Abstract
Asymmetric cell division is a fundamental mechanism for the generation of body axes and cell diversity during early embryogenesis in many organisms. During intrinsically asymmetric divisions, an axis of polarity is established within the cell and the division plane is oriented to ensure the differential segregation of developmental determinants to the daughter cells. Studies in the nematode Caenorhabditis elegans have contributed greatly to our understanding of the regulatory mechanisms underlying cell polarity and asymmetric division. However, much remains to be elucidated about the molecular machinery controlling the spatiotemporal distribution of key components. In this review we discuss recent findings that reveal intricate interactions between translational control and targeted proteolysis. These two mechanisms of regulation serve to carefully modulate protein levels and reinforce asymmetries, or to eliminate proteins from certain cells.
Keywords: Asymmetric division, Caenorhabditis elegans, PAR proteins, Translational control, Ubiquitin proteolysis
INTRODUCTION
The early stage of metazoan development is a crucial period during which body axes are determined and the blueprints of cell fates are established. In most species these early events are carried out with little or no de novo transcription, relying instead on regulated translation of stored maternal mRNAs and modulation of protein activity (1–4). Such spatiotemporal regulation of maternal factors generates polarized distributions of cell fate determinants in the zygote and early blastomeres, and in many organisms asymmetric cleavage promotes unequal segregation of these determinants into daughter cells. A leading model for studies of asymmetric division is the nematode Caenorhabditis elegans (5–10). In this organism, early blastomeres attain distinct fates through the differential inheritance of cell fate determinants during a series of asymmetric cleavages.
During asymmetric division, the first step is to break the symmetry of cellular components in the mother cell. Symmetry breaking is initiated at fertilization in C. elegans (5–8, 10). An unidentified signal from the sperm with its associated centrosome and microtubule aster triggers a cortical flow of acto-myosin away from the site of the sperm entry, which results in the formation of anterior and posterior cortical domains containing distinct PAR polarity proteins (Fig. 1A). By the end of this “establishment phase”, the two PDZ-domain proteins, PAR-3 and PAR-6, localize to the anterior, where they form a complex with an atypical protein kinase C (aPKC-3). On the other hand, a RING-finger protein PAR-2 and a serine-threonine kinase PAR-1 are placed at the posterior cortex. The anterior cortical movements are controlled by a Rho family GTPase and its regulators, and PAR-3 itself is needed for a robust establishment period through feedback on acto-myosin movement. PAR-5, a 14-3-3 protein family member, is also required for establishment of PAR polarity, in particular to prevent overlapping of the anterior PARs with PAR-2. Once established, the anterior and posterior domains maintain the partition through mutual exclusion. A serine/threonine kinase PAR-4, although not asymmetric itself, participates in the maintenance of PAR domains as well as the establishment of downstream asymmetries.
Fig. 1.
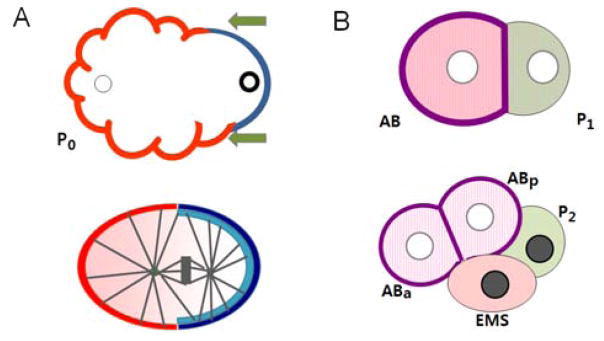
PAR polarity and cell fate determination. (A) Establishment of PAR polarity and spindle displacement. In the newly fertilized zygote (P0), the position of the sperm (bold circle at right) defines the posterior pole of the cell. A sperm associated signal triggers a directed flow of acto-myosin cortical contraction towards the future anterior (arrows), accumulating PAR-3, PAR-6 and PKC-3 proteins (red) at the anterior half of the cortex. The absence of these PARs at the posterior allows cortical association of PAR-2 (dark blue) and PAR-1 (light blue). Establishment of PAR polarity domains results in the asymmetric distribution of MEX-5 and MEX-6 (pink) in the cytoplasm. PAR domains also regulate alignment of the spindle along the A/P axis and its displacement towards the posterior. (B) Distribution of Cell Fate Determinants. The first asymmetric cleavage produces the larger anterior AB cell and the smaller posterior P1 cell. The amount of MEX-5/6 is higher in the AB cell. MEX-3 is present in both cells (pink hatch marks), while POS-1 and PIE-1 (light green) are more abundant in the P1 cell. The anterior determinant GLP-1 (purple) is localized to the periphery of the AB cell and its descendents. By the 4-cell stage, MEX-3 is restricted to the anterior cells and the posterior determinant PAL-1 (grey) appears in the nuclei of the two posterior cells. MEX-5 is actually dynamic, with higher levels in the EMS daughter, but decreasing levels in the anterior by the end of the 4-cell stage.
The polarization of PAR proteins instigates a panoply of downstream events (1, 8). First, PAR polarization results in the differential localization of a number of molecules that influence cell fate, including MEX-5, MEX-6, MEX-3 and GLP-1 to anterior cells and PIE-1, POS-1, and PAL-1 to posterior cells (Fig. 1B). Some of these asymmetries are visible in the one-cell, others are manifest slightly later, as further asymmetric divisions reinforce their unequal distribution among progeny cells. Second, the PAR proteins regulate the position of the mitotic spindle through asymmetric localization of cortical components of a G protein signaling pathway (6–8). Consequently the cleavage plane is formed perpendicular to the polarity axis and off center, dividing the P0 zygote into the larger anterior AB cell and the smaller posterior P1 cell.
While the morphological aspects of early asymmetric divisions have been extensively studied, the molecular nature of their regulation has only begun to be understood. Several recent reviews describe the regulation of PAR localization by Rho family GTPases, and other aspects of asymmetric division such as the role of the conserved G protein signaling pathway in cleavage plane specification in C. elegans (5–8, 10). In this review, we focus on recent findings in the regulation of early asymmetric division that reveal an intriguing interplay of translational repression and targeted protein degradation within a given pathway. In the absence of de novo transcription, these modes of regulation provide a means to fine tune the levels of key modulators in rapidly dividing early blastomeres.
