Summary
A clear understanding of myocardial development is essential not only for understanding of the molecular basis of congenital heart disease and its prevention but also for successful regeneration after cardiac injury. A recent study employed a novel Cre/LoxP-based lineage tracing approach with a multi-color reporter in zebrafish to examine the fates of populations of developing cardiomyocytes. The results showed that a remarkably few number of clones of cardiomyocytes are involved in the formation of adult zebrafish heart. Furthermore, a striking difference in the mechanism of myocardial compaction was described involving the creation of a completely new layer of cortical myocardium.
Cardiac development is a complex process involving multiple meticulously orchestrated molecular, cellular, and morphogenetic events to give rise to a properly functioning organ. While detailed investigation in model organisms such as flies, frogs, chicks, and mice have greatly informed our understanding of the early regulators in this process, there has been a relative lack of data addressing the later stages of myocardial formation and the cell origins that give rise to maturing cardiomyocytes.
To identify the native myocardial architecture, pioneering studies from Mikawa et al have attempted to clonally-label myocardial cell populations during cardiac development in the chick by employing a single-cell retroviral tagging strategy (Figure 1) [1]. These studies showed that one single-labeled cardiomyocyte can give rise to a wedge-shaped cluster of descendent cardiomyocytes that migrate inward from the outer subepicardial layer toward the inner trabecular layer. It was proposed that each of these cell clusters represent individual units of clonally-related myocardium that function in synchrony [2]. Consistent with this model, the development of the trabecular myocardium in mice appears to involve the delamination of myocardial cells away from their neighbors along the epicardial wall, a cell population that eventually gives rise to the compact myocardial layer (Figure 1) [3].
Figure 1. Comparison of early myocardial development in chick, mouse, and zebrafish.
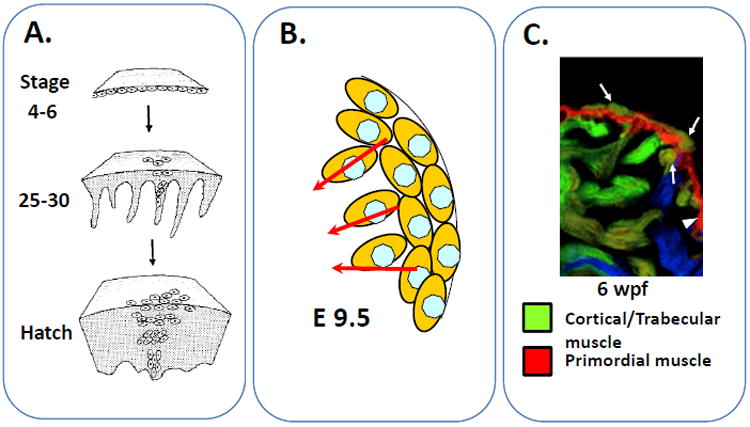
A. Clonal analysis of cardiomyocyte precursor in chick by single cell retroviral tagging. B. Expansion of murine myocardial precursors cells at embryonic day 9.5. C. Development of cortical myocardial layer from trabecular cardiomyocytes in the zebrafish heart. Arrows represent cortical cardiomyocytes and their putative trabecular origin. Adapted with permission from references #1, 3, and 5.
While these descriptions of myocardial development in chick and mice have provided a conceptual framework for this process, the clonality issue at an organ-wide level was not addressed since the number of independent cardiomyocyte clones needing to be tracked in a single mouse or avian embryo is far greater than the number that the existing technology is able to support. Taking advantage of a novel Cre/LoxP-based lineage-reporting tool that was originally developed for studies in neural patterning [4], Gupta and Poss examined the fate of a major proportion of the developing cardiomyocyte precursors during zebrafish cardiac development [5]. In traditional cell lineage tracing experiments using the Cre/LoxP strategy, a single population of cells that expresses the Cre recombinase can be followed in an embryo during organogenesis [6]. Although such a technique has already been employed in heart development studies in mice, and more recently, in zebrafish, it is inadequate to track multiple heart forming cells simultaneously because of the limited number of color discriminations in the reporter. Gupta and Poss circumvented this problem by harnessing a new technology called Brainbow that allows for the simultaneous labeling of multiple cells where each cell has its own distinct color tag [4]. By combining the power of a tamoxifen inducible cardiomyocyte lineage-specific Cre recombinase with a modified Brainbow system that contains a β–actin2 promoter-driven reporter cassette carrying three fluorescent protein-expressing transgenes (RFP, CFP, YFP) each flanked by distinct LoxP recognition sites, Gupta and Poss were able to follow the individual fates of a majority of the cardiomyocytes present at the time of cre induction [5]. When these transgenic zebrafish were treated briefly with tamoxifen at 2 days post fertilization (dpf), Cre-mediated recombination events occurred in each embryonic cardiomyocyte that led to random excision of one of these three lox-flanked fluorescent reporter transgenes on each copy of the introduced transgene [5]. With the application of appropriate excitation frequency and assuming that multiple copies of the reporter transgene were incorporated into each cardiomyocyte, a range of fluorescent colors will be displayed in discrete patches of lineage-related cardiomyocytes. This allows for the simultaneous tracking of multiple clones of embryonic cardiomyocytes during zebrafish development.
