Hematopoietic stem cells (HSCs) primarily reside in the bone marrow where signals generated by stromal cells regulate their self-renewal, proliferation, and trafficking. Endosteal osteoblasts1,2 and perivascular stromal cells including endothelial cells3, CXCL12-abundant reticular (CAR) cells4,5, leptin-receptor positive stromal cells6, and nestin-GFP positive mesenchymal progenitors7 have all been implicated in HSC maintenance. However, it is unclear if specific hematopoietic progenitor cell (HPC) subsets reside in distinct niches defined by the surrounding stromal cells and the regulatory molecules they produce. CXCL12 (stromal-derived factor-1, SDF-1) regulates both HSCs and lymphoid progenitors and is expressed by all of these stromal cell populations7–11. Here, we selectively deleted Cxcl12 from candidate niche stromal cell populations and characterized the effect on HPCs. Deletion of Cxcl12 from mineralizing osteoblasts has no effect on HSCs or lymphoid progenitors. Deletion of Cxcl12 from osterix-expressing stromal cells, which includes CAR cells and osteoblasts, results in constitutive HPC mobilization and a loss of B lymphoid progenitors, but HSC function is normal. Cxcl12 deletion in endothelial cells results in a modest loss of long-term repopulating activity. Strikingly, deletion of Cxcl12 in nestin-negative mesenchymal progenitors using Prx1-Cre is associated with a marked loss of HSCs, long-term repopulating activity, HSC quiescence, and common lymphoid progenitors. These data suggest that osterix-expressing stromal cells comprise a distinct niche that supports B lymphoid progenitors and retains HPC in the bone marrow, while expression of CXCL12 from stromal cells in the perivascular region, including endothelial cells and mesenchymal progenitors, support HSCs.
CXCL12 plays a crucial role in maintaining HSC function, including retention in the bone marrow8,12–14, quiescence15,16, and repopulating activity16. To test the hypothesis that CXCL12 production by different stromal cell populations has distinct effects on HSCs and lineage-committed HPC, we generated a floxed allele of Cxcl12 (Cxcl12fl) to conditionally delete Cxcl12 from candidate niche cells in the bone marrow (Suppl. Fig. 2). Deletion of Cxcl12 in endothelial cells and mature osteoblasts was mediated by the Tie2-Cre recombinase (Cre) and osteocalcin (Oc)-Cre transgenes, respectively. To target Cxcl12 deletion in osteoprogenitors, we used the osterix (Osx)-Cre transgene, which mediates efficient recombination in mature osteoblasts and osteoblast progenitors17. It also targets CAR cells, a perivascular stromal cell population implicated in HSC and B lymphoid progenitor maintenance5,11. Finally, we used the Prx1-Cre transgene to target multipotent mesenchymal progenitors in the appendicular skeleton. Prx1 is a transcription factor expressed early during limb bud mesoderm development, and Prx1-Cre targets all cells derived from limb bud mesoderm18. Lineage mapping studies were performed using a Cxcl12gfp knock-in mouse to define CAR cells11. These studies showed that both the Osx- and Prx1-Cre transgenes efficiently targeted recombination in mature osteoblasts, osteocytes, and CAR cells in long bones (Fig. 1a–d and Suppl. Fig 3).
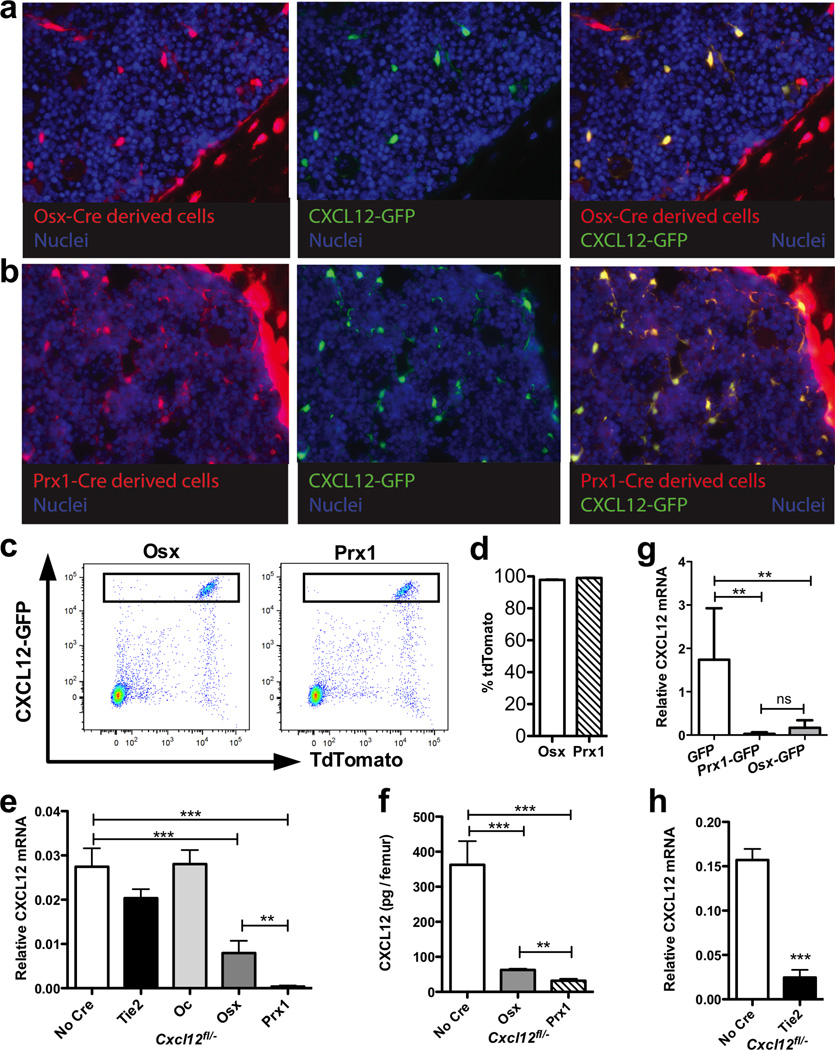
Figure 1. Targeting Cxcl12 deletion in bone marrow stromal cell populations.