Regulation of PAR protein levels by the worm homologs of Nanos, Pumilio and Brat
The anterior PAR complex proteins exclude PAR-2 from the cortex. Functional studies showed that this exclusion depends on its phosphorylation by PKC-3 (11). By analogy to other systems, phosphorylation of PAR-2 could generate a binding site for the 14-3-3 protein PAR-5, which would cause PAR-2 to dissociate from the cortex. Similarly, exclusion of the anterior PARs from the posterior PAR-1/PAR-2 domain during the maintenance phase could involve phophorylation of anterior PARs by PAR-1 and binding by PAR-5. Such mutual exclusion via phosphorylation has been demonstrated for PAR proteins in other systems (8–10) but these ideas have not yet been tested in the early C. elegans embryo. In addition, the role of PAR-2 in the maintenance of PAR polarity at the molecular level remains to be elucidated. PAR-2’s identity as a putative E3 ligase raises the possibility that it could cause degradation of the anterior PARs in a localized fashion, but it could also act indirectly by affecting modulators of PAR proteins.
Recently, novel insight into the regulation of PAR polarity via protein degradation and translation has come from a genome-wide search for suppressors of par-2 mutations (12). In par-2 mutants, posterior expansion of PAR-3/PAR-6/PKC-3 results in polarity defects and embryonic lethality. Yet the disrupted equilibrium can be restored if function of the anterior complex is compromised. For example, RNAi knock-down of par-3, par-6 or pkc-3 can all suppress the lethal phenotype of par-2 mutant. Interestingly, the list of recently identified par-2 suppressors includes the homologs of Drosophila nanos (nos-3), pumilio (FBF-1/2) and brat (Cebrat) (12, 13). The Nanos-Pumilio-Brat complex controls the development of posterior structures in Drosophila embryos, by inhibiting the translation of hunchback mRNA in the posterior (14). However, the effect of their worm homologs in early C. elegans embryos is not directly on repressing translation of posterior determinants, but rather on modulating PAR protein levels through an intersection of translational repression and ubiquitin-mediated degradation.
NOS-3 regulates PAR-6 protein level through the CBCFEM-1 ubiquitin ligase
In theory, suppressor mutations of par-2 can reverse the lethality in two ways; by reducing the level of PAR-3/PAR-6/PKC-3 in the anterior, or by increasing the residual activity of PAR-2 in the posterior. NOS-3 appears to regulate polarization mainly by modulating the levels of PAR-6 protein (15). The levels of PAR-6 protein, but not mRNA, were decreased in nos-3 mutant worms to 60% of the wild type levels. Moreover, restoring PAR-6 protein levels by the expression of a transgene in the nos-3; par-2 double mutant reversed the suppression of the par-2 phenotype. What then would be the contribution of NOS-3 in this process? NOS-3, together with a Pumilio homolog FBF-1, has been proposed to bind the fem-3 3′-UTR and repress its translation in the germ line (16). FEM-1, FEM-2 and FEM-3 are subunits of the CBC FEM-1 complex, a CUL-2 based ubiquitin ligase (17). The ubiquitin ligase, also called E3 complex, marks the target protein for degradation by attaching a poly-ubiquitin tag. In the CBC FEM-1 complex, CUL-2 serves as the main scaffold protein, while FEM-1 is the target-binding adaptor subunit. Interestingly, suppression of par-2 lethality in nos-3; par-2 double mutants required not only the presence of CUL-2 and FEM-1, but also FEM-2 and FEM-3 (15). Moreover, a physical interaction between FEM-1 and PAR-6 has been demonstrated, suggesting that PAR-6 is subjected to degradation by the CBC FEM-1 E3 ligase. A model emerging from these data is that the NOS-3/FBF-1 complex may prevent the assembly of CBC FEM-1 E3 ligase by inhibiting the translation of the FEM-3 subunit, and as a result increase PAR-6 protein levels (Fig. 2A).
Fig. 2.
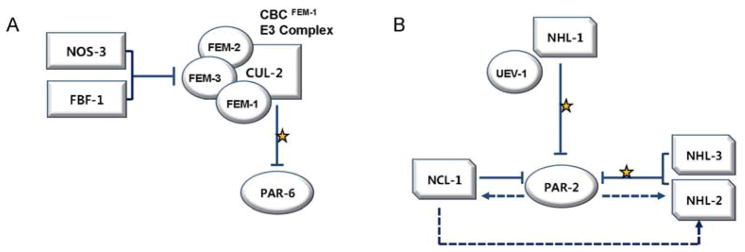
Proposed Regulation of Anterior and Posterior PAR Proteins by Ubiquitin Proteolysis. (A) Regulation of PAR-6 protein levels by NOS-3 and FBF-1. A Nanos homolog NOS-3, together with the Pumillio homolog FBF-1, inhibit translation of the FEM-3 subunit of the CBC FEM-1 ubiquitin ligase complex. The FEM-1 subunit of the complex recognizes PAR-6 for ubiquitin-mediated proteolysis. (B) Regulation of PAR-2 by CeBRATs. 4 out of the 5 CeBRATs affect PAR-2 protein levels. Three RING finger proteins (NHL-1, NHL-2, NHL-3) may target PAR-2 for ubiquitin-mediated proteolysis. PAR-2 itself affects the level of NCL-1 and NHL-2 through unknown mechanisms, and NHL-2 may also be positively regulated by NCL-1, presumably to ensure prompt adjustment of PAR-2 protein levels. (arrows: activation, blocked lines: suppression, yellow stars: ubiquitination, dashed lines: potential activation/suppression).
Because FEM-1 has only been shown to be present during oogenesis thus far (17), it is possible that regulation of PAR-6 protein levels mainly occurs in oocytes. However, other CUL-2 based E3 complexes that utilize ZIF-1 and ZYG-11 as substrate- recruiting adaptors participate in polarity establishment by degrading proteins in the zygote and 2-cell embryo (18, 19). Thus, it would be worthwhile to determine if CBC FEM-1 can persist beyond fertilization, and whether its own localization takes an asymmetric pattern. In any case, the consecutive actions of the NOS-3/FBF-1 and CBC FEM-1 complexes in PAR polarity establishment demonstrate a novel regulatory circuit where the levels of a key protein are modulated by targeted degradation, which in turn is under the control of translational repression.
