Abstract
The LIM-only protein, LMO4, is a transcriptional modulator overexpressed in breast cancer. It is oncogenic in murine mammary epithelium and required for G2/M progression of ErbB2-dependent cells as well as growth and invasion of other breast cancer cell types. However, the mechanisms underlying the oncogenic activity of LMO4 remain unclear. Herein, we show that LMO4 is expressed in all breast cancer subtypes examined and its expression level correlates with the degree of proliferation of such tumors. In addition, we have determined that LMO4 silencing induces G2/M arrest in cells from various breast cancer subtypes, suggesting LMO4 action in the cell cycle is not restricted to a single breast cancer subtype. This arrest was accompanied by increased cell death, amplification of centrosomes and formation of abnormal mitotic spindles. Consistent with its ability to positively and negatively regulate the formation of active transcription complexes, overexpression of LMO4 also resulted in an increase in centrosome number. Centrosome amplification has been shown to prolong the G2/M phase of the cell cycle and induce apoptosis, thus we conclude that supernumerary centrosomes mediate the G2/M arrest and cell death in LMO4-deficient cells. Furthermore, the correlation of centrosome amplification with genomic instability suggests that the impact of dysregulated LMO4 on the centrosome cycle may promote LMO4-induced tumor formation.
Keywords: Breast Cancer, LMO4, centrosome, cell cycle, proliferation
Introduction
LMO4 (LIM-only protein 4) is a transcriptional modulator that is overexpressed in several epithelial cancers including prostate, pancreas and breast (1–3). In breast cancer, increased LMO4 expression is observed in >50% of primary tumors (2). The functional significance of this increase has been demonstrated in transgenic mice where LMO4 overexpression induced mammary gland hyperplasia and tumors (4). Conversely, deletion of LMO4 or expression of dominant negative LMO4 decreases proliferation and invasion of epithelial cells in the mammary gland (5,6). These data implicate LMO4 in the pathogenesis of breast cancer through its regulation of several cellular events including proliferation and invasion. However, the specific molecular mechanisms by which LMO4 contributes to breast cancer progression remain unknown.
LMO4 is comprised of tandem, non-DNA binding LIM domains that interact with multiple proteins and thus acts as a bridging factor in multiprotein complexes (7,8). LMO4 regulates transcription via several mechanisms. It can suppress transcription by recruiting co-repressors such as MTA1 and CtIP to transcription factors (9,10). Alternatively, LMO4 can promote transcription by recruiting activating factors or by decreasing recruitment of co-repressors as described for the BMP7 gene (6,11). Lastly, LMO4 modulates the stoichiometry and functional availability of components of various transcriptional complexes, which results in an inverted U-shaped dose response curve for LMO4 on target gene expression (12). Upregulation of LMO4 can repress gene transcription by squelching or sequestering co-regulators and similar perturbations can be observed when the levels of LMO4 are low relative to the overall pool of co-regulators and transcription factors. The specific mechanism(s) employed by LMO4 to modulate transcription is thus context-dependent and contingent upon available binding partners.
LMO4 regulates factors such as Cyclin E (13), p21 (11), p27 (14) and BRCA1 (10), which are essential for cell cycle progression as well as the centrosome cycle (15–22). While these LMO4 targets may contribute to an essential network that maintains the integrity of the genome through regulation of DNA segregation, the role for LMO4 in this process has yet to be elucidated. In addition, because abnormal centrosome replication is thought to contribute to the aggressiveness of some tumors (23,24), determining whether LMO4 regulates the centrosome cycle will be essential for understanding the mechanisms of carcinogenesis in response to changes in LMO4 activity.
Several subtypes of breast cancer have been molecularly identified including luminal-like, ErbB2-overexpressing and basal-like (25). These subtypes are characterized by distinct clinical behaviors with ErbB2-overexpressing and basal-like cancers having a much poorer prognosis compared to luminal-like tumors (25). The aim of this study focused on determining the role of LMO4 in various breast cancer subtypes and elucidating the mechanisms by which LMO4 contributes to tumor aggressiveness. Previous studies have shown LMO4 expression is increased in invasive, poorly differentiated breast tumors and that it regulates cell cycle progression in ErbB2-dependent breast cells (2,13). However, the role that LMO4 has in regulating proliferation of other breast cancer subtypes and the molecular mechanisms employed by LMO4 that contribute to poor prognosis remain unknown. Herein, we explore the impact of aberrant LMO4 expression on the centrosome and cell cycle in luminal-like and basal-like breast cancer cells.