Lineage mapping was by performed by generating Osx-Cre ROSA26Ai9/+ Cxcl12gfp/+ mice (a) or Prx1-Cre ROSA26Ai9/+ Cxcl12gfp/+ mice (b). Cells that had undergone Cre-mediated recombination express tdTomato (red). Cells that express CXCL12 also express GFP (green). Counterstaining with DAPI highlights nuclei (blue). Shown are representative photomicrographs of femur sections. The left panel shows tdTomato fluorescence, the middle panel GFP fluorescence, and the right panel both. Original magnification 40X. (c) Representative dot plots showing GFP and tdTomato expression in CD45− lineage− stromal cells harvested from Osx-Cre ROSAAi9/+ Cxcl12gfp/+ mice (left) or Prx1-Cre ROSA26Ai9/+ Cxcl12gfp/+ mice (right). GFPbright CAR cells are boxed. (d) Shown is the percentage of CAR cells that express tdTomato (n=5). (e) CXCL12 mRNA expression relative to β-actin mRNA is shown on total bone marrow RNA (n=6–13). (f) CXCL12 protein in bone marrow extracellular fluid as measured by ELISA (n=3–5). (g) CAR cells were sorted from Osx-Cre Cxcl12gfp/fl (Osx-GFP), or Prx1-Cre Cxcl12gfp/fl (Prx1-GFP) mice and RNA prepared. Shown is CXCL12 mRNA expression relative to β-actin mRNA (n=5–7). (h) CD31+ lineage− CD45− endothelial cells were sorted from Tie2-Cre-targeted and control mice, and RNA was prepared. Shown is CXCL12 mRNA expression relative to β-actin mRNA (n=3). *P < 0.05, **P < 0.01; ***P < 0.001.
Triple transgenic mice were generated containing one floxed Cxcl12 allele, one null allele (Cxcl12fl/−), and a Cre-recombinase transgene. Total CXCL12 mRNA expression in the femoral bone marrow of Oc- and Tie2-Cre-targeted mice was similar to that observed in control mice (Fig. 1e). In contrast, CXCL12 mRNA expression was reduced by 70% in Osx-Cre-targeted mice and nearly undetectable in Prx1-Cre-targeted mice. A similar decrease in CXCL12 protein levels was observed (Fig. 1f). To confirm Cxcl12 deletion in CAR cells, mice containing Cxcl12fl/gfp and either the Osx- or Prx1-Cre transgenes, were generated (the Cxcl12gfp allele is a null allele). Indeed, CXCL12 mRNA was nearly undetectable in CXCL12-GFPbright CAR cells that were sorted from these mice (Fig. 1g). As expected, CXCL12 mRNA was nearly undetectable in endothelial cells sorted from Tie2-Cre-targeted mice (Fig. 1h). Together these data suggest that, under basal conditions, the majority of CXCL12 is produced by CAR cells, while mature osteoblasts and endothelial cells are only minor contributors.
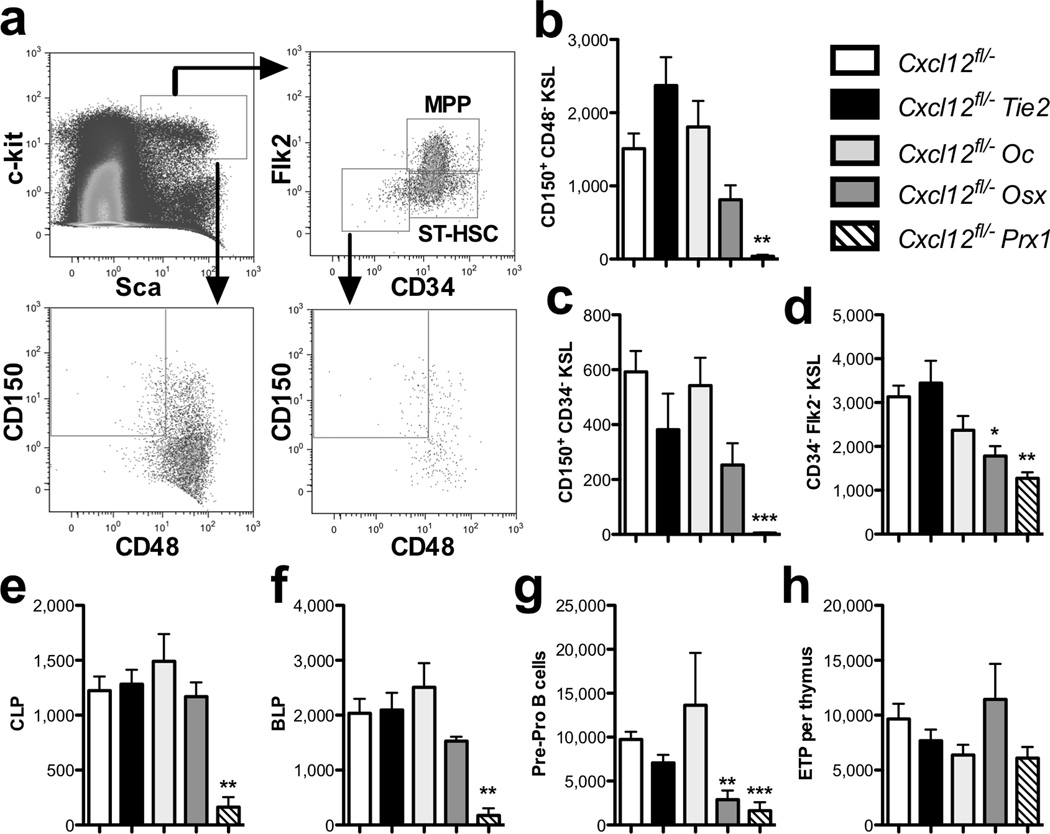
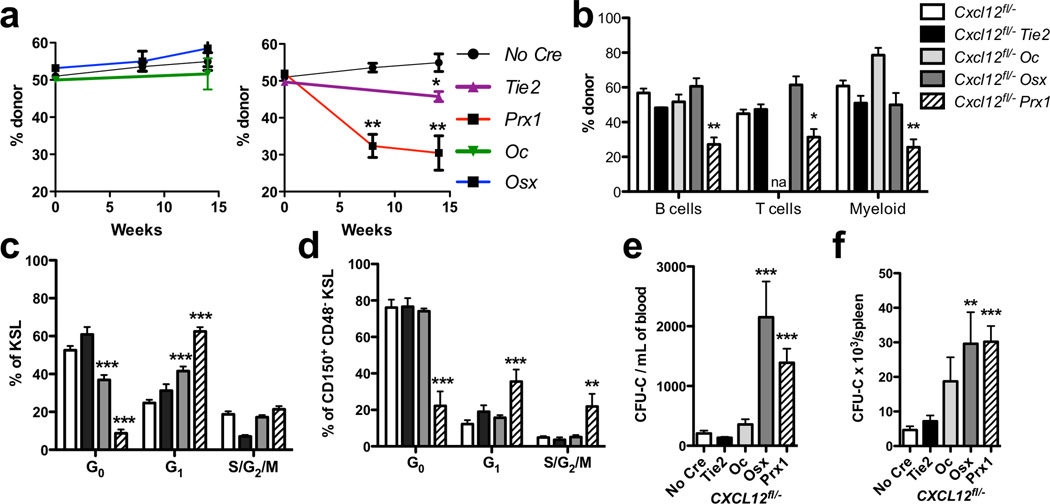