C. elegans Brat homologs may regulate PAR-2 polarity independently of NOS-3
C. elegans has 5 homologs of Brat, the third component of the Nanos-Pumilio-Brat complex in Drosophila. Mutations in 4 out of the 5 Brat homologs (cebrats; ncl-1, nhl-1, nhl-2 and nhl-3) can individually suppress the lethality of par-2 mutants to varying degrees (13). Interestingly, none of these mutations affected the level of PAR-6 protein as the nos-3 mutant did (15). However, disruption of the cortical PAR-2 domain in a par-2 strain was partially restored in double mutants with nos-3, ncl-1, nhl-2 pnhl-3. Given the interplay between the anterior PARs and PAR-2 in restricting each other’s domain, these results raise the possibility that CeBRATs act via regulation of PAR-2 levels. Significantly, double mutant analysis suggested that at least NHL-2 functions in a separate pathway from NOS-3 and FEM-3 (13). It is possible that CeBRATs are responsible for the modulation of PAR-2 protein levels independently of NOS-3, while the main job of NOS-3 is to control PAR-6 levels. Functional segregation of Brat from Nanos and Pumilio has also been noticed in Drosophila. For example, Nanos and Pumilio inhibit the translation of Cyclin B mRNA without the participation of Brat; and Brat alone is known to play a role in asymmetric division of the neuroblast without Nanos/Pumilio (20).
What could be the nature of modulation exerted by CeBRATs on the polarity of C. elegans zygote? With the exception of NCL-1, which functions as a cell growth inhibitor and has been suggested as the ortholog of Drosophila Brat (21), the other three (NHL-1, NHL-2, NHL-3) all contain a RING finger domain found in most E3 ligases. Among these, the ubiquitinating activity of NHL-1 has been verified in vitro (22). Also, an ubiquitin-conjugating enzyme UEV-1 bound to NHL-1 and PAR-2 in a yeast 2-hybrid assay. These data suggest the possibility that PAR-2 protein levels are regulated directly by NHL-1 mediated ubiquitin proteolysis. The presence of RING finger domains in NHL-2 AND NHL-3 suggest that they may affect PAR-2 protein levels through a similar mechanism (Fig. 2B). Interestingly, expression of NCL-1 and NHL-2 was decreased in par-2 mutants, and ncl-1 mutants exhibited reduced levels of NHL-2 (13). Such complex feedback among PAR-2 and CeBRATs may be required for prompt and accurate adjustment of PAR-2 levels within the cell.
Translating the PAR polarity cue into AP-axis determination
One major outcome of PAR protein segregation in the zygote is the interpretation of this polarity into the asymmetric distribution of downstream cell fate determinants (Fig. 1) (1, 8). These asymmetries are mediated in large part through the action of MEX-5 and MEX-6, two very similar CCCH Zn-finger proteins that are enriched in the anterior cytoplasm of the one-cell in response to the PAR proteins. MEX-5 and MEX-6 are highly redundant and will be referred to hereafter as MEX-5/6 where their localizations or functions appear to be the same. Although the precise molecular function of MEX-5/6 remain to be determined, recent work implicates these proteins in a network of protein degradation and translational regulation that leads to asymmetric localization of the cell-fate determinants GLP-1 and PAL-1 to specific blastomeres by the 4-cell stage (1, 4, 23, 24).
Mutual regulation between MEX-5/6 proteins and their targets sharpens the asymmetric distribution of cell fate determinants
The best studied role of MEX-5/6 is in triggering the asymmetric protein degradation of the posterior acting factors PIE-1 and POS-1. Like MEX-5/6, PIE-1 and POS-1 are tandem CCCH Zn finger proteins of the TTP family, but they are required for different aspects of posterior development. In particular PIE-1 inhibits the transcription of somatic genes in the germ lineage cells and has other germ-line roles, while POS-1 is a key translational regulator of the anterior determinant, GLP-1 (1, 25).
The levels of PIE-1 and POS-1 become enriched in the posterior during the first cell cycle, and this gradient is further sharpened during subsequent divisions. Degradation by CUL-2ZIF-1 is responsible, at least in part, for differential accumulation of these proteins. MEX-5/6 are required for the activation of the CUL-2ZIF-1 E3 ligase. Because MEX-5/6 are themselves enriched in the anterior, this results in the degradation of any remaining PIE-1 and POS-1 in the progeny of anterior blastomeres. Degradation requires the first CCCH finger of each of these proteins, which mediates binding to the ZIF-1 substrate adaptor for CUL-2 (18, 26). How MEX-5/6 activate degradation of the posterior CCCH proteins remains to be elucidated, but the Zn-finger domains of MEX-5 were shown to bind mRNA non-specifically (27). Thus, an intriguing model to test is that MEX-5/6 activates the translation of ZIF-1 or other components of the CUL-2ZIF-1 E3 ligase asymmetrically in the anterior.
Interestingly, MEX-5 is itself subjected to both degradation and translational control. The initial anterior enrichment of MEX5/6 established by the PAR proteins may be re-enforced by translational control by POS-1. Although loss of pos-1 in a wild-type background does not affect MEX-5/6 asymmetry, reduction of pos-1 gene function rescued the phenotype of mex-5 single mutants (28). Furthermore, POS-1 bound the 3′-UTR of mex-6 (28). Whether binding of POS-1 inhibits the translation of these mRNAs was not tested directly, but POS-1 has been shown to act as a translational repressor (25). By the 4-cell stage, the levels of MEX-5/6 begin to decrease in the anterior cells. This change is mediated by the second Zn-finger of MEX-5 and the CUL-2ZIF-1 E3 ligase (18). The fact that MEX-5/6 are both activators and targets of the CUL-2ZIF-1 is reminiscent of the mutual regulation between MEX-5/6 and POS-1. Perhaps such a complex circuit of regulation serves to precisely tune the activities of these proteins during rapid cleavage divisions.
