SUMMARY
A central enigma in epigenetics is how epigenetic factors are guided to specific genomic sites for their function. Previously, we reported that a Piwi-piRNA complex associates with the piRNA-complementary site in the Drosophila genome and regulate its epigenetic state. Here, we report that Piwi-piRNA complexes bind to numerous piRNA-complementary sequences throughout the genome, implicating piRNAs as a major mechanism that guide Piwi and Piwi-associated epigenetic factors to program the genome. To test this hypothesis, we demonstrate that inserting piRNA-complementary sequences to an ectopic site leads to Piwi, HP1a, and Su(var)3–9 recruitment to the site as well as H3K9me2/3 enrichment and reduced RNA Polymerase II association, indicating that piRNA is both necessary and sufficient to recruit Piwi and epigenetic factors to specific genomic sites. Piwi deficiency drastically changed the epigenetic landscape and Polymerase II profile throughout the genome, revealing the Piwi-piRNA mechanism as a major epigenetic programming mechanism in Drosophila.
Keywords: piRNA, Piwi, chromatin modification, HP1a, RNA Polymerase II, gene expression, epigenetic regulation
INTRODUCTION
Epigenetics refers to the regulation of gene expression that is heritable to daughter cells without genetic variations. Epigenetic programming, occurring in a gene- and sequence-specific fashion, is commonly achieved via DNA methylation, covalent modification of histones, and association of epigenetic factors with chromatin (Kouzarides, 2007). However, most epigenetic factors, such as DNA methyltransferases/demethylases and histone modification enzymes, do not recognize specific DNA sequences. Then, what is the molecular mechanism that guides these epigenetic factors to their target sites in the genome? This question is central to epigenetics yet remains largely unexplored.
Recent studies indicate that certain transcription factors and non-coding RNAs can recruit epigenetic factors to their target sites. For example, the leukemia-promoting PML-RAR fusion protein induces gene hypermethylation and silencing by recruiting DNA methyltransferases to target promoters (Di Croce et al., 2002). During mouse pituitary development, ZEB1, a Kruppel-like repressor, can recruit the CoREST-CtBP co-repressor complex that contains the histone lysine demethylase LSD1 to target genes (Wang et al., 2007). In muscle satellite cells, transcription factor Pax7 recruits MLL2/Ash2L/Wdr5 complex to target promoters (McKinnell et al., 2008). In Drosophila, the Ultrabithorax (Ubx) transcripts containing three Trithorax Response Elements (TREs) can recruit the epigenetic regulator Ash1 to the template TRE to activate transcription (Sanchez-Elsner et al., 2006). Most notably, heterochromatin formation in the fission yeast S. pombe is RNAi-induced, in which the Argonaute1 (Ago1) protein and its associated small interfering RNAs (siRNAs0 serve as a guidance mechanism of RNA-induced transcriptional silencing (RITS) complex. The base-pairing between siRNAs and nascent non-coding transcripts from the pericentromeric heterochromatin recruits RITS complex into this region to initiate the heterochromatin formation (Grewal, 2010; Iida et al., 2008; Zofall and Grewal, 2006). Despite these exciting findings, transcription factors are unlikely to play a major role in epigenetic targeting in a genome-wide scale given their relatively small number of binding sites and their low sequence specificity in the genome. In contrast, hundreds of thousands of Piwi-interacting RNAs (piRNAs), each with a sufficient length to recognize any sequence specifically in the genome (see below), are logical candidates for sequence-specific targeting in the genome.
Here, we report that the Piwi protein and its associated piRNAs represent a major epigenetic guidance mechanism in Drosophila. Piwi is the founding member of the evolutionarily conserved Ago/Piwi protein family (Cox et al., 1998), which can be divided into Ago and Piwi subfamilies. The Ago proteins associate with siRNAs and hundreds of microRNAs (miRNAs) of 20~22 nucleotides (nt); whereas the Piwi proteins interact with hundreds of thousands of piRNAs of mostly 24~32 nt (Hock, 2008; Juliano et al., 2011). Several lines of evidence strongly implicate that Drosophila Piwi-piRNA complex plays an important role in epigenetic regulation. First, piwi mutations are suppressors of position effect variegation towards transgenic tandem arrays of Adh-white fusion genes (Pal-Bhadra, 2002), suggesting that Piwi functions as an epigenetic repressor towards these transgenes. Second, Piwi frequently co-localizes with nuclear Polycomb Group (PcG) bodies and promotes PcG-dependent inter-chromosomal associations (Grimaud, 2006). Third, Piwi is a nuclear protein in both germline and somatic cells (Cox et al., 2000), and has been further shown in salivary glands to be a chromatin-binding factor that binds to centromeres and more than one hundred distinct bands on polytene chromosomes (Brower-Toland et al., 2007).
Most relevantly, we have shown that a Piwi-piRNA complex specifically binds to the piRNA complementary sequence in a subtelomeric region and regulates the epigenetic status of the target sequence, implicating piRNA as a sequence-recognition and guidance molecule for Piwi (Yin and Lin, 2007). Furthermore, we have shown that Piwi directly binds to Heterochromatin Protein 1a (HP1a) and co-localizes with HP1a in many bands on polytene chromosomes (Brower-Toland et al., 2007). These observations led us to hypothesize that different piRNAs guide Piwi to numerous piRNA-complementary sites in the genome, which serves as an epigenetic guidance mechanism to recruit epigenetic factors such as HP1a to their target sites (Lin and Yin, 2008). Because Piwi associates with more than 14,000 piRNAs that correspond to many more target sequences in the genome due to the fact that many piRNAs arise from repetitive sequences (Brennecke et al., 2007; Cox et al., 2000; Saito, 2006; Vagin, 2006; Yin and Lin, 2007), this hypothesis, if proven to be true, should provides an effective answer to epigenetic programming in Drosophila.
To test this hypothesis, here we report high-resolution whole-genome mapping of Piwi, which reveals that Piwi-piRNA complexes indeed bind to numerous sites throughout the Drosophila genome. Furthermore, we provide direct in vivo evidence that piRNA is both necessary and sufficient to guide Piwi and HP1a to specific sites in the genome. Moreover, we report high-resolution whole-genome mapping of key epigenetic marks and RNA polymerase II (Pol II) in wildtype and piwi mutant flies, which indicates that Piwi is required for HP1a and other epigenetic factors to bind to their target sites in the genome. Taken together, these findings demonstrate that the Piwi-piRNA mechanism is a major epigenetic guidance and programming mechanism in Drosophila.
RESULTS
Piwi-piRNA Complexes Bind to Numerous piRNA-Complementary Sites Throughout the Genome
Although we have previously shown that a Piwi-piRNA complex can bind to a piRNA-complementrary site in heterochromatin to regulate its epigenetic state (Yin and Lin, 2007), it is not clear how generally this mechanism is utilized by the genome. To assess the impact of the Piwi-piRNA-mediated epigenetic regulation, we first investigated how many sequences in the genome are bound by Piwi-piRNA complexes by ChIP-Seq of nuclei isolated from adult flies using a Piwi antibody (Table S1). We have previously demonstrated that such a ChIP-Seq method can readily unveil key epigenetic paradigms shared by major types of cells (Yin et al. 2011). To test the specificity of the Piwi antibody, we performed Piwi ChIP-qPCR for six representative genes and heterochromatic regions that are bound by Piwi at high levels to see if these loci no longer have Piwi signal in the piwi1/piwi2 mutant. This indeed is the case (Figure S1), validating the specificity of the Piwi antibody. We further tested the validity of using nuclei isolated from adult flies by comparing Piwi binding at the six representative regions in whole flies versus in the ovary. ChIP-qPCR analysis indicated that the levels of Piwi binding at all six regions are very similar in both samples (Figure S1), validating the use of adult flies in our analysis.
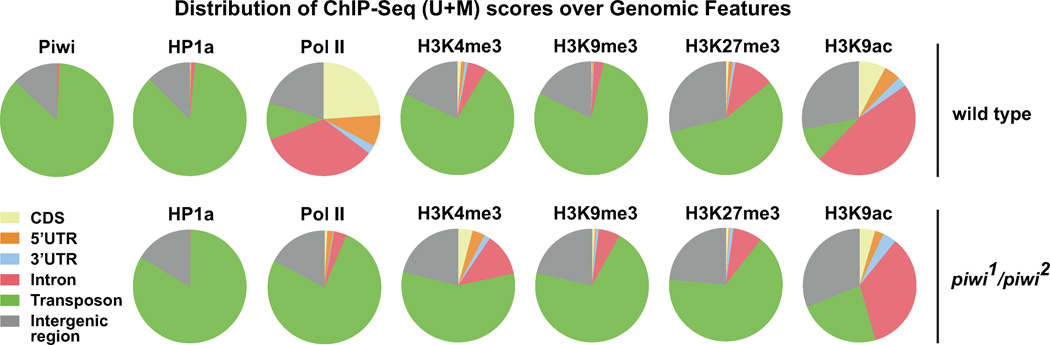
We used our Piwi ChIP-Seq data to map Piwi distribution in the genome at a 50-bp resolution with unbiased representation of both euchromatin and heterochromatin using an established bioinformatic method (Yin et al. 2011). Interestingly, we found ~86% of Piwi ChIP-Seq signals overlap with transposons and repetitive sequences (Figure 1), in line with the fact that ~92% of Piwi-associated piRNAs are derived from transposons and repetitive sequences (Brennecke et al., 2007; Yin and Lin, 2007). Expectedly, Piwi signals are evidently lower along gene-rich chromosome arms and higher along unallocated supercontigs (U, Uextra) that are abundant with transposons and repetitive sequences (Figure S2C). The remaining Piwi signals are within non-repetitive regions of the genome, and apparently do not correspond to any obvious sequence feature or particular gene expression pattern (Figure S2A–B).