All conditional knockout mice exhibited normal peripheral blood counts and the same relative percentage of granulocytes, monocytes, B cells, and T cells (Suppl. Table 1). However, bone marrow cellularity in femurs was reduced by approximately 50% in both the Osx-Cre- and Prx1-Cre-targeted mice, which was due, in part, to a loss of B cells. HPC subsets in the bone marrow were quantified by flow cytometry (Fig. 2a). The number of c-Kit+ Sca+ Lineage− (KSL) cells, short-term HSCs, multipotent progenitors, and myeloid-committed progenitors was similar in all mice with the exception of a two-fold decrease in common myeloid progenitors in Prx1-Cre-targeted mice (Suppl. Fig. 4). Loss of CXCL12 expression in endothelial cells or mature osteoblasts had no effect on the number of phenotypic HSCs (Fig. 2 b–d). The frequency of phenotypic HSCs in the bone marrow of Osx-Cre-targeted mice was comparable to control mice (data not shown); however, since bone marrow cellularity was reduced, a modest decrease in the absolute number of HSCs was observed. In contrast, a significant decrease in both the frequency and absolute number of phenotypic HSCs in Prx1-Cre-targeted mice was observed, with nearly undetectable dormant HSCs (Flk2− CD34− CD150+ CD48− KSL cells). Consistent with these findings, competitive repopulation assays showed a significant multi-lineage long-term repopulating defect using bone marrow from Prx1-Cre- but not Osx-Cre-targeted mice (Fig. 3a–b). Despite the normal number of phenotypic HSCs, a small, but significant, decrease in long-term repopulating activity also was observed using Tie2-Cre-targeted bone marrow. Serial transplantation of bone marrow from Prx1-Cre- or Tie2-Cre-targeted mice showed no further decrease in repopulating activity in secondary recipients, suggesting that self-renewal capacity may be restored when HSCs are exposed to a normal stromal microenvironment (Suppl. Fig. 5). Quiescence is a fundamental property of HSCs, which is closely related to long-term repopulating activity19. Increased cycling of HSCs was observed in Prx1-Cre- but not Osx-Cre- or Tie2-Cre-targeted cells. In contrast, increased cycling of more mature KSL progenitors was observed in both Prx1-Cre- and Osx-Cre-targeted cells (Fig. 3c–d). Collectively, these data show that CXCL12 production from Prx1-Cre-targeted stromal cells and, to a lesser extent, endothelial cells is required for maintenance of HSC repopulating activity and quiescence. Consistent with results from the companion paper by Ding et al., our data suggest that CXCL12 production from mature osteoblasts and osteoblast precursors is dispensable for HSC maintenance.