Localization of PAL-1 and GLP-1 by a circuit of translational regulation
The selective degradation of posterior CCCH proteins via MEX-5/6 leads to further asymmetries in several cell fate regulators, the best studied of which are GLP-1 and PAL-1 (1, 3, 4). GLP-1 is one of the two members of the C. elegans LIN-12/Notch family, and it is expressed on plasma membrane of the AB cells beginning at the 2-cell stage (29). glp-1 mRNA is found in all cells, thus the anterior confinement of GLP-1 protein must involve selective translation or degradation. In this case, GLP-1 accumulation is governed by several RNA binding proteins that function as translational regulators. POS-1 and GLD-1 suppress GLP-1 translation in the posterior blastomeres by binding to distinct 3′-UTR elements (25, 30). At the same time, SPN-4, an RNP-type RNA binding factor that also binds the glp-1 3′UTR, is required for the activation of GLP-1 translation in the anterior (25). SPN-4 is present throughout the embryo at the two-cell stage when GLP-1 translation is first seen. Because POS-1 is being degraded in the anterior via the MEX-5/6 pathway at this point, the ratio of SPN-4 to POS-1 is higher and thus activation prevails (25).
In contrast, the homeobox protein PAL-1 is localized to the posterior nuclei of the 4-cell embryo where it activates transcription of genes involved in specifying muscle cell fates (31, 32). The posterior localization of PAL-1 in early C. elegans embryo resembles its Drosophila homolog Caudal in many ways (33). While pal-1 mRNA is evenly distributed in the oocyte and early embryos, PAL-1 protein is not synthesized until the 4-cell stage and then, only in posterior blastomeres (34, 35). As with Caudal, posterior expression of PAL-1 is placed under a multistep translational control. First, translation of pal-1 is repressed by a STAR/Maxi-KH domain protein GLD-1 in the germ line (36). After fertilization, MEX-5/6 and MEX-3, a KH domain RNA binding protein, are all required for restriction of PAL-1 to the posterior (37).
Since MEX-3, MEX-5 and MEX-6 all contain RNA-binding motifs, a plausible hypothesis would be that they bind directly to pal-1 mRNA and repress translation. Indeed the translation of pal-1 mRNA was dictated by its 3′-UTR in a lacZ reporter assay (34), and recent work identified MEX-3 binding sites in the pal-1 3′UTR, as well as in the 3′UTRs of other developmentally regulated genes (38, 39). However, MEX-5/6 act through MEX-3, at least in part, because MEX-5/6 are required for the enrichment of MEX-3 to the anterior (Fig. 3). In addition, PAL-1 protein was ectopically present in all blastomeres of mex-5; mex-6 double mutants whether MEX-3 was present or not, suggesting that MEX-5/6 facilitate MEX-3 activity. Further, MEX-6 bound to MEX-3. Together these results suggest that MEX-5/6 could either function as an upstream activator of MEX-3 or as a co-repressor on the pal-1 3′UTR, or both (37). How MEX-5/6 localize MEX-3 has been unclear, but recent studies show that POS-1 can bind to the 3′UTR of mex-3 mRNA, consistent with translation repression in the posterior (40). Thus a simple model to test is that MEX-5/6 affects MEX-3 accumulation through restriction of POS-1 to the posterior. At the same time, reporter assays revealed that POS-1 binding to the mex-3 3′UTR was not sufficient for repression (40). Therefore, as seen for GLP-1 regulation above, it is likely that there is a complex interplay among RNA binding proteins to generate different translational outputs.
Fig. 3.
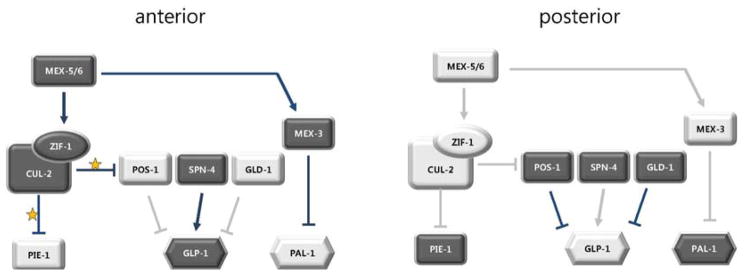
Regulation of anterior vs. posterior fates by CCCH proteins. Posterior PAR proteins are responsible for the localization of MEX-5/6, resulting in higher concentrations of these CCCH proteins in the anterior where they activate CUL-2ZIF-1. The major targets of ubiquitination by CUL-2ZIF-1 in the anterior are two other CCCH proteins PIE-1 and POS-1. MEX-5/6 also activate a KH domain protein MEX-3, which inhibits translation of the posterior determinant PAL-1 in the anterior. The RNA binding protein SPN-4 activates GLP-1 translation in the anterior, while the GLD-1 RNA binding protein represses GLP-1 in the posterior. (arrows: activation, blocked lines: suppression, yellow stars: ubiquitination, dark shapes and lines: high amount of proteins and robust regulation, faint shapes and lines: low amount or absence of proteins and regulation).
Positioning of the mitotic spindle relative to the polarity axis
Once PAR polarity is established in the one-cell embryo, it instructs the orientation and asymmetric movement of the mitotic spindle along the AP axis to ensure proper segregation of cell fate determinants into daughter cells. The mechanisms by which the PAR proteins regulate spindle positioning are an active area of study (6, 7). PAR polarization results in asymmetric activation of a heterotrimeric G protein signaling pathway, which creates unequal pulling on the astral microtubules of the mitotic spindle. Asymmetries in the pulling forces serve first to orient the spindle along the polarity axis, then displace it towards the posterior end of the embryo (Fig. 1). In addition to cortical pulling, proper movement of mitotic spindles also requires regulation of microtubule length and dynamics so that the microtubules can interact with the cortex. A well studied case is the targeted proteolysis of the microtubule severing protein MEI-1/MEI-2 (katanin) after fertilization (23). Interestingly, recent studies suggest that the levels of MEI-1 and other microtubule associated proteins are also controlled at the translational level.