Figure 1. Distribution of ChIP-Seq (U+M) scores over genomic features.
Distribution of ChIP-Seq (U+M) scores over CDS, 5’ UTR, 3’ UTR, introns, transposons, repetitive sequences, and intergenic regions within the whole genome (X, 2L, 2R, 3L, 3R, 4, XHet, 2LHet, 2RHet, 3LHet, 3RHet, YHet, U, Uextra). The top and bottom rows show the distributions in wild type and piwi1/piwi2 flies, respectively. See also Figures S1 and Table S1.
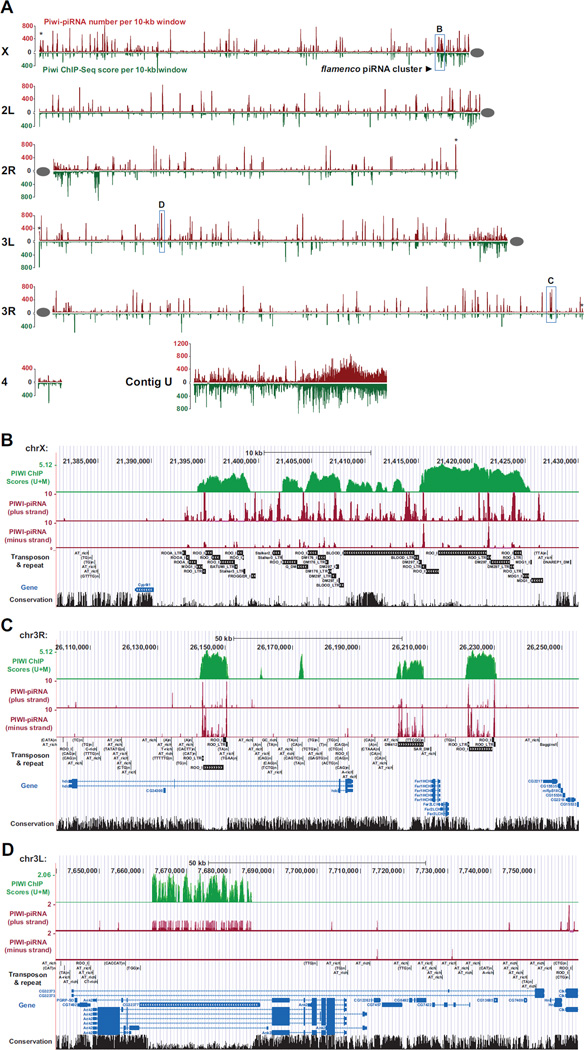
To determine whether Piwi binds to piRNA-complementary genomic sites, we plotted the high-resolution localization of Piwi and Piwi-bound piRNAs along euchromatic chromosome arms and a supercontig (U) representative of heterochromatic genome (Figure 2). Remarkably, Piwi and piRNAs peaks co-localize at vast majority of genomic sites (98.5% within 1-Kb window), covering numerous euchromatic genomic loci as well as heterochromatic genome, especially within pericentromeric regions and subtelomeric regions (Figures 2 and S2D-F).
Figure 2. Genome-wide co-localization of Piwi and Piwi-associated piRNAs.
(A) Genome-wide localization of Piwi and Piwi-associated piRNAs are shown for X, 2L, 2R, 3L, 3R, 4 and unassembled Contig U. The horizontal line shows the length of each chromosome arm proportionally. The red peak above the chromosomal line represents Piwi ChIP scores [sum of ChIP-Seq (U+M) scores per 10-kb window; both unique-mapping and multiple-mapping reads were considered] and the green peak below the chromosomal line represents the abundance of Piwi-associated piRNAs (numbers of piRNAs per 10-kb window). Grey ovals indicate centromeres. Asterisks denote enrichment of Piwi in telomere regions. Boxed regions labeled as B, C, and D are 260-Kb region containing the 42AB piRNA cluster, a 150-Kb region of sporadic transposons, and a 75-Kb region containing a gene CG32377, respectively. See also Figures S2. (B–D) Zoomed-in views of localization of Piwi and Piwi-associated piRNAs at the 42AB piRNA cluster (B), a sporadic transposon region (C), and CG32377(D).
To quantitatively measure the co-localization of Piwi and Piwi-associated piRNAs, we divided the whole genome into 168,724 one-kb windows and separately calculated Piwi ChIP-Seq scores and copy number-weighted Piwi-bound piRNA scores for every 1-kb windows. The Pearson correlation coefficient between Piwi scores and piRNA scores is extremely high (R2 = 0.954) for Piwi-located windows (Piwi scores > 0, representing 20.8% of all windows; Figure S2F). These observations indicate that the binding of Piwi is highly correlated to piRNA-complementary sequences throughout the genome, even in the context of all types of cells in adult flies.
Notably, at some of the Piwi-piRNA co-localized sites, Piwi signals are not proportional to that of Piwi-associated piRNAs. For example, only medium levels of Piwi signals are present within the most abundant piRNA cluster in cytological 42AB region in chromosome 2R (Figure 2A and B). This phenomenon also exists outside piRNA clusters (e.g., Figure 2A, C and D). These observations likely reflect that the piRNA peaks, determined by Piwi-piRNA co-immunoprecipitation, represent the total abundance of individual Piwi-piRNA complexs; yet Piwi peaks, determined by Piwi ChIP, represent only a subset of these complexes that are associated with chromatin. It is likely that different Piwi-piRNA complexes have different chromatin-binding efficacies, which would cause the observed discrepancy. In addition, the same Piwi-piRNA complex may have variable affinities towards different target sequences, yet by necessity, bioinformatically we can only follow the conventional method to evenly distribute a non-unique-mapping piRNA to all of its target sites, which would further skew the relative abundance. Therefore, we do not expect to see the abundance of Piwi peaks to be proportional to their cognate piRNA peaks. Despite of this, the Piwi and piRNA peaks are precisely co-localized in the genome (e.g. Figure 2B–D). These data reveal that Piwi-piRNA complexes bind to numerous sites in the genome, and implicate that piRNAs guide Piwi to all these sites.
piRNA Is both Necessary and Sufficient to Recruit Piwi to a piRNA-Complementary Site in the Genome
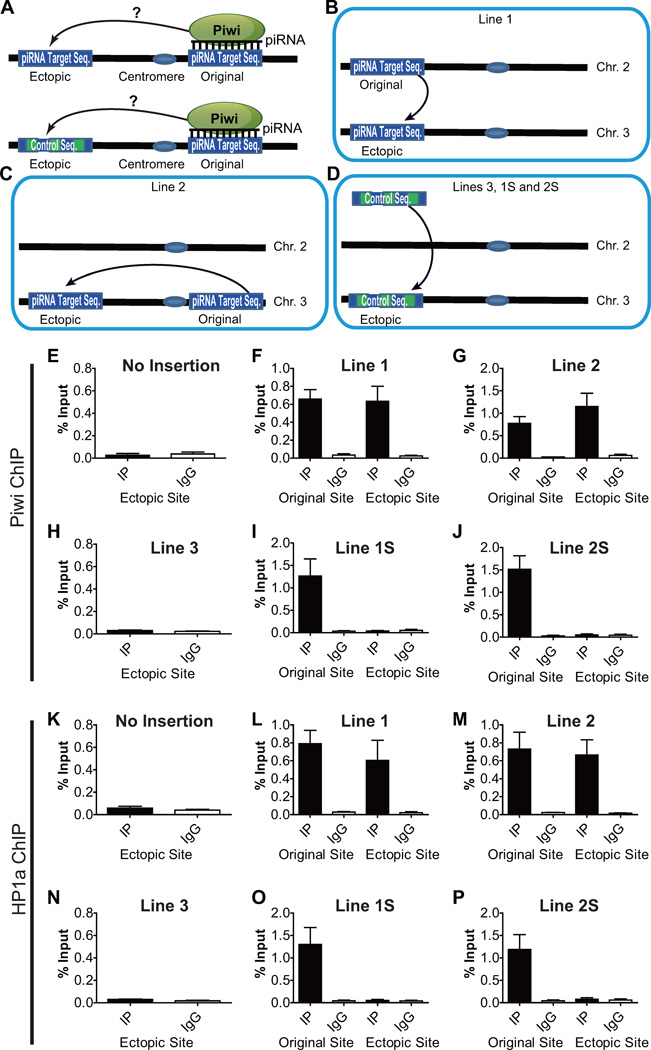
To investigate whether piRNA can indeed guide Piwi to piRNA-target sites in the genome, we examined if inserting a piRNA-complementary sequence to an ectopic site can lead to the recruitment of Piwi to that site (Figure 3A). We identified a 20-bp genomic sequence on the second chromosome that possesses unique complementarity to a specific Piwi-associated piRNA, and inserted eight tandem copies of this sequence into an ectopic site on the left arm of the third chromosome via the PhiC31 site-specific recombination system (Line 1, Figure 3B). Similarly, in another transgenic line, we inserted into the same ectopic site another six tandem copies of 27-bp unique piRNA-complementary sequence from the right arm of the third chromosome (Line 2, Figure 3C). As a negative control for the insertion of piRNA-complementary sequences, in a third transgenic line, we inserted a non-complementary sequence, a lacI sequence of similar length (158 bp), to the same ectopic site (Line 3, Figure 3D). To further rule out the possibility that 6–8 copies of tandem repeats per se might induce silencing and Piwi recruitment independent of piRNA, we also generated a transgenic line containing eight tandem copies of a scrambled 20-bp sequence (Line 1S, where S stands for scrambled) as a parallel control of Line 1, and another transgenic line containing six tandem copies of a scrambled 27-bp sequence (Line 2S) as a parallel control of Line 2 (Figure 3D).
Figure 3. Ectopic piRNA target sequences recruit Piwi and HP1a de novo.