Figure 2. Deletion of Cxcl12 in defined stromal cell population results in the selective loss of HSCs and lymphoid progenitors.
(a). Representative dot plots showing the gating strategy to identify HPC populations. Data are gated on lineage− cells. (b) The number of CD150+ CD48− CD41− c-Kit+ Sca+ Lineage− (KSL) cells, (c) CD150+ CD41− CD48− CD34− Flk2− KSL (dormant HSCs), (d) CD34− Flk2− KSL cells, (e) common lymphoid progenitors (CLP), (f) B lymphoid progenitors (BLP), and (g) pre-pro B cells per femur is shown. (h) The number of earliest thymic progenitors (ETP) per thymus is shown. Data represent the mean ± SEM of 6–13 mice. *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 3. Deletion of Cxcl12 in defined stromal cell populations results in HPC mobilization and a selective loss of repopulating activity and HSC quiescence.
(a) Competitive repopulation assays were performed with a one to one ratio of donor and wild-type competitor bone marrow. Shown is the percentage of donor peripheral leukocytes over time. Data represent the mean ± SEM of 10–16 mice. (b) The percentage of donor cells in the bone marrow of the indicated lineage 14 weeks after transplantation is shown. Data represent the mean ± SEM of 5–6 mice. (c & d) The percentage of cells in the indicated stage of the cell cycle is shown for KSL (c) and CD150+ CD48− CD41− KSL cells (d). The number of colony-forming cells (CFU-C) in the blood (e) or spleen (f) is shown. Data represent the mean ± SEM of 5–15 mice. *P < 0.05; **P < 0.01; ***P < 0.001. na = not available.
Since CXCL12 has been shown to play an important role in the retention of HPC within the marrow5,20, we next quantified HPCs in the blood and spleen. In Osx-Cre-targeted mice, the number of colony-forming cells and KSL cells was increased in the blood and spleen, demonstrating constitutive HPC mobilization (Fig. 3e–f and Suppl. Fig. 6). Interestingly, though CXCL12 expression in the bone marrow is significantly lower, Prx1-Cre-targeted mice displayed a similar magnitude of HPC mobilization. Thus, our data suggest that, although CXCL12 production from Osx-Cre-targeted stromal cells is largely dispensable for HSC maintenance, it is required for the efficient retention of HPCs in the bone marrow.
CXCL12 is required for normal B and T cell development19,21. Pre-pro-B cells are found in close association with CAR cells11, and ablation of CAR cells is associated with a loss of CLPs and pro-B cells4. Here, we show that deletion of Cxcl12 in mineralizing osteoblasts or endothelial cells has no effect on CLPs, B lymphoid progenitors (BLPs), or pre-pro-B cells (Fig. 2e–h, Suppl. Figure 7). In contrast, Cxcl12 deletion in CAR cells using Osx-Cre results in a marked loss of pre-pro B cells, and a trend towards a loss of BLP. However, CLPs and earliest thymic progenitors (ETPs) in the thymus are normal. Deletion of Cxcl12 in Prx1-Cre-targeted stromal cells results in a similar phenotype but also results in a marked loss of CLPs. In the companion paper, Ding et al show that deletion of Cxcl12 in osteoblasts using Col2.3-Cre also results in a modest decrease in CLP and lymphoid-primed multipotential progenitors (LMPP). Together, these data suggest that CXCL12 production from CAR cells or osteoblast precursors, but not mineralizing osteoblasts or endothelial cells, is required for the maintenance of B-lymphoid committed progenitors, while CLP maintenance is supported by CXCL12 production from both endosteal osteoblasts and a Prx1-Cre-targeted perivascular stromal cell population. The normal CLP in Osx-Cre-targeted mice may be secondary to compensatory changes related to the severe loss of pre-pro-B cells.
