Ruepp A, Graml W, Santos-Martinez ML, et al. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407(6803):508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- 112.Ettema TJG, Brinkman AB, Lamers PP, Kornet NG, de Vos WM, van der Oost J. Molecular characterization of a conserved archaeal copper resistance (cop) gene cluster and its copper-responsive regulator in Sulfolobus solfataricus P2. Microbiology. 2006;152(7):1969–1979. doi: 10.1099/mic.0.28724-0. [DOI] [PubMed] [Google Scholar]
- 113.Villafane AA, Voskoboynik Y, Cuebas M, Ruhl I, Bini E. Response to excess copper in the hyperthermophile Sulfolobus solfataricus strain 98/2. Biochemical and Biophysical Research Communications. 2009;385(1):67–71. doi: 10.1016/j.bbrc.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Villafane A, Voskoboynik Y, Ruhl I, et al. CopR of Sulfolobus solfataricusrepresents a novel class of archaeal-specific copper-responsive activators of transcription. Microbiology. 2011;157(10):2808–2817. doi: 10.1099/mic.0.051862-0. [DOI] [PubMed] [Google Scholar]
- 115.DasSarma S, Capes M, DasSarma P. Haloarchaeal Megaplasmids. In: Schwartz E, editor. Microbial Megaplasmids. Berlin, Germany: Springer-Verlag; 2009. pp. 3–30. [Google Scholar]
- 116.Ng WV, Ciufo SA, Smith TM, et al. Snapshot of a large dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Research. 1998;8(11):1131–1141. doi: 10.1101/gr.8.11.1131. [DOI] [PubMed] [Google Scholar]
- 117.Mukhopadhyay R, Rosen BP, Phung LT, Silver S. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiology Reviews. 2002;26(3):311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 118.Suzuki Y, Matsushita H. Interaction of metal ions and phospholipids monolayers as a biological membrane model. Industrial Health. 1968;6(3):128–133. [Google Scholar]
- 119.Stolz JF, Basu P, Oremland RS. Microbial transformation of elements: the case of arsenic and selenium. International Microbiology. 2002;5(4):201–207. doi: 10.1007/s10123-002-0091-y. [DOI] [PubMed] [Google Scholar]
- 120.Dey S, Rosen BP. Dual mode of energy coupling by the oxyanion-translocating ArsB protein. Journal of Bacteriology. 1995;177(2):385–389. doi: 10.1128/jb.177.2.385-389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Butcher BG, Deane SM, Rawlings DE. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli . Applied and Environmental Microbiology. 2000;66(5):1826–1833. doi: 10.1128/aem.66.5.1826-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cullen WR, Bentley R. The toxicity of trimethylarsine: an urban myth. Journal of Environmental Monitoring. 2005;7(1):11–15. doi: 10.1039/b413752n. [DOI] [PubMed] [Google Scholar]
- 123.Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yuan C, Lu X, Jie Q, Rosen BP, Le XC. Volatile arsenic species released from Escherichia coli expressing the aslll S-adenosylmethionine methyltransferase gene. Environmental Science and Technology. 2008;42(9):3201–3206. doi: 10.1021/es702910g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Osborn AM, Bruce KD, Strike P, Ritchie DA. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiology Reviews. 1997;19(4):239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 126.Begley TP, Walts AE, Walsh CT. Mechanistic studies of a protonolytic organomercurial cleaving enzyme: bacterial organomercurial lyase. Biochemistry. 1986;25(22):7192–7200. doi: 10.1021/bi00370a064. [DOI] [PubMed] [Google Scholar]
- 127. http://archaea.ucsc.edu/lists/haloTurk1/refSeq-list.html.