(A) A diagram depicts the experimental design of the ectopic recruitment assay for Piwi-piRNA complexes.
(B–C) Two piRNA target sequences (originally located in 2L and 3R) were inserted to the same genomic location (cytoband 68A4 in 3L) via Phi31C recombination-based site-specific transgenic system. The resulting transgenic strains were referred as Line 1 and Line 2, respectively.
(D) An exogenous LacI sequence of similar length or same tandem-arrayed copies of scrambled sequences for Line 1 or 2 was also inserted to the same ectopic site as negative controls. The resulting transgenic strains were referred as Line 3, 1S, and 2S, respectively.
(E–J) Piwi is ectopically recruited to piRNA target insertion site. Piwi is not present in the ectopic site before the insertion (E), but is recruited to ectopic piRNA target insertion sites at a comparable level of enrichment as in the original sites in Line 1 (F) and Line 2 (G). Negative control lines (H–J) did not show any Piwi recruitment. IP: ChIP using antibody against Piwi. IgG: ChIP using non-specific IgG. Error bars in E-P denote S.D.
(K–P) HP1a is not enriched in the ectopic site before the insertion (K), but is also recruited to ectopic piRNA target sequence insertion sites in both Line 1 (L) and Line 2 (M), but not in negative control lines (N-P). IP: ChIP using antibody against HP1a. IgG: ChIP using non-specific IgG.
To examine whether the inserted piRNA-complementary sequences can indeed recruit Piwi–piRNA complexes to the ectopic site, we performed ChIP-qPCR assays with DNA primers specific to the original sites and the ectopic site with the inserted sequences. Prior to the insertion of piRNA target sequence, Piwi was not detected above the background level at the ectopic site yet the original site was associated with marked level of Piwi (Figure 3E). In contrast, when piRNA-complementary sequences originated from a different or the same chromosome were inserted into the ectopic site (Line 1 and Line 2, respectively), Piwi protein was evidently recruited to this ectopic site at levels comparable to that of the original site (Figures 3F–G). This indicates that the inserted piRNA-complementary sequences recruit Piwi to the ectopic site. Parallel ChIP experiments in Line 3, carrying a non-complementary sequence, did not exhibit Piwi enrichment at the ectopic site (Figure 3H). Furthermore, parallel ChIP experiments in Line 1S and Line 2S, containing tandem repeats of scrambled sequences for Line 1 and 2, respectively, did not show ectopic Piwi recruitment, either (Figure 3I and J). These results indicate that piRNA is both necessary and sufficient to recruit Piwi to a piRNA-target site.
HP1 Is Also Recruited to the piRNA-Target Site
Previously, we showed that Piwi directly binds to HP1a, and that Piwi colocalizes with HP1a at many of HP1a-positve cytological bands on polytene chromosomes (Brower-Toland et al., 2007). This led us to propose that Piwi–piRNA complexes recruit HP1a and possibly other epigenetic factors to Piwi-binding sites (Lin and Yin, 2008). We tested this hypothesis by investigating whether Piwi binding to the ectopic site also leads to the recruitment of HP1a to this site by ChIP-qPCR for HP1a. No HP1a binding was detected at the ectopic site before the insertion of the piRNA-complementary sequences (Figure 3K). As expected, HP1a is present at the ectopic insertion sites in Line 1 and 2, at similar levels to the original genomic sites (Figure 3L–M). No HP1a accumulation at the ectopic site was observed in Lines 3, 1S, or 2S (Figure 3N–P). Together, these observations indicate that HP1a is recruited to the ectopic piRNA-complementary site by Piwi-piRNA complexes instead of by other mechanisms such as tandem-repeat-induced silencing.
The Recruitment of Piwi and HP1a to piRNA Target Site Is RNase-Sensitive
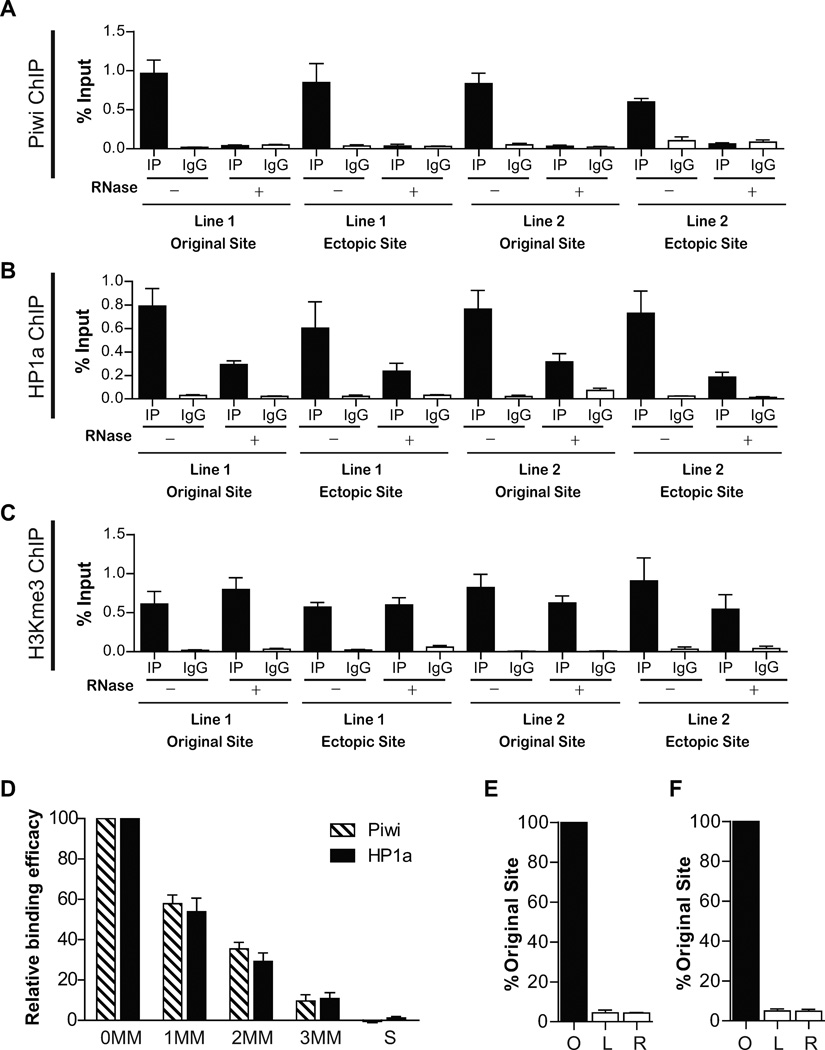
To further test the requirement of piRNA in Piwi and HP1a recruitment, we examined whether the binding of Piwi and HP1a to the ectopic site is RNase-sensitive. Isolated nuclei were treated with RNase A before crosslinking, so that RNA-dependent binding of epigenetic factors to chromatin would be affected. As expected, the binding of Piwi to both the original sites and the ectopic site was abolished by RNase A treatment (Figure 4A). These results further support the notion that Piwi binding to these sites are mediated by piRNAs. Likewise, HP1a binding to the ectopic site is also significantly reduced by RNase A treatment, although not completely abolished (Figure 4B). This indicates that HP1a binding and recruitment to these genomic sites depend, at least in part, on Piwi-piRNA complexes. To test whether the RNase treatment perturbed chromatin organization, we conducted ChIP-qPCR on H3K9me3 at both the original and the ectopic sites. H3K9me3 binding is not affected by the nuclease treatment (Figure 4C), validating that the nuclease treatment procedure did not disturb general chromatin structure.
Figure 4. The ectopic recruitment of Piwi and HP1a is sensitive to RNase treatment and is achieved by piRNA from the original site.
(A) Piwi ChIP in the presence or absence of RNase A digestion. In both Line 1 and Line 2, Piwi binding to the original site and the ectopic site is abolished by RNase treatment. Error bars in A-F denote S.D.
(B) HP1a ChIP in the presence or absence of RNase A. Similar to (A), HP1a binding to original site and the ectopic site is impaired in both Line1 and Line 2 by RNase treatment.
(C) H3K9me3 ChIP in the presence or absence of RNase A. Unlike (A) and (B), H3K9me3 signal in the original site and the ectopic site is not affected in both Line1 and Line 2 by RNase treatment.
(D) The impact of mismatches (MM) on the efficacy of piRNA biding to its target sequence. S: scrambled sequence. See also Figure S3.
(E–F) The ectopic sites in Line 1 (C) and Line 2 (D) produce negligible levels of RNA transcripts comparing to their original piRNA-generating sites. O: original site; L and R: sequences spanning the left and right insertion junctions at the ectopic site.
The RNase-resistant portion of HP1a binding to chromatin may reflect the presence of other, RNA-independent, HP1a recruitment mechanisms such as binding to H3K9me3 and H3K9me2. Indeed, we proposed that initial HP1a binding to chromatin leads to recruitment of histone methyltransferase Su(var)3–9 to the Piwi-piRNA binding sites, which methylates H3K9 at these sites and provides a substrate for subsequent HP1a binding that becomes independent of Piwi (Lin and Yin, 2008). This hypothesis is supported by our present study (see below).
piRNA Binding to Its Target Sites Is Highly Sequence-Specific
We next examined the degree of sequence-specificity of piRNA binding to its target site, which has not been investigated for either DNA or RNA targets in any system. We generated transgenic strains in which all of the six tandem copies of the ectopic piRNA binding sites in Line 1 carried identically a transverional mutantion and all of the eight tandem copies of the ectopic piRNA binding sites in Lines 2 carried identically another transversional mutation. These mutations are denoted as A, B, and C, respectively, when they are at the 25% (5’ side), 50% (mid-point), or 75% positions (3’ side) of the piRNA target sequence (see Experimental Procedures and Figure S3). Furthermore, we generated transgenic strains carrying double mutations of A and B (denoted as D), A and C (denoted as E), B and C (denoted as F), triple mutations of A, B, and C (denoted as G) and scrambled sequences (denoted as S, Figure S3). These mutant sequences would allow us to test not only the impact of mismatches on piRNA binding to its target site, but also possible position-dependence of such impact.