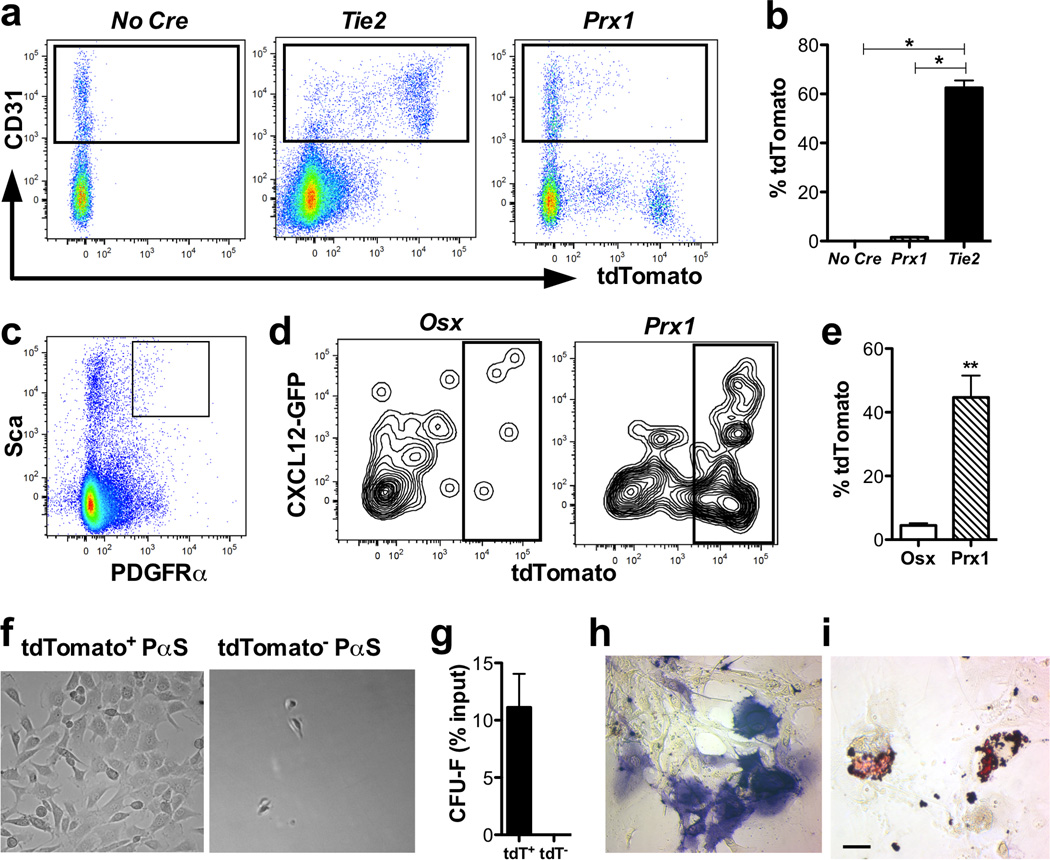
We next performed studies to define the stromal cell population(s) differentially targeted by Prx1-Cre and Osx-Cre. We first considered the possibility that Prx1-Cre may target endothelial cells in the bone marrow. However, we detected no tdTomato expression in bone marrow endothelial cells from Prx1-Cre reporter mice (Fig. 4a–b). Moreover, expression of CXCL12 mRNA from sorted CD31+ endothelial cells from Prx1-Cre-targeted mice was comparable to control mice (Suppl. Fig. 8). Thus, loss of CXCL12 from bone marrow endothelial cells does not account for the loss of HSCs in Prx1-Cre-targeted mice.
Figure 4. Prx1-Cre differentially targets a PDGFRα+ Sca+ CXCL12 expressing mesenchymal progenitor cell population.
(a) Representative dot plots showing tdTomato and CD31 expression in bone marrow cells from the indicated mice. Data are gated on CD45− lineage− cells. (b) The percentage of CD31+ CD45− lineage− cells that express tdTomato is shown. (c) Representative dot plot showing the gating strategy to identify CD45− lineage− PDGFRα+ Sca+ cells (boxed). Data are gated on CD45− lineage− cells. (d) Representative dot plots showing GFP and tdTomato expression in CD45− lineage− PDGFRα+ Sca+ (PαS) cells from Osx-Cre ROSAAi9/+ Cxcl12gfp/+ (left panel) or Prx1-Cre ROSA26Ai9/+ Cxcl12gfp/+ mice (right panel). (e) Shown is the percentage of lineage− PDGFRα+ Sca+ cells that express tdTomato (n=3). *P < 0.001; **P < 0.01. (f) TdTomato+ and tdTomato− PαS cells were sorted from Prx1-Cre ROSA26Ai9/+ Cxcl12gfp/+ mice and colony-forming fibroblast (CFU-F) assays performed. Shown are representative photomicrographs of the cultures on day 14. (g) Number of CFU-F per 100 sorted PαS cells. Data represent the mean ± SEM of three independent experiments. Cells were harvested from the CFU-F cultures on day 14 and replated under osteogenic (h) or adipogenic (i) culture conditions. Shown are representative photomicrographs of cells stained for alkaline phosphatase or oil red O, respectively. Scale bar = 50 µM.
We extended the lineage mapping studies to the CD45− lineage− PDGFRα+ Sca+ (PαS) cell population, which is enriched for mesenchymal stem cells22. Whereas Osx-Cre did not target this cell population, approximately 50% of cells were targeted by Prx1-Cre, including a subpopulation that expressed intermediate levels of CXCL12 (Fig. 4c–e). To evaluate the mesenchymal progenitor activity of the Prx1-Cre-targeted cells, we sorted Prx1-Cre-targeted (tdTomato+) and non-targeted PαS cells and assessed their clonogenic capacity. All of the colony-forming unit-fibroblast (CFU-F) activity was contained with the Prx1-targeted PαS cell population, with greater than 10% of these cells having CFU-F activity (Fig 4f–g). This compares to a CFU-F frequency of approximately 4% in unselected PαS cells22 and less than 1% in nestin-GFP+ stromal cells7. The Prx1-targeted PαS cells have osteogenic and adipogenic differentiation potential in vitro, consistent with a mesenchymal stem cell phenotype (Fig 4h–i). RNA expression profiling of Prx1-targeted PαS cells is notable for the lack of nestin7, CD1469, or leptin receptor6, all of which have been used to mark stromal cells contributing to HSC maintenance (Suppl. Table 2). Interestingly, other than moderate CXCL12 expression, these cells do not express genes classically associated with HSC maintenance, including kit ligand6 and angiopoietin-119, though high expression of several matrix proteins (e.g., proteoglycan 423 and osteonectin24) implicated in HPC regulation is present.
Collectively, these data suggest that distinct stromal cell niches in the bone marrow exist that regulate specific HPC populations (Suppl. Fig. 1). Osterix-expressing stromal cells comprise a niche that supports B lymphoid progenitors and retains HPC in the bone marrow, while CXCL12 production from nestin− leptin receptor− mesenchymal progenitors is required for HSC and CLP maintenance.