Timely degradation of MEI-1 by a network of protein degradation pathways
One of the critical prerequisites for the asymmetric cleavage of the C. elegans one-cell embryo is the switch from a small meiotic spindle to a larger mitotic spindle with astral microtubules that contact the cortex. Meiotic spindle microtubules are maintained short by the action of the MEI-1/MEI-2 katanin complex, while the prolonged presence of MEI-1/MEI-2 after fertilization causes shorter mitotic spindles that are mispositioned with respect to the polarity axis (41–44). Since meiotic and mitotic spindles are assembled in the same cytoplasm, it is a challenge to achieve rapid inactivation of the MEI-1/MEI-2 complex after fertilization without attenuating its activity during the second division of meiosis. C. elegans solves this problem by subjecting the MEI-1 subunit to multiple levels of regulation, including ubiquitin proteolysis (23, 42, 45–51).
In the zygote, MEL-26 binds and targets MEI-1 for degradation by a CUL-3 based E3 ligase (Fig. 4A) (50). The levels of MEL-26 protein are kept low in the oocyte by the action of an unknown CUL-2 based E3 ligase, although it is not known if this interaction is direct (45). Equally unclear is the mechanism of keeping CUL-3MEL-26 turned off during meiosis, but one candidate for the switch is RFL-1 which activates E3 ligases by neddylation. RFL-1 is required for CUL-3MEL-26 to degrade MEI-1 (49), and it may also activate the CUL-2 based E3 ligase in the oocyte since abnormal accumulation of MEL-26 was observed in both rfl-1 and cul-2 mutants (45). Perhaps RFL-1 alternates its activation target from CUL-2 in the oocyte to CUL-3 in the zygote, postponing degradation of MEI-1 until after fertilization.
Fig. 4.
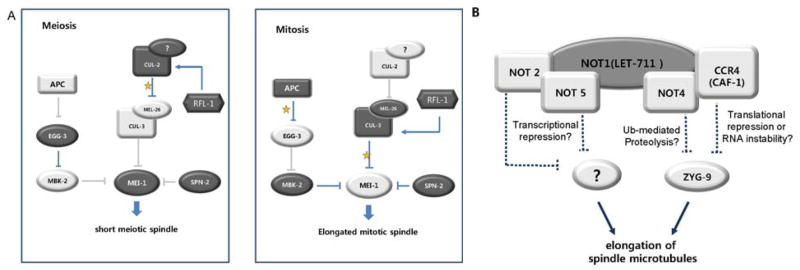
Regulation of microtubule length during asymmetric division. (A) Differential regulation of MEI-1 during meiosis and mitosis. The microtubule severing activity of MEI-1 is required for a short meiotic spindle but is removed before the first mitotic cleavage. RFL-1 may serve as a switch in this transition by activating an unknown E3 ligase in the oocyte which degrades CUL-3MEL-26, thus preventing ubiquitination of MEI-1. During mitosis, RFL-1 activates CUL-3MEL-26, and robust degradation of MEI-1 ensues. Removal of MEI-1 is also achieved via translational repression by SPN-2 as well as MBK phosphorylation. Interaction with MBK-2 is prevented in the oocyte because this kinase is sequestered at the cortex by EGG-3, which is degraded once APC is activated after fertilization. (arrows: activation, blocked lines: suppression, yellow stars: ubiquitination, dark shapes and lines: high amount of proteins and robust regulation, faint shapes and lines: low amount or absence of proteins and regulation). (B) Opposing effects of LET-711 and ZYG-9 LET-711 negatively regulates the localization of ZYG-9 at centrosomes. As a scaffold protein of the CCR4-NOT complex, LET-711 may affect ZYG-9 and thus microtubule organization through any of several mechanisms as indicated. (dashed lines: potential suppression).
Another pathway of inactivation targets MEI-1 exclusively in the zygote. Substrates of E3 ligases are often phosphorylated before ubiquitination, and the MBK-2 kinase was shown to be required for timely degradation of MEI-1 (47, 48, 51, 52). Initially phosphorylation of MEI-1 by MBK-2 was believed to prime MEI-1 for degradation by CUL-3MEL-26, but it appears that MBK-2 and MEL-26 act in parallel pathways (46). While the nature of MBK-2 mediated degradation remains to be identified, it nonetheless serves as an elegant case of a molecular timer (Fig. 4A). During meiotic arrest before fertilization, MBK-2 is sequestered at the cortex by EGG-3 and is thus unable to phosphorylate MEI-1 in the cytoplasm (53). When meiosis is resumed upon fertilization, the meiotic APC degrades EGG-3 and MBK-2 is released into the cytoplasm. Thus the inactivation of microtubule severing complex during first mitosis appears to be insured by dual regulatory pathways, one through the degradation of MEI-1 by CUL-3MEL-26 and the other by an MBK-2 associated degradation pathway. In each case, the activity of the degradation machinery itself is regulated by targeted protein degradation to confer temporal control.
Negative regulation of MEI-1 translation by a novel 4E-BP, SPN-2
In addition to the elaborate panoply of protein degradation, the activity of MEI-1 was recently proposed to be controlled by translational repression after fertilization. Because mei-1 mRNA persists in embryos after meiosis (44), a means to prevent its translation would be required in addition to degradation of existing protein. A regulator suitable for this function was identified from the recent characterization of a spindle orientation defective mutant, spn-2 (54). The spn-2 gene encodes a novel eIF4E binding protein (4E-BP); 4E-BPs in other systems disrupt the translation initiation complex by competing with the eIF4G initiation factor (1, 2). The abnormal nuclear and spindle positioning phenotypes observed in early spn-2 embryos were caused by ectopic expression of MEI-1 (54). SPN-2 bound in vitro to the RNA binding protein OMA-1, which interacted with the 3′-UTR of the mei-1 in an RNA pull-down assay. Further, RNAi knock-down of oma-1, together with functionally redundant oma-2, similarly caused ectopic MEI-1 in early cleavage divisions (54). In Drosophila, translational repression by a 4F-BP (Cup) is mediated by gene-specific adaptors, for example Bruno for oskar mRNA (55) and Smaug for nanos mRNA (56). In the case of C. elegans SPN-2, OMA-1 may serve as a gene-specific adaptor for mei-1.