Materials and Methods
Antibodies
The following antibodies were used: LMO4 (26), cleaved caspase-3 (#9691 Cell Signaling), cleaved caspase-7 (#9491, Cell Signaling), Ki67 (M7240, DAKO), γ-tubulin (T6557, Sigma), FITC-conjugated α-tubulin (F2168, Sigma), and s-actin (A1978, Sigma). Secondary antibodies include: horseradish peroxidase-conjugated donkey anti-rat (712-035-153, Jackson ImmunoResearch), goat anti-rabbit (sc-2054, Santa Cruz Biotechnology), goat anti-mouse (sc-2005, Santa Cruz Biotechnology) as well as Alexa-Fluor 488 conjugated donkey anti-rat (A21208, Invitrogen) and Alexa-Fluor 594 conjugated rabbit anti-mouse (A11062, Invitrogen).
Immunostaining
De-identified, paraffin-embedded breast cancer tissue sections were obtained from University Hospitals - Case Medical Center Department of Pathology with Case Cancer Institutional Review Board approval (CASE 1108-CC504) and in accordance with the ethical standards of the committee. Sections were immunostained for LMO4 as previously described (4). Antigen retrieval was performed for 20 min in 10 mM sodium citrate utilizing the Digital Decloaking Chamber (Biocare Medical). Slides were incubated overnight with 20 μg/mL LMO4 primary antibody. After incubation with biotinylated rabbit α-rat IgG (DAKO) and peroxidase-conjugated Streptavidin (DAKO), bound antibody was detected with DAB substrate (DAKO). Incubation with rat gamma-globulin (Jackson ImmunoResearch) in place of α-rat LMO4 antibody at an equivalent concentration yielded no staining.
Immunohistochemical Scoring
LMO4 immunostained tissue was scored based on two criteria. Intensity of nuclear staining was scored on a scale of 0–3 with “0” indicating absence and “3” indicating abundance. In addition, the percentage of cancerous cells stained positively for LMO4 was quantified. For the final score, the signal intensity was multiplied by the percentage of positive cells (Score = Percentage × Intensity). Two pathologists, blinded to the receptor status of each carcinoma, independently evaluated each slide.
LMO4 immunofluorescence
For detection of Ki67 and LMO4, antigen retrieval was performed as described above. Sections were incubated overnight with either α-human Ki67 (1:100 dilution) followed by anti-mouse AlexaFluor-594 (1:800) or 20 μg/mL LMO4 antibody followed by anti-rat AlexaFluor-488 (1:800). Slides were counterstained with DAPI. Five images of each invasive carcinoma were captured at 40X magnification at equal exposure times.
For co-localization experiments, cells were fixed in 100% methanol and blocked with 1.5% normal goat serum/1% Triton X-100/PBS. Cells were incubated with an anti-γ-tubulin antibody for 8 hrs followed by anti-mouse AlexaFluor-594 (1:500) for 1 hr. LMO4 antibody was then added for 8–10 hrs. Anti-rat AlexaFluor-488 (1:500) was used for detection of LMO4, and DAPI was used as counterstain. At least 3 independent experiments were performed in two different breast cancer cell lines. To analyze LMO4 and γ-tubulin expression during the cell cycle, cells were synchronized with 1.5 μg/mL aphidicolin (Sigma) for 14 hrs, washed and harvested after an additional 10–12 hrs. To control for antibody cross-reactivity, γ-tubulin staining was examined in the presence of the secondary antibody used to detect LMO4; however, no cross-reactivity was observed. In addition, the immunohistochemistry protocol was repeated without addition of the LMO4 primary antibody and no cross-reactivity was detected.
Statistical Analyses
Unless otherwise stated, statistical analyses were performed using one or two-tailed Student’s t-test with p-values less than 0.05 considered significant. The square of the Pearson product moment correlation (R2) was used to determine the correlation coefficient.