METHODS SUMMARY
Targeting of the Cxcl12 locus was accomplished by conventional techniques. Conditional knockouts were accomplished by interbreeding with Osx-Cre25, Prx1-Cre18, Oc-Cre mice26, Tie2-Cre27, and Cxcl12+/− mice21. Lineage mapping was accomplished using Ai928 and Cxcl12gfp mice12. All mice with the exception of Cxcl12gfp mice were maintained on the C56Bl6/J background. Unless indicated otherwise, data are presented as mean ± SEM and were analyzed with the Student's t-test, one-way ANOVA, or two-way ANOVA.
METHODS
Mice
With the exception of Cxcl12gfp mice, all transgenic strains had been backcrossed at least 10 generations onto a C57BL/6 background. Osx-Cre1, Prx1-Cre2, Tie2-Cre3, and Ai9 (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J)4 mice were obtained from The Jackson Laboratory. EIIa-Cre mice5 were a gift from Monica Bessler (University of Pennsylvania, Pennsylvania). Oc-Cre mice6 were a gift from Thomas Clemens (Johns Hopkins University, Maryland), and Cxcl12gfp mice7 were a gift from Takashi Nagasawa (Kyoto University, Japan). Cxcl12+/− mice8 were obtained through the RIKEN BioResource Center (Ibaraki, Japan). Mice were maintained under standard pathogen free conditions according to methods approved by the Washington University Animal Studies Committee.
Generation of Cxcl12fl mice
A floxed allele of Cxcl12 was generated containing LoxP sites flanking exon 2 of Cxcl12; a third LoxP was inserted 3’ of the neomycin selection cassette (Suppl. Fig. 2). Generation of targeted embryonic stem cells and blastocyst injections were performed as previously described9. Excision of the neomycin cassette was accomplished through partial recombination by intercrossing mice with mice expressing EIIa-Cre. Mice were genotyped using PCR primers: Cxcl12flox forward, 5’-CTACACCTCCTCTAGGTAAACCAGTCAGCC-3’; Cxcl12flox reverse 5’-GGACACCAGAACCTTGAAACTGACA-3’.
Bone marrow transplantation
Bone marrow from WT Ly5.1/5.2-expressing mice was mixed at a 1:1 ratio with marrow from experimental or control mice expressing the Ly5.2 locus. A total of 2 × 106 cells injected retro-orbitally into lethally irradiated (1,000 cGy) WT Ly5.1-expressing mice. Since Prx1 is expressed primarily in limb-bud derived long bones, only tibias and femurs were used for transplant and other analyses.
Blood, bone marrow, spleen, and thymus analysis
Blood, bone marrow, and spleen cells and thymocytes were harvested using standard techniques and quantified using a Hemavet automated cell counter (CDC Technologies).
Flow cytometry
Cells were stained by standard protocols with the following antibodies (eBiosciences unless otherwise noted): Lineage analysis and chimerism was assessed using peridinin chlorophyll protein complex (PerCP)-Cy5.5–conjugated Ly5.1 (A20, CD45.1), allophycocyanin (APC)-conjugated Ly5.2 (104, CD45.2), and one or more of the following lineage markers: APC-conjugated CD115 (AFS98, monocytes), fluorescein isothiocyanate (FITC)-conjugated Ly6C/G (RB6-8C5, Gr-1, myeloid), phycoerythrin (PE)-conjugated CD3e (145-2C11, T lymphocytes), and APC-eFluor780–conjugated CD45R (RA3-6B2, B220, B lymphocytes).
For HSPC analysis, cells were stained with a cocktail of biotin-conjugated B220, TER-119, CD3e, Gr1, and CD41 (MWReg30), PE-conjugated CD150 (TC15-12F12.2, Biolegend), PE-Cy7-conjugated CD48 (HM48-1, BD Biosciences), PerCP-Cy5.5-conjugated Sca1 (D7), APC eFluor780-conjugated c-kit (2B-8), FITC-conjugated CD34 (RAM34), APC-conjugated Flk2 (A2F10), eFluor450-conjugated CD16/32 (93), and eFluor605NC-conjugated streptavidin. For HSC cell cycle staining, cells were stained with the biotin-conjugated lineage panel, PE-conjugated CD150, PE-Cy7-conjugated CD48, PerCP-Cy5.5-conjugated Sca, APC-conjugated c-kit, and APC-eFluor780-conjugated streptavidin. Cells were then fixed using the Cytofix/Cytoperm kit (BD Biosciences), stained with FITC-conjugated Ki-67 (B56, BD Biosciences), and resuspended in 1 mg/mL of 4',6-diamidino-2-phenylindole (DAPI). Doublets were gated out using FSC vs. FSC-W. Data were collected on a Gallios 10-color, 3-laser flow cytometer (Beckman Coulter). Data were analyzed with FlowJo (Treestar).
For CLP/BLP analysis, bone marrow cells were stained with a cocktail of PE-Cy7-conjugated B220, TER-119, CD3e, and Gr-1, APC-conjugated CD27 (LG.7F9), biotinylated IL-7Ra (gift of Deepta Bhattacharya, Washington University), PE-conjugated Flk2, FITC-conjugated Ly6D (49-H4, BD Biosciences), and eFluor450-conjugated streptavidin. CLP were defined as B220- TER-119- CD3e- Gr-1- CD27+ IL-7Ra+ Flk2+ Ly6D- cells, and BLP were defined as B220- TER-119- CD3e- Gr-1- CD27+ IL-7Ra+ Flk2+ Ly6D+ cells.