Using this modified Brainbow technology, Gupta and Poss evaluated the development of the zebrafish ventricle at different time points post-fertilization, starting at 10 dpf. Two distinct muscle types have been shown previously to comprise the zebrafish ventricle: an outer wall of compact myocardium and an inner layer known as the trabecular myocardium. The authors reported that at 10 dpf, the compact layer was only a single cell layer thick and that the inner trabecular myocardium appears to arise from this so-called “primordial” layer. They next examined ventricular development at 30 dpf, coinciding with the juvenile stage of zebrafish development. In this set of experiments, Cre-recombination was again induced at 2 dpf and approximately 55 distinct clones of cardiomyocytes labeled the ventricular wall at this stage of development, implying that roughly 110 founder cardiomyocytes (since only half of the myocardial cells underwent Cre-recombination at the given tamoxifen dose) gave rise to the primordial layer of the juvenile zebrafish. Intriguingly, the number of cardiomyocytes in each patch varies significantly despite their presumed origin from a single founder cardiomyocyte clone at 2 dpf when labeling occurred, suggesting the possibility of clonal dominance in ventricular myocardial development [5].
To further pursue this, Gupta and Poss extended their analysis to adult zebrafish hearts at 6-10 weeks post fertilization. Surprisingly, they uncovered a previously unknown third cardiac muscle layer, which they termed the “cortical layer”, lying outside of the primordial layer (Figure 1). This cortical layer is comprised of cardiomyocyte from only ~8 precursor clones, again suggesting the presence of clonal dominance during myocardial development. While the existence of these unusual and elite cardiomyocyte precursors is interesting, the molecular mechanism involved in the asymmetric development of cardiomyocytes remains to be uncovered. Nevertheless, when these zebrafish hearts were subjected to a previously described apical resection injury model [7], the newly identified cortical myocardial layer exhibited superior capacity for regeneration when compared with the underlying primordial cell layer [5]. Finally, to elucidate the origins of the cortical layer myocardium, the authors performed additional cell lineage tracing experiments and found, unexpectedly, that the cardiomyocytes in the cortical layer arose from the migration and subsequent proliferation of cardiomyocyte precursors in the trabecular myocardium. These founder cardiomyocytes appear to migrate a great distance from the trabecular layer through the primordial layer in selected spots before spreading outwards onto the exterior of the primordial ventricle. This is the first report in the cardiac development literature where trabecular myocardial cells invade and migrate through the compact (i.e. primordial) myocardium before finally settling along the external myocardial surface as the cortical layer.
This study by Gupta and Poss illustrate a complex system of cardiac development by which three distinct cardiac muscle layers are formed. While highly novel and potentially paradigm shifting, these findings also raise intriguing questions regarding the growing number of differences in the developmental regulation and the regenerative potential of zebrafish versus mammalian heart. In particular, given the lack of an obvious cortical myocardial layer in the mammalian heart, does this imply that an entirely different morphogenetic mechanism is employed by the zebrafish to generate a working myocardium or have we just failed to observe this mechanism at work in other species? Furthermore, does the fact that the zebrafish but not mammalian heart can regenerate completely after experimentally-induced injury imply that this missing cortical myocardial layer in mammalian hearts is the key to the differences in the capacity for a zebrafish heart to regenerate compared with mammalian hearts?