Results
LMO4 mRNA is highly expressed in BRCA1 mutant and ER-negative breast cancers
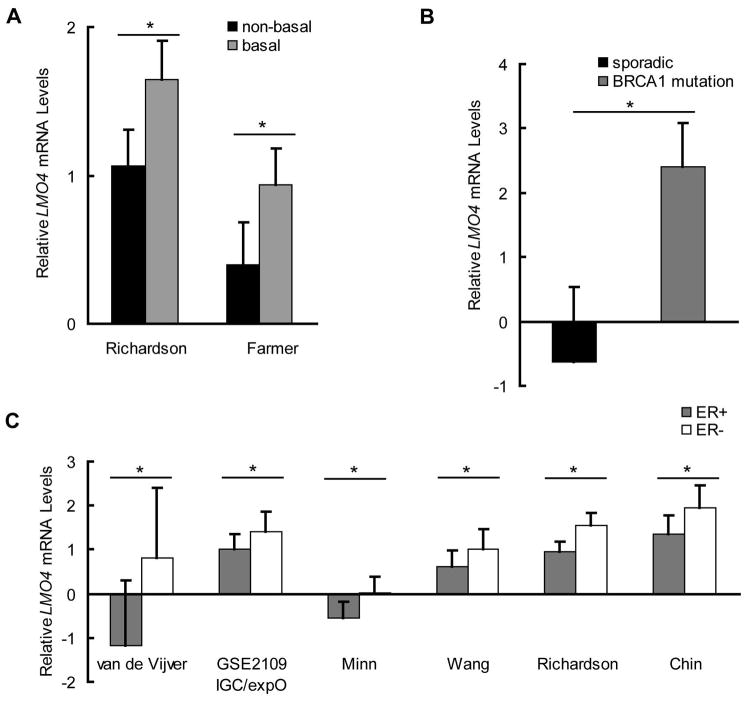
While upregulation of LMO4 has been observed in breast cancer (4), the pattern of LMO4 expression across subtypes is unknown. We compared LMO4 mRNA expression across subtypes using all available gene expression profiles with a minimum of 20 breast samples being curated in Oncomine through 2008 (27). LMO4 expression is elevated in basal-like breast cancers compared to non-basal tumors (Figure 1A) (28,29). LMO4 mRNA levels were also elevated in tumors with mutations in the breast cancer susceptibility gene 1 [BRCA1] (Figure 1B), which also exhibits a basal-like molecular signature (30). In addition, LMO4 expression is lower in estrogen receptor (ER) positive tumors compared to those lacking ER in several independent studies (Figure 1C) (29,31–34). Stratification of tumors based on their level of LMO4 mRNA within individual studies revealed that tumors with the highest quartile of LMO4 expression are exclusively basal-like cancers, typically ER-negative or enriched for BRCA1 mutations (Supplemental Table 1).
Figure 1.
LMO4 mRNA is highly expressed in BRCA1 mutant and ER-negative breast cancers. Expression of LMO4 mRNA was assessed in cohorts of breast cancers using publicly available gene expression profiles. Graphs depict the mean LMO4 mRNA levels ± standard deviation. (A) Comparison of LMO4 mRNA levels in two independent studies showed a 1.5-fold and 2.4-fold increase in LMO4 in basal-like cancers compared to non-basal-like tumors in the Richardson (n=38) (29) and Farmer (n=43) (28) studies, respectively. (B) LMO4 mRNA was also evaluated in sporadic breast cancers (n=97) and tumors associated with BRCA1 mutations (n=18). When compared to sporadic cancers, LMO4 mRNA was increased 3.9-fold in tumors with BRCA1 mutations (30). (C) LMO4 expression was assessed in ER-positive (ER+) and ER-negative (ER−) breast cancers in several independent studies: van de Vijver (n=295), GSE2109 IGC/expO (n=232), Minn (n=99), Wang (n=286), Richardson (n=39) and Chin (n=118) (29,31–34). LMO4 was consistently upregulated 1.4- to 1.6-fold in ER- tumors when compared to ER+ cancers. * p<0.05.