For Pre-pro B cell analysis, bone marrow cells were stained with APC-eFluor780-conjugated B220, a cocktail of PerCP-Cy5.5-conjugated CD3e, CD11c (N418), and NK1.1 (PK136), APC-conjugated IgM (II/4), eFluor450-conjugated IgD (11–26c), PE-Cy7-conjugated CD19 (eBio1D3), PE-conjugated CD43 (S7, BD Biosciences), and FITC-conjugated Ly6D (BD Biosciences). Pre-Pro B cells were defined as B220+ CD3e− CD11c− NK1.1− IgM− IgD− CD19− CD43+ Ly6D+ cells.
For ETP analysis, thymocytes were stained using a cocktail of FITC-conjugated CD4 (RM4-5), CD8 (53-6.7), and CD11b (M1/70), a cocktail of PE-Cy7-conjugated B220, TER-119, CD3e, and Gr-1, PE-conjugated CD44 (IM7), eFluor450-conjugated CD25 (PC61.5), and APC-eFluor780-conjugated c-kit. ETP were defined as CD4− CD8− CD11b− B220− TER-119− CD3e− Gr−1− CD44+ CD25− c-kit+ thymocytes.
Stromal cell analysis and sorting
To extract bone marrow stromal cells, intact bones were crushed in PBS by mortar and pestle. Crushed fractions in PBS were collected and stored on ice. The bone chips were digested using subjected to enzymatic digestion by collagenase type II (3mg/mL, Worthington Biochemical) and dispase (4mg/mL, Roche) at 37°C for 45 minutes at 37°C in a shaking water bath. Both crushed and digested fractions were pooled. Following RBC lysis, endothelial cells were stained with APC-conjugated CD45 (30-F11), FITC-conjugated lineage cocktail (CD3e, Gr-1, B220, and TER-119), and biotinylated anti-mouse CD31 (PECAM-1) followed by streptavidin PE. Dead cells were excluded using 7-AAD (EMD Biosciences). Perivascular mesenchymal progenitor cells were stained with APC-eFluor780-conjugated CD45 and APC-eFluor780-conjugated lineage cocktail, APC-conjugated Sca-1, biotinylated CD140a (PDGRFα), and streptavidin Brilliant Violet 421 (Biolegend). FACS analyses were performed using FACScan (BD Biosciences), LSRII (BD Biosciences), or Gallios (Beckman Coulter) flow cytometers. Cell sorting was done on Reflection (iCyt) or Aria (BD Biosciences) flow cytometers. RNA of sorted cells was extracted using NucleoSpin RNA XS kit (Macherey-Nagel) per manufacture recommendations.
Quantitative RT-PCR
For total bone marrow RNA, femurs were flushed with 1 mL of Trizol (Invitrogen). RNA was prepared according to manufacturer’s specification. One-step qRT-PCR was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems) using no template and no RT controls. Data was collected on a 7300 Real-Time PCR System (Applied Biosystems). Primers were: CXCL12 forward, 5’-GAGCCAACGTCAAGCATCTG-3’; CXCL12 reverse, 5’-CGGGTCAATGCACACTTGTC-3’; CXCL12 dT-FAM/TAMRA probe, 5’-TCCAAACTGTGCCCTTCAGATTGTTGC-3’; β-actin forward, 5’-ACCAACTGGGACGATATGGAGAAGA-3’; β-actin primer; and β-actin dT-VIC/TAMRA probe, 5′-AGCCATGTACGTAGCCATCCAGGCTG-3′.
Immunofluorescence
Mice were lethally sedated and perfused with PBS followed by 4% paraformaldehyde. Hind limbs were removed and post-fixed in 4% paraformaldehyde overnight at 4°C. Bones were washed in water, decalcified in 14% EDTA pH 7.4, and cryoprotected in 30% sucrose in PBS. Bones were then snap frozen in OCT using liquid nitrogen-cooled 2-methylbutane, and blocks were sectioned at 7 µM. For lineage mapping, sections were washed in PBS and mounted with Prolong Gold Antifade Reagent with DAPI (Invitrogen). Slides were imaged using an ApoTome fluorescent microscope (Zeiss).
Colony-forming unit cell (CFU-C) assay
25,000 bone marrow cells or 50,000 spleen cells were plated in 2.75 ml methylcellulose media (MethoCult 3434; Stemcell Technologies). 20 µL of whole peripheral blood was RBC lysed and plated in methylcellulose. Duplicate cultures were incubated at 37°C for 7 days, after which colonies containing at least 100 cells were counted in a blinded fashion.
Colony-forming unit-fibroblast (CFU-F) assay
PDGFRα+ Sca+ cells were sorted by flow cytometry and directly plated on tissue culture plates containing alpha MEM and 10% fetal bovine serum (Atlanta Biologicals). Media exchanges were performed every 3–4 days for a total of 14 days, after which colonies containing at least 50 cells were counted. On day 14, cells were harvested from the cultures and replated in osteogenic or adipogenic media and cultured for an additional 14 days. Osteogenic media: alpha MEM with 10% fetal bovine serum, 50µM ascorbic acid (Sigma) and 10µM of β-glycerophosphate (Sigma). Adipogenic media: alpha MEM with 10% fetal bovine serum, 5µg/mL insulin, 100µM indomethacin, and 100nM dexamethasone. Osteoblast differentiation was assessed using the Leukocyte Alkaline Phosphatase Kit (Sigma), per manufacture’s recommendations. Adipocyte differentiation was assessed by staining with Oil Red O, as reported previously10.