We subjected all of the 18 transgenic strains individually to the same ChIP-PCR analysis as described above to quantify the efficacy of these mutated ectopic target sequences in recruiting Piwi and HP1a. Single mutations decreased the recruiting efficacy of Piwi and HP1a to (57.8±4.3)% and (53.9±6.7)% of the wildtype level, respectively, without significant position-dependent variation (Figure 4D). Double mutations further decreased the recruiting efficacy for Piwi and HP1a to (35.4±3.2)% and (29.2±4.2)% of the wildtype level, respectively, again without significant position-dependent variation. The triple mutation drastically decreased the recruiting efficacy of Piwi and HP1a to only (9.5±1.1)% and (10.7±1.0)% of the wildtype level, and the scrambled sequence completely abolished the recruitment. These results indicate that mismatches have additive effect in reducing piRNA binding to its target sequences, and that three mismatches can effectively abolish piRNA binding to its target sequence. In addition, the position of a mismatch does not have obvious effect on piRNA binding efficacy, in contrast to miRNAs that have a 6–7 bp sub-sequence in their 5’ region (the “seed sequence”) that are more important than the remaining sequences (Huang and Lin, 2012).
The piRNAs That Recruit Piwi and HP1a to the Ectopic Site Are Produced from the Original Sites
We then examined whether the piRNAs responsible for the ectopic Piwi-HP1a recruitment are produced by the original sites and then act in trans at the ectopic site. Since these piRNAs are uniquely mapped in the genome, there are two possible origins and modes of action: (1) they are produced at their original genomic sites and then guide Piwi to the ectopic site in trans; or (2) they are also produced ectopically at the insertion site and assist the Piwi binding in situ. To distinguish between these two possibilities, we measured the transcription activity at the ectopic site. Site-specific primers covering both the upstream and downstream boundaries of the insertion site were designed to specifically measure potential RNA transcription activity at the ectopic site. cDNA libraries were prepared from both Line 1 and Line 2, and transcriptional activity at the ectopic site was measured at both the upstream and downstream of the insertion site as well as at the original piRNA production sites. Although the original site has robust transcriptional activity, no RNA was detected at the insertion site (Figure 4E–F). This suggests that these piRNAs are produced from the original sites and then guide Piwi to the ectopic site in trans.
Piwi and HP1a Further Recruit Histone Methyltransferase Su(Var)3–9 to the Ectopic Site and Induce a Suppressive Chromatin State at the Site
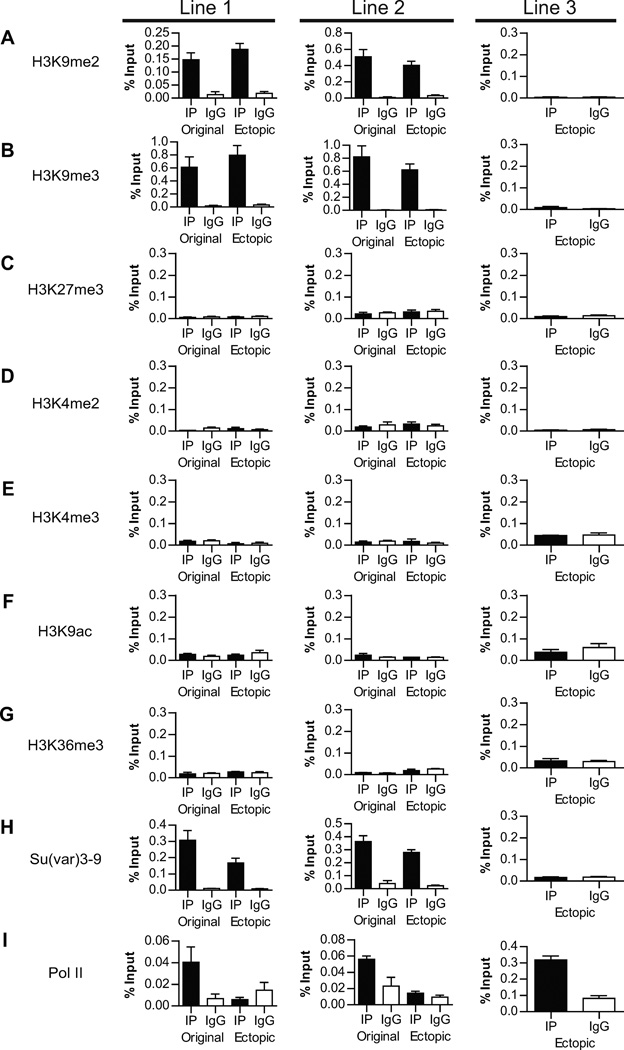
We then assessed the effect of ectopic Piwi recruitment on the chromatin state of the insertion site by examining a panel of chromatin marks in all three lines. ChIP-qPCR assays showed that repressive chromatin marks H3K9me2 and H3K9me3 are enriched at the ectopic site (Lines 1 and 2), which supports our hypothesis that RNase-resistant portion of HP1a is recruited by these histone marks (Figure 5A–B). In contrast, the active chromatin marks H3K4me2, H3K4me3, H3K9ac, H3K36me3 are not enriched to the ectopic site, indicating that Piwi induces a repressive chromatin state at the ectopic site (Figures 5D–G). The increase in these repressive marks is echoed by the reduction of elongating Pol II (Ser 2-phosphorylated) at the ectopic site to the background level (Figure 5I). Interestingly, a repressive mark, H3K27me3, is not enriched at the ectopic site, either (Figure 5C). This suggests that the repressive state of the ectopic site is likely independent of the PRC2 complex, which is known to specifically bind to PRE/TRE sequences.
Figure 5. The change of chromatin state at the ectopic site upon recruitment of Piwi.
ChIP-qPCR using antibodies specific to the histone marks (A–H) and RNA Pol II (I) indicate that the repressive histone marks H3K9me2/3 but not active histone marks are enriched at the ectopic sites. In addition, RNA Pol II binding is reduced upon ectopic Piwi recruitment. Error bars in denote S.D.
As both HP1a and its binding substrates H3K9me2/3 (Bannister et al., 2001; Lachner et al., 2001; Nakayama et al., 2001) are enriched at the ectopic site, we further examined whether piRNA can also recruit histone methyltransferase (HMTs) responsible for the H3K9 methylation to the ectopic site. In Drosophila, H3K9 is mainly methylated by Su(var)3–9, especially in pericentromeric heterochromatin (Ebert et al., 2004; Schotta et al., 2002). In addition, dSetDB and G9a are potential HMTs for methylation of other regions (Seum et al., 2007; Stabell et al., 2006; Tzeng et al., 2007; Yoon et al., 2008). Although it is known that Su(var)3–9 binds to heterochromatin via its SET and chromo domains (Schotta et al., 2002), what recruits Su(var)3–9 to specific genomic sites remains unknown. Since Su(var)3–9 has been demonstrated to interact with HP1a by yeast two-hybrid assays and co-immunoprecipitation (Schotta et al., 2002), we tested whether Su(var)3–9 is also recruited to the ectopic site through the piRNA-Piwi-HP1a- Su(var)3–9 interaction. ChIP-qPCR experiments were performed to measure Su(var)3–9 binding to chromosomes. Remarkably, Su(var)3–9 is also recruited to the ectopic piRNA-target-insertion sites (Figure 5H), supporting this hypothesis.