- 128.Fagan MJ, Saier MH., Jr. P-type ATPases of eukaryotes and bacteria: sequence analyses and construction of phylogenetic trees. Journal of Molecular Evolution. 1994;38(1):57–99. doi: 10.1007/BF00175496. [DOI] [PubMed] [Google Scholar]
- 129.Bublitz M, Preben Morth J, Nissen P. P-type ATPases at a glance. Journal of Cell Science. 2011;124(15):2515–2519. doi: 10.1242/jcs.088716. [DOI] [PubMed] [Google Scholar]
- 130.Saier MH., Jr. Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiological Reviews. 1994;58(1):71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Snavely MD, Florer JB, Miller CG, Maguire ME. Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. Journal of Bacteriology. 1989;171(9):4761–4766. doi: 10.1128/jb.171.9.4761-4766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiology Reviews. 2003;27(2-3):313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 133.Klein JS, Lewinson O. Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence. Metallomics. 2011;3(11):1098–1108. doi: 10.1039/c1mt00073j. [DOI] [PubMed] [Google Scholar]
- 134.Raimunda D, González-Guerrero M, Leeber BW, III, Argüello JM. The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals. 2011;24(3):467–475. doi: 10.1007/s10534-010-9404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. http://halo4.umbi.umd.edu/
- 136.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chemico-Biological Interactions. 2006;160(1):1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 137.Fan B, Rosen BP. Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. Journal of Biological Chemistry. 2002;277(49):46987–46992. doi: 10.1074/jbc.M208490200. [DOI] [PubMed] [Google Scholar]
- 138.Franke S, Grass G, Nies DH. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology. 2001;147(4):965–972. doi: 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 139.Tong L, Nakashima S, Shibasaka M, Katsuhara M, Kasamo K. A novel histidine-rich CPx-ATPase from the filamentous cyanobacterium Oscillatoria brevis related to multiple-heavy-metal cotolerance. Journal of Bacteriology. 2002;184(18):5027–5035. doi: 10.1128/JB.184.18.5027-5035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tottey S, Rich PR, Rondet SAM, Robinson NJ. Two menkes-type ATPases supply copper for photosynthesis in synechocystis PCC 6803. Journal of Biological Chemistry. 2001;276(23):19999–20004. doi: 10.1074/jbc.M011243200. [DOI] [PubMed] [Google Scholar]
- 141.Solioz M, Stoyanov JV. Copper homeostasis in Enterococcus hirae . FEMS Microbiology Reviews. 2003;27(2-3):183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 142.Paulsen IT, Saier MH., Jr. A novel family of ubiquitous heavy metal ion transport proteins. Journal of Membrane Biology. 1997;156(2):99–103. doi: 10.1007/s002329900192. [DOI] [PubMed] [Google Scholar]
- 143.Sharma R, Rensing C, Rosen BP, Mitra B. The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)- translocating ATPase from Escherichia coli . Journal of Biological Chemistry. 2000;275(6):3873–3878. doi: 10.1074/jbc.275.6.3873. [DOI] [PubMed] [Google Scholar]
- 144.Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M. Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics. 2007;8:p. 107. doi: 10.1186/1471-2164-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Haney CJ, Grass G, Franke S, Rensing C. New developments in the understanding of the cation diffusion facilitator family. Journal of Industrial Microbiology and Biotechnology. 2005;32(6):215–226. doi: 10.1007/s10295-005-0224-3. [DOI] [PubMed] [Google Scholar]
- 146.Anton A, Große C, Reißmann J, Pribyl T, Nies DH. CzcD is a heavy metal ion transporter involved in regulation of heavy metal resistance in Ralstonia sp. strain CH34. Journal of Bacteriology. 1999;181(22):6876–6881. doi: 10.1128/jb.181.22.6876-6881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wei Y, Fu D. Selective metal binding to a membrane-embedded aspartate in the Escherichia coli metal transporter YiiP (FieF) Journal of Biological Chemistry. 2005;280(40):33716–33724. doi: 10.1074/jbc.M506107200. [DOI] [PubMed] [Google Scholar]
- 148.Hou Z, Mitra B. The metal specificity and selectivity of ZntA from Escherichia coli using the acyl phosphate intermediate. Journal of Biological Chemistry. 2003;278(31):28455–28461. doi: 10.1074/jbc.M301415200. [DOI] [PubMed] [Google Scholar]
- 149.Stumpe S, Bakker EP. Requirement of a large K+-uptake capacity and of extracytoplasmic protease activity for protamine resistance of Escherichia coli . Archives of Microbiology. 1997;167(2-3):126–136. [PubMed] [Google Scholar]
- 150.Tame JRH, Murshudov GN, Dodson EJ, et al. The structural basis of sequence-independent peptide binding by OppA protein. Science. 1994;264(5165):1578–1581. doi: 10.1126/science.8202710. [DOI] [PubMed] [Google Scholar]