Both SPN-2 and OMA-1 are present in oocytes and meiotic embryos (57), and thus it is necessary to suppress their activity on mei-1 mRNA during these stages. Because 4E-BPs are often activated upon phosphorylation, a specific kinase may trigger translational repression by SPN-2 after fertilization. Alternatively, the interaction between SPN-2 and OMA-1, or the binding of OMA-1 to mei-1 3′-UTR, may be blocked until after meiosis. Interestingly OMA-1/OMA-2 were shown to repress the translation of the nanos homolog nos-2 in the oocyte (39). Since SPN-2 is also required for the translational repression of nos-2 in the oocyte (Li and Rose, unpublished data), a likely hypothesis is that SPN-2/OMA-1 is active in the oocyte but somehow is prevented from binding to the 3′-UTR of mei-1 mRNA. SPN-2 may also have functions independent of OMA-1, because OMA-1 is phosphorylated by MBK-2 and begins to be degraded after the one cell stage (52, 57, 58), while the level of SPN-2 protein remains high until 8-cell stage (54). Taken together, timely elimination of the microtubule severing protein MEI-1 in the mitotic embryo is achieved through a combined effort of ubiquitin-mediated degradation pathways and translational repression by SPN-2 (Fig. 4A).
LET-711, the C. elegans ortholog of NOT-1, may promote the degradation of ZYG-9
Spindle positioning is thought to be defective in mei-1 over-expression mutants due to shorter astral microtubules that fail to interact with the cortex. A similar spindle mispositioning defect is seen with loss of function mutations in zyg-9, which encodes the C. elegans homolog of the microtubule polymerization protein, XMAP215 (6). On the other hand, abnormally long microtubules appear to be equally detrimental because let-711 mutant embryos also have misoriented spindles, but astral microtubules were found to be longer and more resistant to depolymerization by cold temperatures than in wild type (59). The level of ZYG-9 protein was increased at centrosomes in let-711 embryos. In addition, the spindle orientation phenotype was partially rescued in let-711; zyg-9 double mutants, suggesting that LET-711 and ZYG-9 have opposing functions. Surprisingly though, LET-711 is the C. elegans homolog of the NOT1, the core component of the CCR4/NOT complex, which is involved in a wide range of transcriptional and post-transcriptional control (60). In yeast and humans, the NOT4 component of the complex has been demonstrated to function as an E3 ligase, while the CCR4 subcomplex functions in dead-enylation of mRNA. Other roles in transcription and phosphorylation have also been implicated for this versatile complex (60–63). It is not known which of these activities is important for the role of let-711, although a clear possibility is that degradation modulates ZYG-9 levels (Fig. 4B). In contrast to MEI-1, ZYG-9 is needed for meiosis and mitosis. Further, during early mitotic cell divisions, the size of cells and thus astral microtubules get smaller over time, which would necessitate the modulation of ZYG-9 levels in proportion. In this regard, partial degradation would be required in order to fine tune protein levels, rather than eliminating ZYG-9 altogether.
An equally possible scenario is that the CCR4 subunit of the LET-711/NOT1 complex could serve to degrade the zyg-9 mRNA or repress its translation. In support of this role, let-711 (RNAi) causes derepression of nos-2 translation in the oocyte (64). However, the localization of proteins to microtubules and centrosomes has many interdependencies, and thus it is also possible that the effects of LET-711 on ZYG-9 are through an unidentified microtubule-associated protein. It will be important in the future to test for regulation of zyg-9 at the mRNA level and to determine if it binds the CCR4 subunit of the NOT complex or any of the known translational repressors.
Summary and perspectives
During early C. elegans embryogenesis, translational regulation and targeted proteolysis play important roles in cell fate determination among rapidly dividing blastomeres. The intersection of these two modes of post-transcriptional modulation is observed in the asymmetric localization of PAR proteins and downstream determinants, and in the positioning of the spindle along the polarity axis. However, many of the molecular details remain to be elucidated. An important area for future study will be determining how the timing of these various pathways are controlled, as mentioned already for the effect of the degradation pathway on PAR-6, and the effect of SPN-2 on mei-1 mRNA. In particular, the timing of degradation of the CCCH proteins is an important mystery to be solved. PIE-1 and POS-1 are present in maturing oocytes as are their activators of degradation, MEX-5/6. However, degradation of the posterior determinants is not apparent until late in the first cell cycle. Clearly, there must be activation of the pathway in response to fertilization or cell cycle advance. While this could be accomplished through modulation of protein activity, it will not be surprising if translational control is involved. Indeed, it has been proposed that OMA-1/2 could regulate the translation of zif-1 mRNA (58). The nature of this regulation could be repression, as observed for the effect of OMA-1/2 on nos-2 mRNA. In this scenario, phosphorylation of OMA-1/2 by MBK-2 could act as the switch that inactivates and/or removes OMA-1/2, allowing ZIF-1 translation and thus degradation of PIE-1 and POS-1. With the broad range of techniques available for use in this system, C. elegans will continue to pave the way for probing the interplay of translational and degradation pathways that act during early asymmetric divisions and will provide a model to understand corresponding processes in higher organisms.
Acknowledgments
We regret that we were unable to cite all the primary references for the work described here due to space considerations. We refer the reader to the excellent reviews cited for more detail. We thank Monica Gotta and Alan Rose for critical reading of the manuscript. This work was supported by an NIH ARRA award (R01GM068744) to L. S. R. and a sabbatical research award from Hankyong National University to S-Y. H.