The Binding of Piwi-piRNA Complexes to Euchromatin and Heterochromatin Might be Achieved by piRNA-RNA and piRNA-DNA Pairing, Respectively
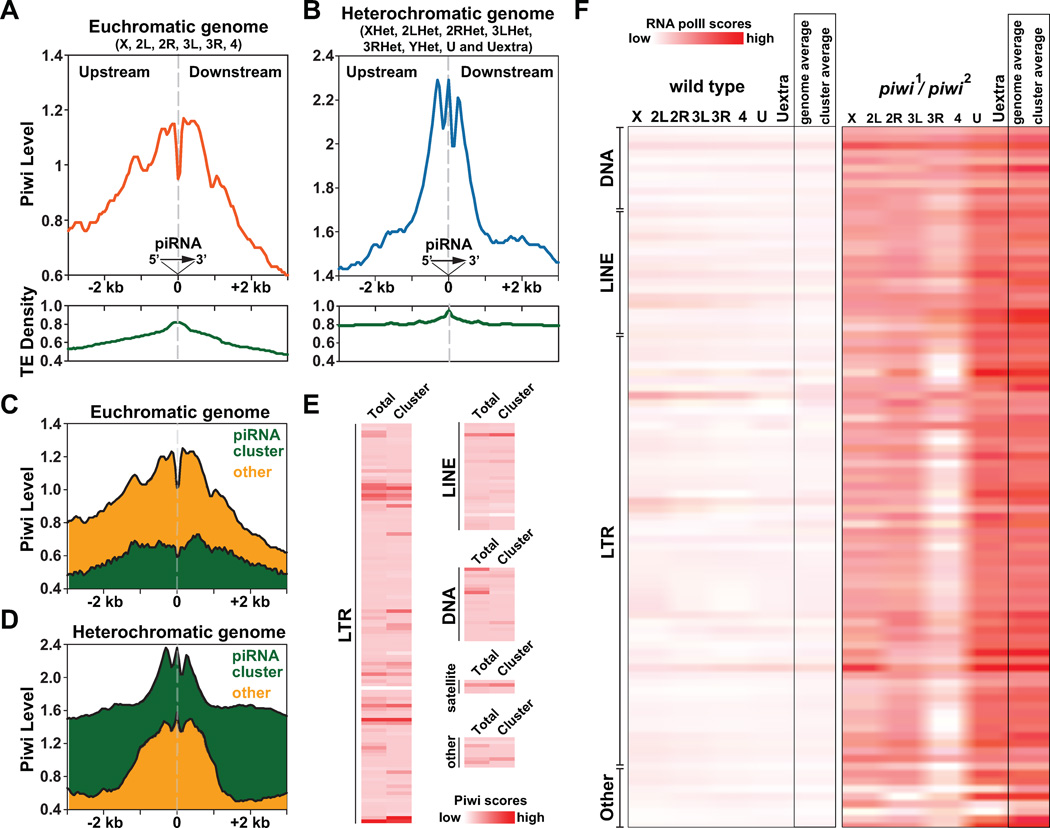
Having demonstrated the ability of piRNA to recruit Piwi and epigenetic factors to a specific site, we further explore the molecular basis of the Piwi-piRNA binding to their target sites at the genomewide scale. We aligned euchromatic and heterochromatic sequences cognate to Piwi-associated piRNAs at the base-pair resolution corresponding to the 5' ends of piRNAs, and calculated surrounding Piwi ChIP-Seq scores and transposon densities (±3-Kb range) at 10-bp resolution (Figure 6A–B). We found concentrated Piwi signals and transposon densities centered around piRNA cognate sequences in both euchromatic and heterochromatic genomes, especially within the central ±1-Kb regions. Intriguingly, euchromatic and heterochromatic regions show distinct patterns of Piwi distribution in the immediate vicinity of the piRNA-target sites. In euchromatin, Piwi signals show two peaks flanking the piRNA-target sites: one at - 800~−100 bp upstream of piRNA cognate sequences, the other at +100~+800 bp downstream of piRNA cognate sequences (Figure 6A). Notably, Piwi signals are remarkably low at a narrow region spanning the piRNA-target sequences (−100~+100 bp). In addition, the overall Piwi signals upstream of piRNA target sequences are notably higher than those downstream. In contrast, in heterochromatin, Piwi signals manifest a three-peak pattern, with the middle peak precisely centered at the piRNA-target sequences (Figure 6B). The middle peak is the highest, spans from −100~+100 bp, and is flanked by two other peaks upstream (−500~-200 bp) and downstream (+200~+500 bp) of the piRNA-target sequences, respectively.
Figure 6. Distinct co-localization patterns of Piwi and Piwi-associated piRNAs in euchromatin and heterochromatin suggest two modes of Piwi-piRNA guidance mechanism.
(A–B) Relative positions of Piwi, Piwi-associated piRNA and transposons within euchromatic genome (X, 2L, 2R, 3L, 3R, 4) and heterochromatic genome (XHet, 2LHet, 2RHet, 3LHet, 3RHet, YHet, U and Uextra). Piwi-associated piRNAs were aligned at their 5' ends in the same direction. Grey dash lines indicate positions of piRNAs. Piwi ChIP-Seq scores within upstream and downstream regions surrounding piRNA-transcribing regions (± 3-kb) were separately plotted for euchromatic genome (orange) and heterochromatic genome (blue), together with the transposon density (TE density; green). See also Figures S4.
(C–D) Relative positions of Piwi with piRNAs derived from piRNA clusters (green) and other sporadic piRNAs (orange).
(E) Heat maps depict Piwi ChIP-Seq scores over various types/classes of transposons within genome. Average Piwi ChIP-Seq scores of all same types of transposons within genome (total) or only within piRNA clusters (cluster) were separately calculated.
(F) Heat maps depicts levels of chromatin-associated RNA Pol II over various types/classes of transposons within wildtype flies (left) and piwi1/piwi2 mutants (right). Average RNA Pol II ChIP-Seq scores were separately calculated for all same types of transposons on chromosomal arms (X, 2L, 2R, 3L, 3R, 4), on contigs (U, Uextra) as well as for all same types of transposon within genome (genome average) or within piRNA clusters (cluster average).
The above two-peak vs. three-peak patterns of Piwi-piRNA localization holds true even when we separately analyzed unique-mapping piRNAs derived from piRNA clusters and from sporadic transposons (Figure 6C and D).
To examine if the two-peak vs. three-peak patterns are caused by possibly different phasing patterns of piRNA localization in euchromatin vs. heterochromatin, we calculated the relative distance between a piRNA and its neighboring piRNAs in euchromatin and heterochromatin. The distance between piRNAs does not show phasing in either euchromatin or heterochromatin (Figure S4A–C).
To examine if the two-peak vs. three-peak patterns are caused by different affinity of different types of transposons towards Piwi, we plotted the averaged levels of Piwi signals over 181 classes of transposons in the genome, including those specifically located within piRNA clusters (Figure 6E). Piwi signals are present in almost all types/classes of transposons; yet some transposons show differential Piwi levels within piRNA clusters. This implicates that Piwi levels are affected by local chromatin environment rather than by transposon sequences per se.
Thus, the above observations implicate that piRNA might associates with euchromatin and heterochromatin by two distinct piRNA-dependent mechanisms: In euchromatin, piRNA likely base-pairs with nascent RNA that is tethered to chomatin, with tethering arm ranging from 100~800bp. Such base-pairing can only occur after RNA polymerases traverse the piRNA complementary sequences, which explains observed more Piwi enrichment in the upstream region but not at the piRNA target sequences. In heterochromatin, Piwi-piRNA complexes might directly bind to the DNA target sequence, creating a middle Piwi peak centered precisely at the piRNA-target site. Additionally, the condensed structure of heterochromatin would allow the crosslinking of Piwi to the two adjacent nucleosomes, leading to the observed three-peak pattern.
Functions of Piwi in Global Distribution of Epigenetic Marks
Since Piwi-piRNA complexes bind to numerous sites in the genome and can guide HP1a to piRNA-complementary sites, we expected the Piwi-piRNA guidance mechanism to play a major role in epigenetic programming. To assess the impact of the Piwi-piRNA mechanism to the genome, we examined the global epigenetic states of Drosophila genome in piwi mutants. First, we compare the distributions of epigenetic marks and RNA Pol II in wildtype and piwi mutants in a genome-wide scale (Figure 1). In the piwi−/− genome, RNA Pol II signals are drastically redistributed from non-repetitive sequences (CDS, intron, 5’ UTR) into transposons/repetitive sequences. Similar trends were also observed for H3K9ac (Figure 1).
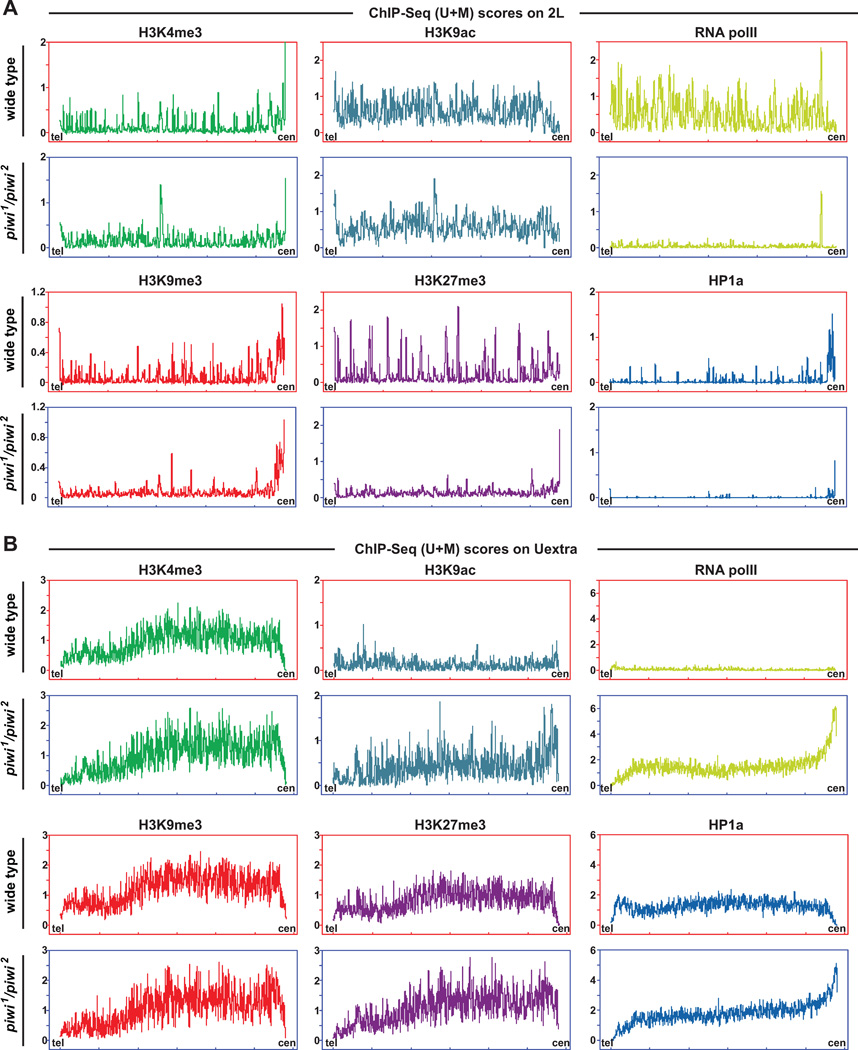
We then plotted the signals for these epigenetic marks along chromosome arm 2L (Chr2L) and supercontig Uextra, as representative distributions of these marks on euchromatic chromosomal arms and heterochromatic centromeric regions (Figures 7 and S5). Signals were calculated separately based on unique-plus-multiple mapping tags (U+M scores, Figure 7) or exclusively unique mapping tags [U scores, which yielded similar results to U+M scores; see Experimental Procedures and Yin et al. (2011)]. All signals were normalized to the total score of each mark in each type of genotype and thus represent the relative proportion of the mark as to the whole genome (Figures 7 and S5). By ChIP-Seq (U+M) scores, which reflect the overall changes, we observed that signals of H3K9me3 and HP1a are dramatically decreased over the euchromatic regions on Chr2L in piwi mutants, echoing the requirement of Piwi for HP1 binding to its target sites and H3K9me3 enrichement at these sites (cf. Figures 3–5). H3K27me3 signals in Ch2L euchromatic regions are also reduced. In contrast, pericentromeric regions of Chr2L show increased H3K27me3 signals and decreased HP1a signals. The levels of H3K9me3 are elevated at pericentromeric regions in wildtype flies and such elevation is maintained in piwi mutants. We found H3K4me3 levels appear to spread within euchromatic regions whereas H3K9ac signals are largely not affected by piwi mutation. Consistent with these changes, RNA Pol II is reduced in euchromatic regions but increased in pericentromeric regions enriched with transposons and repeats in piwi mutants (Figures 7 and S5). This trend is most obvious for RNA Pol II and HP1a at the centromere (right) end of this contig, based on both ChIP-Seq (U+M) and (U) scores. Taken together, our data clearly revealed a genome-wide redistribution of epigenetic marks between low-repetitive/euchromatic chromosome arms and high-repetitive/ heterochromatic and centromeric/pericentromeric regions in piwi mutants, which illustrates a profound impact of Piwi-piRNA-mediated guidance mechanism on global epigenetic landscape.