RNA expression profiling
PDGFRα+ Sca+ cells or CAR cells, pooled from 2–6 mice, were sorted directly into lysis buffer and RNA was prepared using the RNA XS column kit (Macherey-Nagel, Bethlehem, PA) according to the manufacturer’s directions. RNA was amplified using the NuGen Ovation system (NuGen, San Carlos, CA), and hybridized to the Affymetrix MoGene 1.0 ST array. Data normalization was performed using the Robust Multichip Average (RMA) algorithm. Submission of this RNA expression data to Gene Expression Omnibus is in progress.
Statistics
Significance was determined using Prism software (GraphPad). Unless otherwise stated, statistical significance of differences was calculated using 1- or 2-way ANOVA. P-values indicate the result of Bonferroni post-testing relative to Cxcl12fl/− control mice unless other comparisons are explicitly shown. P-values less than 0.05 were considered significant. All data are presented as mean ± SEM.
Supplementary Material
Acknowledgements
We thank Jill Woloszynek, Fulu Liu, Alex Khalaf, and Molly Romine for technical assistance; Gayle Callis, Stephen Oh, and Monica Vig for technical advice; Jackie Tucker-Davis for animal care, and Dr. Thomas Clemens for the Oc-Cre mice. This work was supported by NIH grants RO1 HL60772 (DCL) and F30 HL097423 (AMG).
Footnotes
Author Contributions
AMG and YMH designed and performed the research, analyzed the data, and wrote the manuscript. LGS, JNB, and RBD performed experiments characterizing hematopoiesis in the conditional Cxcl12 deficient mice. MJC designed and cloned the CXCL12 conditional knockout construct. DCL supervised all of the research and edited the manuscript, which was approved by all co-authors. The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Calvi LM, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 3.Hooper AT, et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 2009;4:263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Omatsu Y, et al. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33:387–399. doi: 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peled A, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 9.Sacchetti B, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 10.Semerad CL, et al. G-CSF potently inhibits osteoblast activity and CXCL12 mRNA expression in the bone marrow. Blood. 2005;106:3020–3027. doi: 10.1182/blood-2004-01-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokoyoda K, Egawa T, Sugiyama T, Choi BI, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Ara T, et al. A role of CXC chemokine ligand 12/stromal cell-derived factor-1/pre-B cell growth stimulating factor and its receptor CXCR4 in fetal and adult T cell development in vivo. J Immunol. 2003;170:4649–4655. doi: 10.4049/jimmunol.170.9.4649. [DOI] [PubMed] [Google Scholar]
- 13.Bonig H, Priestley GV, Nilsson LM, Jiang Y, Papayannopoulou T. PTX-sensitive signals in bone marrow homing of fetal and adult hematopoietic progenitor cells. Blood. 2004;104:2299–2306. doi: 10.1182/blood-2004-04-1605. [DOI] [PubMed] [Google Scholar]
- 14.Kawabata K, et al. A cell-autonomous requirement for CXCR4 in long-term lymphoid and myeloid reconstitution. Proc Natl Acad Sci U S A. 1999;96:5663–5667. doi: 10.1073/pnas.96.10.5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie Y, Han YC, Zou YR. CXCR4 is required for the quiescence of primitive hematopoietic cells. J Exp Med. 2008;205:777–783. doi: 10.1084/jem.20072513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzeng YS, et al. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood. 2011;117:429–439. doi: 10.1182/blood-2010-01-266833. [DOI] [PubMed] [Google Scholar]
- 17.Maes C, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Developmental cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 19.Arai F, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Foudi A, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagasawa T, et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 22.Morikawa S, et al. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J Exp Med. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhan M, Han ZC. Hemangiopoietin inhibits apoptosis of MO7e leukemia cells through phosphatidylinositol 3-kinase-AKT pathway. Biochemical and biophysical research communications. 2004;317:198–204. doi: 10.1016/j.bbrc.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann S, et al. Common deleted genes in the 5q- syndrome: thrombocytopenia and reduced erythroid colony formation in SPARC null mice. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2007;21:1931–1936. doi: 10.1038/sj.leu.2404852. [DOI] [PubMed] [Google Scholar]
- 25.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M, et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. The Journal of biological chemistry. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 27.Kisanuki YY, et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Developmental biology. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 28.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 29.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 30.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 32.Kisanuki YY, et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Developmental biology. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 32.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M, et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. The Journal of biological chemistry. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 35.Ara T, et al. A role of CXC chemokine ligand 12/stromal cell-derived factor-1/pre-B cell growth stimulating factor and its receptor CXCR4 in fetal and adult T cell development in vivo. J Immunol. 2003;170:4649–4655. doi: 10.4049/jimmunol.170.9.4649. [DOI] [PubMed] [Google Scholar]
- 36.Nagasawa T, et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 37.Semerad CL, Liu F, Gregory AD, Stumpf K, Link DC. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity. 2002;17:413–423. doi: 10.1016/s1074-7613(02)00424-7. [DOI] [PubMed] [Google Scholar]
- 38.Churukian C. Lillie's oil red O method for neutral lipids. Journal of Histotechnology. 1999;4:309–311. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.