References
- 1.Farley B, Ryder S. Regulation of maternal mRNAs in early development. Crit Rev Biochem Mol Biol. 2008;43:135–162. doi: 10.1080/10409230801921338. [DOI] [PubMed] [Google Scholar]
- 2.Vardy L, Orr-Weaver T. Regulating translation of maternal messages: multiple repression mechanisms. Trends Cell Biol. 2007;17:547–554. doi: 10.1016/j.tcb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Evans TC, Hunter CP. WormBook, editor. The C. elegans Research Community. WormBook; 2005. Translational control of maternal RNAs (November 10, 2005) http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuersten S, Goodwin E. The power of the 3′-UTR: translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 5.Cowan C, Hyman A. Acto-myosin reorganization and PAR polarity in C. elegans. Development. 2007;134:1035–1043. doi: 10.1242/dev.000513. [DOI] [PubMed] [Google Scholar]
- 6.Galli M, van den Heuvel S. Determination of the cleavage plane in early C. elegans embryos. Annu Rev Genet. 2008;42:389–411. doi: 10.1146/annurev.genet.40.110405.090523. [DOI] [PubMed] [Google Scholar]
- 7.Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein B, Macara I. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki A, Ohno S. The PAR-aPKC system; lessons in polarity. J Cell Sci. 2006;119:979–998. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 10.Nance J. PAR proteins and the establishment of cell polarity during C. elegans development. BioEssays. 2005;27:126–135. doi: 10.1002/bies.20175. [DOI] [PubMed] [Google Scholar]
- 11.Hao Y, Boyd L, Seydoux G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev Cell. 2006;10:199–208. doi: 10.1016/j.devcel.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labbe JC, Pacquelet A, Marty T, Gotta M. A genomewide screen for suppressors of par-2 uncovers potential regulators of PAR-protein dependent cell polarity in Caenorhabditis elegans. Genetics. 2006;174:285–295. doi: 10.1534/genetics.106.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyenne V, Desrosiers M, Labbe JC. C. elegans Brat homologs regulate PAR protein-dependent polarity and asymmetric cell division. Dev Biol. 2008;321:368–378. doi: 10.1016/j.ydbio.2008.06.037. [DOI] [PubMed] [Google Scholar]
- 14.Sonoda J, Wharton R. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacquelet A, Zanin E, Ashiono C, Gotta M. PAR-6 levels are regulated by NOS-3 in a CUL-2 dependent manner in Caenorhabditis elegans. Dev Biol. 2008;319:267–272. doi: 10.1016/j.ydbio.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Cur Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- 17.Starostina N, Lim JM, Schvarzstein M, Wells L, Spence A, Kipreos E. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell. 2007;13:127–139. doi: 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeRenzo C, Reese K, Seydoux G. Exclusion of germ plasm proteins from somatic lineage by cullin-dependent degradation. Nature. 2003;424:685–689. doi: 10.1038/nature01887.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Vasudevan S, Kipreos E. CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development. 2004;131:3515–3525. doi: 10.1242/dev.01245. [DOI] [PubMed] [Google Scholar]
- 20.Lee CY, Wilkinson B, Siegrist S, Wharton R, Doe C. Brat is a Miranda cargo protein that promotes neuronal differentiation and neuroblast self renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Frank D, Roth M. ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans. J Cell Sci. 1998;140:1321–1329. doi: 10.1083/jcb.140.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudgen M, Chandrasekaran A, Frazier T, Boyd L. Interactions within the ubiquitin pathway of Caenorhabditis elegans. Biochem Biophys Res Comm. 2004;325:479–486. doi: 10.1016/j.bbrc.2004.10.047. [DOI] [PubMed] [Google Scholar]
- 23.Bowerman B, Kurz T. Degrade to create: developmental requirements for ubiquitin mediated proteolysis during early C. elegans embryogenesis. Development. 2006;133:773–784. doi: 10.1242/dev.02276. [DOI] [PubMed] [Google Scholar]
- 24.Schubert C, Lim R, de Vries D, Plastert R, Priess J. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- 25.Ogura KI, Kishimoto N, Mitani S, Gengyo-Ando K, Kohara Y. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development. 2003;140:2495–2503. doi: 10.1242/dev.00469. [DOI] [PubMed] [Google Scholar]
- 26.Reese K, Dunn M, Waddle J, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complimentary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- 27.Pagano J, Farley B, McCoig L, Ryder S. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem. 2007;282:8883–8894. doi: 10.1074/jbc.M700079200. [DOI] [PubMed] [Google Scholar]
- 28.Tenlen J, Schisa J, Diede S, Page B. Reduced dosage of pos-1 suppresses Mex mutants and reveals complex interaction among CCCH zinc-finger proteins during Caenorhabditis elegans embryogenesis. Genetics. 2006;174:1933–1945. doi: 10.1534/genetics.105.052621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mello C, Draper B, Priess J. The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell. 1994;77:95–106. doi: 10.1016/0092-8674(94)90238-0. [DOI] [PubMed] [Google Scholar]
- 30.Marin V, Evans T. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development. 2003;130:2623–2632. doi: 10.1242/dev.00486. [DOI] [PubMed] [Google Scholar]
- 31.Lei H, Liu J, Fukushige T, Fire A, Kraus M. Caudal-like PAL-1 directly activates the bodywall muscle module regulator hlh-1 in C. elegans to initiate the embryonic muscle gene regulatory network. Development. 2009;136:1241–1249. doi: 10.1242/dev.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar L, Carr S, Wang H, Wood W. Zygotic expression of the caudal homolog pal-1 is required for posterior patterning in Caenorhabditis elegans embryogenesis. Dev Biol. 2001;229:71–88. doi: 10.1006/dbio.2000.9977. [DOI] [PubMed] [Google Scholar]
- 33.Mlodzik M, Gibson G, Gehring W. Effects of ectopic expression of caudal during Drosophila development. Development. 1990;109:271–277. doi: 10.1242/dev.109.2.271. [DOI] [PubMed] [Google Scholar]
- 34.Hunter C, Kenyon C. Spatial and temporal controls target pal-1 blastomere specification activity to a single blastomere lineage in C. elegans embryo. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- 35.Bowerman B, Ingran M, Hunter C. The maternal par genes and the segregation of cell fate specification activities in early Caenorhabditis elegans embryos. Development. 1997;124:3815–3826. doi: 10.1242/dev.124.19.3815. [DOI] [PubMed] [Google Scholar]