Figure 7. Chromosome-wide changes of chromatin states in Piwi mutants.
(A) Distribution of various epigenetic regulators/marks over the entire chromosome arm 2L [ChIP-Seq (U+M) scores; both unique-mapping and repetitive sequences were considered] in wildtype and piwi1/piwi2 mutants. cen: the centromere end of 2L. tel: the telomere end of 2L. ChIP-Seq scores were binned and averaged for every 20-kb windows on the plots. See also Figures S5 and S6, and Tables S2 and S3.
(B) Distribution of various epigenetic regulators/marks over the entire contig Uextra [ChIP-Seq (U+M) scores; both unique-mapping and repetitive sequences were considered] in wildtype and piwi1/piwi2 mutants. See also Figures S5 and S6, and Tables S2 and S3.
Piwi Functions in Global Transcriptional Silencing of Transposons
Previous studies have discovered that transcripts from several transposons are dramatically accumulated in the germline of piwi mutants (Kalmykova, 2005; Klenov, 2007; Vagin, 2006). It is proposed that this transposon silencing function of Piwi occurs at both transcriptional (Brower-Toland et al., 2007; Klenov, 2007) and post-transcriptional levels (Brennecke et al., 2007). Although it was found that transposon de-repression in piwi mutants, as referred from increased transposon RNA levels, is accompanied with opening of chromatin structures (Klenov, 2007), no direct evidence, such as RNA Pol II accumulation, is available that demonstrates the increased transposon RNA levels are due to increased transcription. To address this question, we conducted ChIP-Seq using RNA Pol II antibodies to measure RNA Pol II-dependant transcriptional activities over various types of transposons in both newly eclosed wildtype and piwi1/piwi2 mutant flies, which have similar cellular contents in their germline. Although the cellular levels of both total RNA Pol II and the activated RNA Pol II (serine-2-phosporylated) are similar in wildtype and piwi mutant flies (Figure S4D), all types of transposons show drastically increased association of RNA Pol II on chromatin in piwi mutants, with an average of 10.5-fold increase comparing to the wildtype level (Figure 6F). Interestingly, the increase of transcriptional activities occurs unevenly across the genome. Transposons within piRNA clusters and heterochromatic regions show 1.3-fold and 1.5-fold higher levels of transcription than transposons in euchromatic regions. Despite the overall increase of chromatin-associated RNA Pol II, its association with the LTR regions of several transposons (e.g. Burdock_LTR, Gypsy7_LTR) are decreased in euchromatic genomes of piwi mutants. These observations directly demonstrate the overall function of Piwi in global transcriptional silencing of transposons, and implicate that such silencing function is subject to influences from local chromatin states.
Redistribution of Epigenetic Marks within Non-Repetitive Sequences and Around Protein Coding Genes
To further investigate the influence of Piwi-piRNA on protein-coding gene expression, we examined distribution patterns of various epigenetic marks within non-repetitive genomic features [Figure S5A–E for piwi mutants, compared to wildtype profiles in Figure 3A–3E fromYin et al. (2011)] and around protein coding genes [Figure S5F–J for piwi mutants, compared to wildtype profiles in Figure 3F–3J fromYin et al. (2011)]. We found that the associations of H3K4me3 score continuum with various genomic features are similar between wildtype flies and piwi mutants, with 5’ UTR and CDS features over-represented in top 20% windows of the highest H3K4me3 scores [Figure 3A fromYin et al. (2011) and Figure S5A]. Similar to wildtype flies, H3K4me3 signals are also enriched at the 5’ ends (+50~+750 bp) of protein coding genes and positively correlated to gene expression in both types of flies [Figure 3F fromYin et al. (2011) and Figure S5F]. However, this mark notably appears to spread into proximal promoter regions (−1 kb ~ TSS) in piwi mutants (Figure S5F, left panel).
We observed that the associations of H3K9ac scores with various genomic features mimic those of wildtype flies. Over gene regions, H3K9ac levels decrease to ~20% of wildtype levels with similar distribution over gene bodies and transcription end points (Figure S5B). Interestingly, we also noticed that H3K9ac spreads into proximal promoter regions similar to H3K4me3 (Figure S5G).
As a heterochromatic mark, H3K9me3 is present within promoter regions at low levels [see Figure 3C fromYin et al. (2011)] and is completely abolished in piwi mutants (Figure S5C and H), with the remaining signals quite evenly distributed and indistinguishable from background signals. The disappearance of H3K9me3 marks from the promoter region is consistent with the spreading of H3K4me3 and H3K9ac marks into this region and the loss of H3K9me3 in euchromatic region (Figures 7A and S5A).
We found that high H3K27me3 scores (top 20%) are still present within intergenic regions in piwi mutants (Figure S5D). Similar to wildtype, relatively low levels of H3K27me3 in protein coding genes are negatively correlated to gene expression in piwi mutants (Figure S5I).
As a direct indicator of transcription activities, distributions of RNA Pol II over various genomic features are very similar to those of wildtype flies (Figure S5E). Accordingly, RNA Pol II shows almost identical distribution patterns over protein coding genes, with a prominent peak immediately downstream of TSS (Figure S5J, TSS~+100 bp, left panel).
Causal Relationships among Changes of TSS-Bound Epigenetic Regulators/Marks and Adjacent Transposons
The above data reveal profound influences of Piwi-piRNA-mediated epigenetic guidance mechanism on global epigenetic landscapes of transposons and correlative changes of epigenetic states for protein coding genes. To understand the effects of this mechanism on gene expression, we identified 353 activated genes and 2,396 repressed genes in piwi mutants based on the RNA Pol II-association scores and indexes over TSS regions. Gene ontology analyses indicate that the activated genes are enriched for those involved in nuclear functions such as DNA-binding and alternative splicing; whereas genes encoding for proteins related to cytoplasmic functions are significantly enriched among repressed genes (Tables S2 and S3). These data strongly suggest that the global changes of epigenetic marks result in a systematic change of cell functions in piwi mutants.
To reveal the underlying mechanism, we utilized Bayesian network to infer the causal relationships among TSS-bound epigenetic marks and presence of Piwi (Figure S6A). To faithfully represent causal relationships, we visualized the Bayesian network by directional compelled edges, color-coded with correlation between a pair of nodes (Figure S6B). We discovered positive influences of Piwi on epigenetic activation and increased transcriptional activities of nearby genes. Such an influence is not directly due to the proximity of these genes with centromeric heterochromatic regions. Furthermore, we extracted hierarchical decisive influences on RNA Pol II-dependent transcriptional activities (Figure S6C). We found that among those genes with unchanged levels of H3K9ac and increased or unchanged levels of HP1a, genes with high or medium levels of Piwi over adjacent transposons tend to have increased levels of transcriptional activities. These observations strongly infer that the transcriptional activations of transposons in piwi mutants establish transcription favorable epigenetic states for nearby genes and thus affect their expression.
DISCUSSION
In this study, we have systematically demonstrated the existence of the Piwi-piRNA epigenetic guidance mechanism and its function as a major mechanism of guiding epigenetic factors to their target sites in Drosophila. This mechanism provides a clear and effective answer to the long-standing question on how epigenetic factors are recruited to their specific target sites to achieve epigenetic programming throughout the genome. Given that some other Piwi proteins and piRNAs also exist in the nucleus of other organisms including mammals, this mechanism might have profound significance in diverse organisms.
The Piwi-piRNA Complex Is a Major Epigenetic Programming Mechanism in Drosophila
Our whole-genome mapping produced the high-resolution map of Piwi and piRNA binding to the genome. The perfect colocalization of Piwi and piRNA binding sites is expected given their association as molecular complexes. These Piwi-piRNA complexes directly bind to many regions in the genome, exerting epigenetic repression at most of the target sites. This may account for the diverse biological functions of Piwi in different cell types during development. Especially, many Piwi-piRNA complexes bind to transposon sequences, this may be a major mechanism that is responsible for transposon silencing by Piwi as reported in many studies (Juliano et al., 2011).