LMO4 expression is elevated in tumors with high mitotic indices
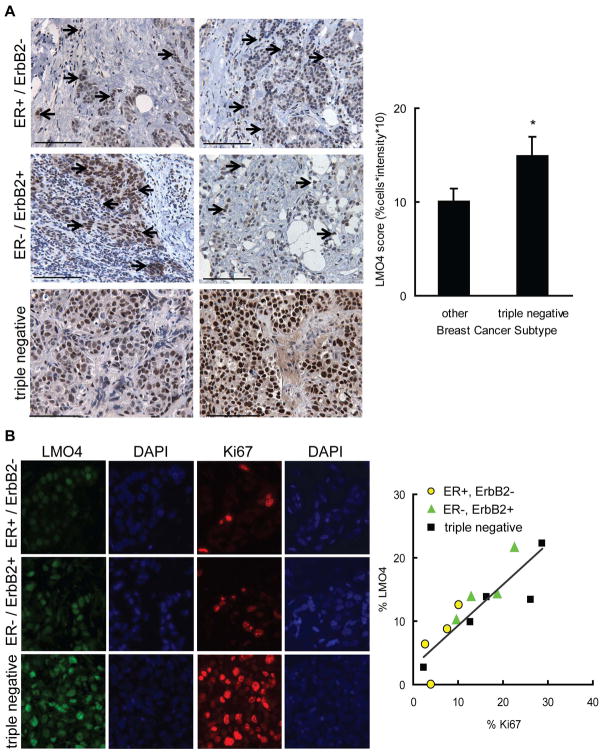
To determine if LMO4 protein levels are differentially expressed in breast cancer subtypes, 48 breast cancers encompassing ER-positive (ER+/ErbB2−), ErbB2/HER2-positive (ER−/ErbB2+) and triple negative tumors, were evaluated for LMO4 expression by immunohistochemistry (IHC). The triple-negative phenotype was used as surrogate for basal-like subtype because both tumor types lack expression of ER, progesterone receptor (PR) and ErbB2 (35). Tumors were scored according to the intensity and quantity of tumor cells with nuclear LMO4 staining. While the majority of tumors displayed nuclear LMO4 expression (96%), several tumors also had distinct cytoplasmic LMO4 staining (Supplemental Figure S1) suggesting LMO4 may shuttle between the nucleus and the cytoplasm. Analysis of the different tumor subtypes showed diffuse LMO4 positive cells in most ER+ and ErbB2+ tumors (Figure 2A). Conversely, the majority of cells in triple negative tumors (~70%) had robust nuclear LMO4 expression, with a 1.5-fold average increase in LMO4 staining relative to all other subtypes combined (Figure 2A). This pattern is consistent with that of LMO4 mRNA expression (Figure 1), indicating that both LMO4 mRNA and protein are elevated in triple negative breast cancers compared to other subtypes.
Figure 2.
LMO4 expression is elevated in tumors with high mitotic indices. (A, left panel) A cohort of triple negative (n=18), ErbB2-positive (ER−/ErbB2+ n=14) and ER-positive (ER+/ErbB2− n=16) breast tumors were evaluated for LMO4 protein expression by immunohistochemistry. Two tumors of each subtype are shown with arrows identifying LMO4 positive cells. In triple negative tumors, nearly all tumor cells were positive for LMO4. Scale bar = 100 μm. (right panel) The intensity (on a scale of 0–3, with 3 being the highest) and extent (percent of LMO4 positive tumor cells) of LMO4 nuclear stain was assessed. The final tumor score was the product of intensity and percentage of positive cells, resulting in a scale that ranged from 0 to 30. Bars are the mean histopathological score for each set of tumors and error bars depict the group’s standard deviation. * p<0.05. (B, left panel) Tumors of each subtype (ER+/ErbB2 n=4; ErbB2+/ER− n=4 and triple negative n=5) were stained for LMO4 (green) and the proliferation marker Ki67 (red). (right panel) Scatter plot analysis of the percent of tumor cells with Ki67 and intense LMO4 staining. Subtypes are represented by different symbols. Correlation coefficient, R2=0.76.
Although triple-negative or basal-like cancers have the highest LMO4 levels, some ER+/ErbB2− and ER−/ErbB2+ tumors showed areas of strong LMO4 positivity. Considering LMO4 expression has been previously associated with proliferating epithelial cells during development (26) and is necessary for proliferation of ErbB2-dependent breast cancer cells (13), we hypothesized that LMO4 expression may be indicative of the rate of tumor proliferation rather than a specific measure of basal features of a tumor. We further proposed that the increase in LMO4 expression observed in basal-like cancers is reflective of their inherently elevated mitotic index (36). To test these hypotheses, a second cohort of tumors from each of the previously examined subtypes (ER+/ErbB2−; ER−/ErbB2+ and triple negative) was immunostained for LMO4 and the proliferation marker Ki67. In each tumor, LMO4 was present with varying degrees of intensity (Figure 2B). Comparison of LMO4 and Ki67 across the breast cancer subtypes revealed that the percent of cells with intense LMO4 staining correlated linearly with the percent of cells positive for Ki67 regardless of subtype. Tumors with low proliferation rate, such as ER+/ErbB2− tumors, had the lowest percent of cells with intense LMO4 staining (Figure 2B, R2=0.76), while LMO4 levels were highest in tumors with high mitotic indices (Figure 2B).