While further studies will be needed to answer these questions, it is worth considering the caveats of the experimental system described here. First, despite the increased number of cardiomyocyte clones that can be tracked by this novel strategy compared with those typical for Cre/LoxP-based lineage tracing approaches, only half (i.e. ~55) of the total number of unique primordial cardiomyocyte clones at 2 dpf are labeled. The limited labeling approach was presumably necessary for this study because of the need to take advantage of the full color diversity (i.e. inclusive of red fluorescence which is the color when the transgenic cassette is not excised); otherwise there would only be two primary color combinations available if every reporter cassette underwent Cre-mediated excision. If differential survival or proliferation exists between cells that have undergone Cre excision and cells that have not, then the extrapolated number of total clones would be incorrect. Second, it is estimated by the authors that at 2 dpf there are an estimated 250-300 total cardiomyocytes in the zebrafish heart and 115 of these are represented on the ventricular surface. What are the fates of these 135-185 non-surface localized cardiomyocytes? Do they contribute into trabecular or primordial layer during development? Third, the exact number of transgenes that have integrated within each cardiomyocyte is unclear. This is an important issue as it determines the upper limit in the diversity of colors that can be generated. Fourth, can any one of the copies of transgene gets silenced or attenuated during cardiomyocyte maturation? The assumption of the Brainbow reporter system is that each transgene cassette is able to maintain the expression of its fluorescent protein at the same level as the next cassette within the same cell. This is required since the final fluorescent color of each cell is a sum of all the contribution from multiple transgene cassettes. However, if one copy of the transgenes is preferentially silenced during development, then the final color will be skewed. Fifth, it is unclear whether the rate of fluorescent protein decay in each cell is uniform since any changes in the number of fluorescent proteins present in each cell will alter the final number of clones counted. Finally, since tamoxifen pulsing is not instantaneous but takes place over a several hours to as late as one or two days after treatment, it is possible that some of the clones are not labeled synchronously thus giving rise to the heterogeneity in clone sizes.
These caveats notwithstanding, the data reported by Gupta and Poss suggests several features of myocardial formation that are unique in zebrafish. First, the persistence of a single cell layer of primordial cardiomyocytes throughout development and into the adult is highly unusual. The formation of both compact and trabecular myocardium in mice is accompanied by raid proliferation of cells within both regions giving rise to structures that are multiple cell layers thick. The formation of multicellular compact layer in mammalian heart may be required for hemodynamic support that is less necessary for zebrafish heart development. Second, the trabecular myocardium in zebrafish appears to arise from a pool of myocardial precursors that are not adjacent to the compact (i.e. primordial or cortical) myocardium. While the exact source of these trabecular myocardial cells was not explored in detail in this study, their abundance from early development into adulthood suggests a greater dependence of the zebrafish heart on trabecular cardiomyocytes for proper function, since the trabecular myocardium in mammalian hearts undergoes significant regression during myocardial maturation. Finally, the identification of a cortical myocardial layer outside of the primordial layer is intriguing and potentially paradigm shifting. This cell population appears to arise from a few cardiomyocyte precursor clones that sprout outward from the trabecular layer in the base of the heart. They also seem to mediate the majority of the regenerative response following apical resection. Could these cortical cardiomyocytes precursors harbor unique capacity for cell expansion during development that is reactivated for regeneration following injury? Can these basal-predominant clones explain the difference between zebrafish and mammalian heart regeneration?
These studies by Gupta and Poss have brought forth numerous new concepts in cardiac development and potential clues for understanding zebrafish cardiac regeneration. It will be of great interest to see how much these concepts can be extended to cardiac development in other model organisms such as mice and humans. By innovative application of novel tools and technology as exemplified by this study from Gupta and Poss, we may finally be able to harness the power of developmental biology for future cardiac regenerative therapies.
Supplementary Material
Acknowledgments
We thank Dr. Benoit Bruneau for helpful discussion and Drs. Caroline and Geoffrey Burns for manuscript critique.
Sources of Funding
This work was supported by NIH grants U01 HL100408 and DP2 OD004411 (to S.M.W).
Footnotes
Disclosures
S.M.W. serves as a consultant/advisor for Silver Creek Pharmaceutical, Inc.
References
- 1.Mikawa T, Borisov A, Brown AMC, Fischman DA. Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus. I: Formation of the ventricular myocardium. Dev Dyn. 1992;193:11–23. doi: 10.1002/aja.1001930104. [DOI] [PubMed] [Google Scholar]
- 2.Mikawa T, Cohen-Gould L, Fischman DA. Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus. III: Polyclonal origin of adjacent ventricular myocytes. Dev Dyn. 1992;193:133–141. doi: 10.1002/aja.1001950208. [DOI] [PubMed] [Google Scholar]
- 3.Pijnappels DA, Gregoire S, Wu SM. Integrated aspects of cardiac cell therapy. Ann N Y Acad Sci. 2010;1188:7–14. doi: 10.1111/j.1749-6632.2009.05077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 5.Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484(7395):479–84. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148(1-2):33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