Furthermore, our analysis here revealed, at the whole-genome scale, the dependence of HP1a and H3K9 methylation on Piwi, which implicates that HP1a and histone methylases are recruited by Piwi-piRNA complexes as a major mechanism to many sites in the genome. It is important to note that Lei and colleagues reported that in several piRNA clusters, HP1 binding is apparently unaffected by Argonaute proteins, including Piwi (Moshkovich and Lei, 2010). We fully anticipate this result because Piwi colocalizes with HP1a at many, but not all, HP1a-containing bands on polytene chromosomes. The binding of HP1a to chromatin at Piwi-free sites must be via a different mechanism, possibly via the canonical H3K9me2/3-medaited mechanism. These data, combined with our findings, indicates that there are at least two different ways for recruiting HP1a to the chromatin, with Piwi-piRNA mechanism as a main way of recruitment, as demonstrated in this study and implicated by the polytene staining data (Brower-Toland et al., 2007).
piRNA Is both Necessary and Sufficient to Recruit Piwi and Epigenetic Factors to Its Complementary Sequences in the Genome
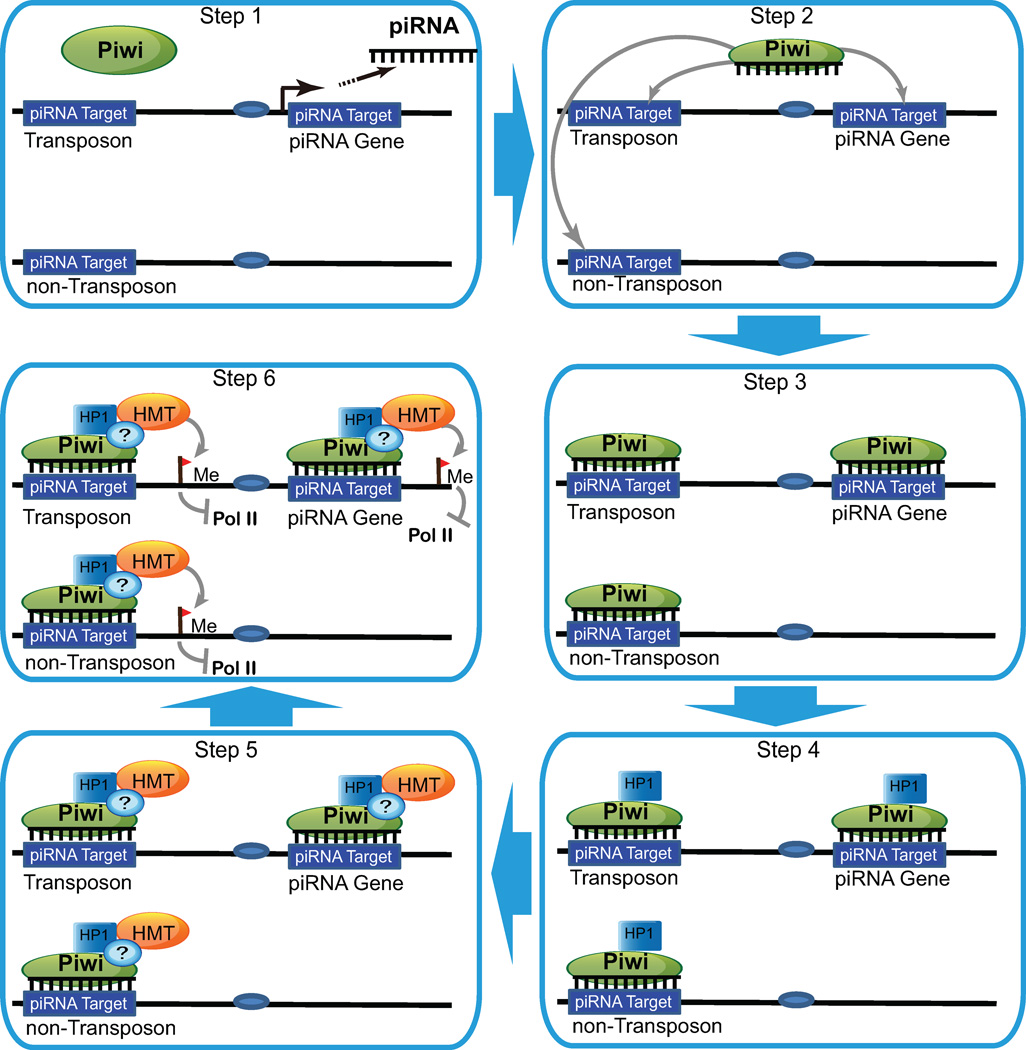
Our results demonstrate that piRNA is both necessary and sufficient to bring Piwi to specific genomic sites in a sequence-specific manner, and reveal a crucial role of piRNA in guiding epigenetic factors to specific sites in the genome. This epigenetic guidance mechanism, as illustrated in Figure 8, is similar to the RNAi-mediated heterochromatin formation in the fission yeast in that both are mediated by small RNAs and Piwi/Ago family proteins (Shiv Is, 2010; Verdel and Moazed, 2005). However, it distinctly differs from the yeast pathway in three major aspects. First, it recruits HP1a without H3K9 methylation, which then leads to recruitment of HMT and H3K9 methylation. This is in stark contrast to the yeast RNAi pathway in which the RITS complex first recruits HMT, which then leads to the methylation of H3K9 and eventual recruitment of HP1 (Iida et al., 2008). In addition, this is also in sharp contrast to the known H3K9 methylation-dependent mechanism of HP1a recruitment in higher eukaryotes, and represents a novel H3K9 methylation-independent mechanism. The recruitment of HMT by HP1a would lead to H3K9 methylation, which would result in further recruitment of HP1a molecules to the site, thereby stabilizing the repressive state of the chromatin. Secondly, the Piwi-piRNA-mediated epigenetic guidance mechanism can lead to transcriptional repression or activation, depending on the genomic context (Lin and Yin, 2008; Yin and Lin, 2007). Lastly, the Piwi-piRNA mechanism involves single-stranded piRNAs and Piwi proteins rather than double-stranded siRNAs and Ago proteins. Given the genomic complexity of the higher eukaryotes, piRNAs, mostly 24–32 nucleotides in length, are ideal candidate molecules for conferring sequence specificity in a genome-wide context. Indeed, the extreme complexity of the identified piRNAs, with more than 20,000 piRNAs associated with Piwi alone in Drosophila (Brennecke et al., 2007; Yin and Lin, 2007) and more than 58,000 piRNAs in mammals (http://pirnabank.ibab.ac.in/stats.html), renders the Piwi-piRNA pathway a likely major epigenetic factor guidance mechanism in Drosophila, and possibly even in mammals.
Figure 8. The Piwi-piRNA epigenetic guidance model.
The steps of epigenetic factor recruitment by the Piwi-piRNA complex. For details, see text.
Our high-resolution mapping analysis suggests that piRNAs might associate with euchromatin by binding to nascent RNA transcripts of 100–800 bp yet with heterochromatin by directly binding to DNA. This is in perfect agreement with our previous observation that Piwi binding to euchromatin and hetertochromatin is sensitive to RNase III and RNase H that selectively digest dsRNA and RNA-DNA hybrid, respectively (Brower-Toland et al., 2007). Given the complexity of heteochromatic context, it is not clear so far exactly how piRNA binds to heterochromatic DNA. However, it is conceivable that such direct binding might occur between piRNA and single-stranded DNA (e.g. during DNA replication or transcription) or between piRNA and DNA duplex. Future studies will resolve these hypotheses.
It is also worthy noting that the sequence-specificity of piRNA binding to its targets is additve with respect to individual base pairs. Each mismatch compromises the piRNA binding efficacy by ~40%, so that a piRNA carrying three point mutations retains only ~10% its binding ability to target sequences. This is in contrast to siRNA targeting that requires perfect complementarity and miRNA targeting that requires a mismatch in the middle position yet requires perfect match in the base 2–7 “Seed sequences” (Huang and Lin, 2012). Indeed, the graded sequence specificity of piRNA binding to its target sequences might create a mechanism of quantitative regulation that allows piRNAs to guide Piwi and epigenetic factors to even more genomic sites with graded effects as well as bestows tolerance to point mutations that frequently occur in heterochromatic and repetitive sequences.
EXPERIMENTAL PROCEDURES
DNA Constructs
Two piRNA targets (six tandem copies of 2L piRNA target 5'- CCA GAG GCA ACT GGA TCC ATG GCT GAA-3' and eight tandem copies of 3R piRNA target 5'-CG CAC CAA ATA ATA AAC TGA-3', for Line 1 and Line 2, respectively) and a 158-bp exogenous lacI sequence (GTGGTGTCGA TGGTAGAACG AAGCGGCGTC GAAGCCTGTA AAGCGGCGGT GCACAATCTT CTCGCGCAAC GCGTCAGTGG GCTGATCATT AACTATCCGC TGGATGACCA GGATGCCATT GCTGTGGAAG CTGCCTGCAC TAATGTTCCG GCGTTATT) were cloned into pENTRTM/D-TOPO entry vectors by pENTRTM/Directional TOPO Cloning Kits (Invitrogen K2400-20). Through the LR reaction, the insertion fragments were introduced into a gateway vector (UASp-Gateway Cassette-mini white-attP-Amp).
To assess the sequence specificity of piRNA binding, eight different transgenic lines with single, double and triple point mutations and with random sequences were generated for both Line 1 and Line 2, respectively. As shown in Figure 4 and listed below, A, B and C denotes the single transversion mutations in the piRNA targets sites, at the 25%, 50% (midpoint), and 75% positions, respectively. D carries double mutations of A and B; E acrries double mutations of A and C; and at 25% and 75% positions; and F carries double mutations of B and C. G carries the A, B, and C triple mutations. H has a non-related random repeat sequence from lacI that exactly matches the length and repeat copy number in either Line 1 or Line 2.
| Insertion | Line1 | Line2 |
|---|---|---|
| wildtype | 6x(ccagagGcaactgGatccatGgctgaa) | 8x(cgcaGcaaaTaataTactga) |
| A | 7G->C | 5C->G |
| B | 14G->C | 10T->A |
| C | 21G->C | 15A->T |
| D | 7G->C, 14G->C | 5C->G, 10T->A |
| E | 7G->C, 21G->C | 5C->G, 15A->T |
| F | 14G->C, 21G->C | 10T->A, 15A->T |
| G | 7G->C, 14G->C, 21G->C | 5C->G, 10T->A, 15A->T |
| H (Scrambled) | 6x(tcgatggtagaacgaagcggcgtcgaa) | 8x(ggcggtgcacaatcttctcg) |
Transgenic Fly Generation
All 19 transgenic lines of flies were generated by site-specific transformation of Drosophila via phiC31 integrase-mediated recombination. The fragments were integrated into attB2 sites and the transgenic flies were screened and balanced.