- 151.Salt DE, Wagner GJ. Cadmium transport across tonoplast of vesicles from oat roots. Evidence for a Cd2+/H+ antiport activity. Journal of Biological Chemistry. 1993;268(17):12297–12302. [PubMed] [Google Scholar]
- 152.Rea PA, Li ZS, Lu YP, Drozdowicz YM, Martinoia E. From vacuolar GS-X pumps to multispecific ABC transporters. Annual Review of Plant Biology. 1998;49:727–760. doi: 10.1146/annurev.arplant.49.1.727. [DOI] [PubMed] [Google Scholar]
- 153.Van Veen HW, Callaghan R, Soceneantu L, Sardini A, Konings WN, Higgins CF. A bacterial antibiotic-resistance gene that complements the human multidrug-resistance P-glycoprotein gene. Nature. 1998;391(6664):291–295. doi: 10.1038/34669. [DOI] [PubMed] [Google Scholar]
- 154.Van Veen HW, Margolles A, Müller M, Higgins CF, Konings WN. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO Journal. 2000;19(11):2503–2514. doi: 10.1093/emboj/19.11.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hartman AL, Norais C, Badger JH, et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PloS One. 2010;5(3):p. e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Falb M, Pfeiffer F, Palm P, et al. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Research. 2005;15(10):1336–1343. doi: 10.1101/gr.3952905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Roth JR, Lawrence JG, Rubenfield M, Kieffer-Higgins S, Church GM. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. Journal of Bacteriology. 1993;175(11):3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Faraldo-Gómez JD, Sansom MSP. Acquisition of siderophores in gram-negative bacteria. Nature Reviews Molecular Cell Biology. 2003;4(2):105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- 159.Schneider S, Paoli M. Crystallization and preliminary X-ray diffraction analysis of the haem-binding protein HemS from Yersinia enterocolitica . Acta Crystallographica Section F. 2005;61(8):802–805. doi: 10.1107/S1744309105023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zumft WG, Viebrock-Sambale A, Braun C. Nitrous oxide reductase from denitrifying Pseudomonas stutzeri .Genes for copper-processing and properties of the deduced products, including a new member of the family of ATP/GTP-binding proteins. European Journal of Biochemistry. 1990;192(3):591–599. doi: 10.1111/j.1432-1033.1990.tb19265.x. [DOI] [PubMed] [Google Scholar]
- 161.Usadel B, Bläsing OE, Gibon Y, et al. Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in arabidopsis rosettes. Plant Physiology. 2008;146(4):1834–1861. doi: 10.1104/pp.107.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mittal D, Madhyastha DA, Grover A. Genome-wide transcriptional profiles during temperature and oxidative stress reveal coordinated expression patterns and overlapping regulons in rice. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0040899.e40899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ercal N, Gurer-Orhan H, Aykin-Burns N. Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Current topics in medicinal chemistry. 2001;1(6):529–539. doi: 10.2174/1568026013394831. [DOI] [PubMed] [Google Scholar]
- 164.Storz G, Imlay JA. Oxidative stress. Current Opinion in Microbiology. 1999;2(2):188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 165.Capes MD, Coker JA, Gessler R, et al. The information transfer system of halophilic archaea. Plasmid. 2011;65(2):77–101. doi: 10.1016/j.plasmid.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 166.Siddaramappa S, Challacombe JF, DeCastro RE, et al. A comparative genomics perspective on the genetic content of the alkaliphilic haloarchaeon Natrialba magadii ATCC, 43099. BMC Genomics. 2012;13:p. 165. doi: 10.1186/1471-2164-13-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Que Q, Helmann JD. Manganese homestasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Molecular Microbiology. 2000;35(6):1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 168.Jensen LT, Ajua-Alemanji M, Culotta VC. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. Journal of Biological Chemistry. 2003;278(43):42036–42040. doi: 10.1074/jbc.M307413200. [DOI] [PubMed] [Google Scholar]
- 169.Hill PJ, Cockayne A, Landers P, Morrissey JA, Sims CM, Williams P. SirR, a novel iron-dependent repressor in Staphylococcus epidermidis . Infection and Immunity. 1998;66(9):4123–4129. doi: 10.1128/iai.66.9.4123-4129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bonneau R, Reiss DJ, Shannon P, et al. The inferelator: an algorithn for learning parsimonious regulatory networks from systems-biology data sets de novo . Genome Biology. 2006;7(5) doi: 10.1186/gb-2006-7-5-r36.R36 [DOI] [PMC free article] [PubMed] [Google Scholar]