- 36.Mootz D, Ho D, Hunter C. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–3272. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- 37.Huang N, Mootz D, Albertha J, Walhout J, Vidal M, Hunter C. MEX-3 interacting proteins link cell polarity to asymmetric gene expression in Caenorhabditis elegans. Development. 2002;129:747–759. doi: 10.1242/dev.129.3.747. [DOI] [PubMed] [Google Scholar]
- 38.Pagano J, Farley B, Essien K, Ryder S. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc Natl Acad Sci USA. 2009;106:20252–20257. doi: 10.1073/pnas.0907916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jadhav S, Rana M, Subramaniam K. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development. 2008;135:1803–1812. doi: 10.1242/dev.013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farley B, Pagano J, Ryder S. RNA target specificity of the embryonic cell fate determinant POS-1. RNA. 2008;14:2685–2697. doi: 10.1261/rna.1256708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNally K, Audhya A, Oegema K, McNally F. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurz T, Pintard L, Willis J, Hamill D, Gonczy P, Peter M, Bowerman B. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science. 2002;295:1294–1298. doi: 10.1126/science.1067765. [DOI] [PubMed] [Google Scholar]
- 43.Srayko M, Buster D, Bazirgan O, McNally F, Mains P. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 2000;14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- 44.Clark-Maguire S, Mains P. Localization of the mei-1 gene product of Caenorhabditis elegans, a meiotic-specific spindle component. J Cell Biol. 1994;126:199–209. doi: 10.1083/jcb.126.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson JL, Lu C, Raharjo E, McNally K, McNally F, Mains P. Levels of the ubiquitin ligase substrate adaptor MEL-26 are inversely correlated with MEI-1/katanin microtubule-severing activity during both meiosis and mitosis. Dev Biol. 2009;330:349–357. doi: 10.1016/j.ydbio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu C, Mains P. The C. elegans anaphase promoting complex and MBK-2/DYRK kinase act redundantly with CUL-3/MEL-26 ubiquitin ligase to degrade MEI-1 microtubule-severing activity after meiosis. Dev Biol. 2007;302:438–447. doi: 10.1016/j.ydbio.2006.09.053. [DOI] [PubMed] [Google Scholar]
- 47.Pang K, Ishidate T, Nakamura K, Shirayama M, Trzepacz C, Schubert C, Priess J, Mello C. The minibrain kinase homolog, mbk-2, is required for spindle positioning and asymmetric cell division in early C. elegans embryos. Dev Biol. 2004;265:127–139. doi: 10.1016/j.ydbio.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 48.Pellettieri J, Reinke V, Kim SK, Seydoux G. Coordinated activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev Cell. 2003;5:451–462. doi: 10.1016/s1534-5807(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 49.Pintard L, Kurz T, Glaser S, Willis J, Peter M, Bowerman B. Neddylation and deneddylation of CUL-3 is required to target MEI-1/katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol. 2003;13:911–921. doi: 10.1016/s0960-9822(03)00336-1. [DOI] [PubMed] [Google Scholar]
- 50.Pintard L, Willis J, Willems A, Johnson J, Srayko M, Kurz T, Glaser S, Mains P, Tyers M, Bowerman B. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- 51.Quintin S, Mains P, Zinke A, Hyman A. The mbk-2 kinase is required for inactivation of MEI-1/ka-tanin in the one-cell Caenorhabditis elegans embryo. EMBO Rep. 2003;4:1175–1181. doi: 10.1038/sj.embor.7400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stitzel M, Pellettieri J, Seydoux G. The C. elegans DYRK Kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation. Curr Biol. 2006;16:56–62. doi: 10.1016/j.cub.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 53.Stitzel M, Cheng K, Seydoux G. Regulation of MBK-2/DYRK kinase by dynamic cortical anchoring during the oocyte-to-zygote transition. Curr Biol. 2007;17:1545–1554. doi: 10.1016/j.cub.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 54.Li W, DeBella L, Guven-Ozkan T, Lin R, Rose L. An eIF4E-binding protein regulates katanin protein levels in C. elegans embryos. J Cell Biol. 2009;187:33–42. doi: 10.1083/jcb.200903003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila Cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 56.Nelson M, Leidal A, Smibert C. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishi Y, Lin R. DYRK2 and GSK-3 phosphorylate and promote the timely degradation of OMA-1, a key regulator of the oocyte-to-embryo transition in C. elegans. Dev Biol. 2005;288:139–149. doi: 10.1016/j.ydbio.2005.09.053. [DOI] [PubMed] [Google Scholar]
- 58.Shirayama M, Soto M, Ishidate T, Kim S, Nakamura K, Bei Y, van den Heuvel S, Mello C. The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans. Curr Biol. 2006;16:47–55. doi: 10.1016/j.cub.2005.11.070. [DOI] [PubMed] [Google Scholar]
- 59.DeBella L, Hayashi A, Rose L. LET-711, the Caenorhabditis elegans NOT1 ortholog, is required for spindle positioning and regulation of microtubule length in embryos. Mol Biol Cell. 2006;17:4911–4924. doi: 10.1091/mbc.E06-02-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collart M, Timmers H. The eukayotic Ccr4-Not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways. Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 61.Cui Y, Ramnarain D, Chiang Y, Ding L, McMahon J, Denis C. Genome wide expression analysis of CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol Genet Genomics. 2008;279:323–337. doi: 10.1007/s00438-007-0314-1. [DOI] [PubMed] [Google Scholar]
- 62.Panasenko O, Landrieux E, Feuermann M, Finka A, Paquet N, Collart M. The yeast Ccr4-Not complex controls uniquitination of the nascent-associated polypeptide (NAC-EGD) complex. J Biol Chem. 2006;281:31389–31398. doi: 10.1074/jbc.M604986200. [DOI] [PubMed] [Google Scholar]
- 63.Albert T, Hanzawa H, Legtenberg Y, de Ruwe M, van den Heuvel F, Collart M, Boelens R, Timmers H. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gallo C, Munro E, Rasoloson D, Merritt C, Seydoux G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryo. Dev Biol. 2008;323:76–87. doi: 10.1016/j.ydbio.2008.07.008. [DOI] [PubMed] [Google Scholar]