Chromatin Immunoprecipitation
The dissected fly ovaries were homogenized and the nuclei were isolated by sucrose cushion, washed by chromatin IP buffer (Tris 10 mM, pH 7.4, NaCl 140 mM, EDTA 1 mM, EGTA 0.5 mM, Sodium deoxycholate 0.1%, Triton X-100 1%, PMSF 1 mM, Roche Protein Inhibitor cocktail 1x, Glycerol 0.5%) and spun down at 5,000 rpm for 1 min at 4°C. The resuspended nuclei were subject to 0.2% formaldehyde crosslinking and sonication. The lysate was further spun down at 10,000 rpm for 1 min at 4°C, and was incubated at 4°C for 8 hours separately with the following antibodies, anti-Piwi (generated in Lin laboratory, raised in guinea pigs against Piwi C-terminal peptide), 1:40; anti-HP1a (Covance), 1:40; RNA Pol II H5 Monoclonal Antibody, (Covance), 1:40; anti-H3K4me3 (Active Motif AM 39159), 1:40; anti-H3K9me2 (Abcam ab1220), 1:40; anti-H3K9me3 (Upstate, 07–442), 1:40; anti-H3K9ac (Active Motif, AM 39137), 1:40; anti-Su(var)3–9 (Abcam ab4811), 1:40; anti-H3K27me3 (Upstate, 07–449), 1:40; anti-H3K36me3 (Abcam ab9050), 1:40. Control IgG immunoprecipitation experiments were conducted in parallel. Salmon sperm DNA coated Protein A/G Agarose beads (Upstate) were added to the sample and were co-incubated at 4°C for 3 hour. After washing, the sample was reverse cross-linked at 65°C for 3 hours. DNA was purified from the beads by eluted and purified for subsequent quantitative PCR reactions. For the RNase treatment, the nuclei were incubated with DNase-free RNase A (Ambion, 10 µg/ml) at 20°C for 1hr.
ChIP-qPCR and RT-qPCR
Genomic DNA fragments purified by ChIP were used as templates in the quantitative PCR reaction on a Bio-rad CFX96 real-time PCR machine. Each primer set was tested on the input fragmented DNA to determine the amplification efficiency and the melting curve. For each qPCR reactions, the percentage of ChIP-DNA and IgG mock ChIP-DNA as a share out of the input was calculated and used to compare the association of various protein factors in different genomic regions in different transgenic lines.
RT-qPCR was conducted to measure the transcription activity. Total RNA was extracted from ovaries and cDNA libraries from Lines 1 and 2 were prepared by Superscript III, and were subjected to qPCR. The primer was designed to bridge over the insertion site. The transcription activity at the ectopic site was measured from both left and right ends. For a summary of primers, see Supplemental Experimental Procedures.
Nuclei Isolation and ChIP-Seq
Isolation of Nuclei and ChIP-Seq were conducted according to methods developed in the lab (Yin et al., 2011).
Western Blotting for the Total and Phospho-Ser2 RNA Pol II Levels
Nuclei isolated from Wildtype and piwi1/piwi2 flies were probed by antibodies against total Pol II and phospho-Ser2 Pol II, respectively. Antibody against unphosphorylated Drosophila CTD (of Rbp1) is affinity-purified goat IgG, with dilution rate 1:1000 (a gift from Dr. A. Greenleaf, Duke University). Antibody against posho-Ser2 Drosophila RNA Polymerase II is H5 Monoclonal Antibody (Catalog # MMS-129R, from Covance). The blots were stained with Ponceau-S solution to visualize the total amount of the lysates on the filters.
ChIP-Seq Analysis Pipeline
Bioinformatic analysis of ChIP-Seq data was according to a method developed in the lab (Yin et al., 2011). Briefly, sequenced 35-nt reads and corresponding quality tracks were mapped to Drosophila melanogaster genome (Dm3) by SOAP. To calculate raw scores, the fly genome was divided into 50-bp windows. Each mappable Illumina read contributed the scores of its mapped windows and downstream windows based on an experimentally determined library length profile. The raw scores, being sums of scores contributed by reads (either all reads or only unique-mapping reads) were normalized based on the background signal levels and total sequenced reads (Yin et al., 2011). For each ChIP-Seq sample, the final adjusted enrichment scores (ChIP-Seq scores) were separately calculated based on unique-mapping reads only [ChIP-Seq (U) scores] or based on all mappable reads (including multiple-mapping reads) [ChIP-Seq (U+M) scores]. ChIP-Seq scores were recorded into files in wiggle track format (WIG) and browser extensible format (BED) for viewing the data in Integrated Genome Browser (Affymetrix) and UCSC Genome Browser.
Alignment of piRNAs with Piwi and Transposons in the Genome
Genomic locations and strandness of Piwi-associated piRNAs and genomic spans of piRNA clusters were collected from previous published dataset (Yin and Lin, 2007). Sporadic piRNAs and piRNAs within piRNA clusters within euchromatic and heterochromatic genomes were aligned at their 5’ end in the same direction. Piwi levels surrounding piRNAs (–3 kb ~ +3 kb) were weighted with piRNA counts and averaged in every 50-bp windows. Transposons were aligned in the same way as Piwi except that every 50-bp windows overlapping with transposons were given score 1 while non-overlapping ones were given score 0.
Bayesian Network Inference
For epigenetic regulators and marks, ChIP-Seq scores of wildtype flies were subtracted by corresponding scores of piwi mutant flies. These differential scores were averaged for 2 kb regions (−1 kb ~ +1 kb) surrounding 7,826 TSS of protein coding genes within the euchromatic genome. For each epigenetic regulator or mark, the averaged scores of TSS were classified into three groups (increased, decreased, unchanged) by unsupervised k-means clustering. For each TSS, Piwi scores over adjacent (−10 kb ~ +10 kb) transposons (TE) were averaged and classified into three groups (high, medium, low) by unsupervised k-means clustering. We employed a previously published method to calculate joint conditional probability and build the preliminary potential Bayesian networks (Yu et al., 2008).
Gene Ontology Analyses
Flybase IDs of grouped genes were analyzed by a web-based Functional Annotation Tool of Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/).
Supplementary Material
Highlights.
-
-
Piwi-piRNA complexes bind to numerous piRNA-complementary sequences in the genome.
-
-
piRNA is necessary and sufficient to recruit Piwi, HP1, and HMT to its target site.
-
-
Piwi deficiency drastically changes the epigenetic landscape in the genome.
-
-
The Piwi-piRNA mechanism is a major epigenetic programming mechanism in Drosophila.
ACKNOWLEDGEMENTS
We thank Drs. Arnold Greenleaf for RNA Pol II antibodies, Lynne Cooley for phiC31 constrcut, Andrew Xiao, Jamy Peng, and Jonathan Saxe for critical reading of the manuscript. This work is supported by the NIH (DP1CA174418) and a G. Harold & Leica Y. Mathers Award to H.L. and the Connecticut Stem Cell Research Foundation Hybrid Grant 065CE01 to M.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes and Development. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HA, Lin H. The miRNA Regulation of Stem Cells. WIRES Dev Biol. 2012;1:83–95. doi: 10.1002/wdev.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Nakayama Ji, Moazed D. siRNA-Mediated Heterochromatin Establishment Requires HP1 and Is Associated with Antisense Transcription. Molecular Cell. 2008;31:178–189. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lin H, Yin H. A novel epigenetic mechanism in Drosophila somatic cells mediated by PIWI and piRNAs. Cold Spring Harb Symp Quant Bio. 2008;73:273–281. doi: 10.1101/sqb.2008.73.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Bontron S, Reo E, Delattre M, Spierer P. Drosophila G9a is a nonessential gene. Genetics. 2007;177:1955–1957. doi: 10.1534/genetics.107.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiv Is G. RNAi-dependent formation of heterochromatin and its diverse functions. Current Opinion in Genetics & Development. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell M, Eskeland R, Bjorkmo M, Larsson J, Aalen RB, Imhof A, Lambertsson A. The Drosophila G9a gene encodes a multi-catalytic histone methyltransferase required for normal development. Nucleic Acids Res. 2006;34:4609–4621. doi: 10.1093/nar/gkl640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng TY, Lee CH, Chan LW, Shen CK. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc Natl Acad Sci U S A. 2007;104:12691–12696. doi: 10.1073/pnas.0705534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Letters. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Yin H, Sweeney S, Raha D, Snyder M, Lin H. A High-Resolution Whole-Genome Map of Key Chromatin Modifications in the Adult Drosophila melanogaster. PLoS Genet. 2011;7:e1002380. doi: 10.1371/journal.pgen.1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Lee KS, Park JS, Yu K, Paik SG, Kang YK. dSETDB1 and SU(VAR)3-9 sequentially function during germline-stem cell differentiation in Drosophila melanogaster. PLoS ONE. 2008;3:e2234. doi: 10.1371/journal.pone.0002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Zhu S, Zhou B, Xue H, Han JD. Inferring causal relationships among different histone modifications and gene expression. Genome Res. 2008;18:1314–1324. doi: 10.1101/gr.073080.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol. 2006;71:487–496. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.