LMO4 regulates G2/M progression in luminal and basal-like breast cancer cells
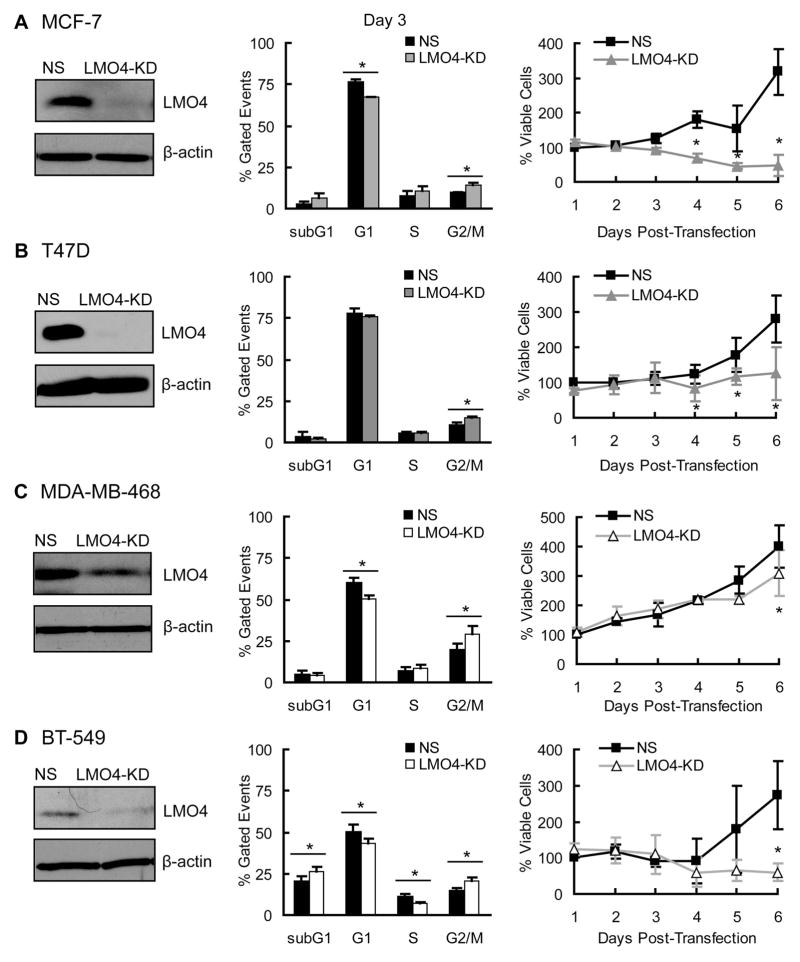
LMO4 regulates G2/M progression in ErbB2-positive breast cancer cells (13) as well as G1/S transition in a luminal breast cancer cell line (4). To determine whether LMO4 globally regulates cell cycle progression in breast cancer cells, we analyzed the cell cycle in a panel of cell lines after transient knock-down of LMO4 with a previously validated siRNA (13). Attenuating LMO4 led to a 28–32% increase in cells in G2/M in cell lines previously defined as luminal-like (MCF-7 and T47D) or basal-like (MDA-MB-468 and BT-549) (37) (Figure 3). The accumulation of cells in G2/M was accompanied by decreased rates of cell growth (Figure 3) and, in some cases, increased apoptotic cell death (Supplemental Figure S2).
Figure 3.
LMO4 regulates G2/M progression in luminal and basal-like breast cancer cells. (left panels) LMO4 expression was transiently silenced in two luminal-like (MCF-7, T47D, Panels A and B, respectively) and two basal-like (MDA-MB-468, BT-549; Panels C and D, respectively) cell lines using a previously validated siRNA (13). A representative western blot is shown for each cell line demonstrating LMO4 silencing in cells transfected with LMO4-specific siRNA (LMO4-KD) compared to cells transfected with a non-silencing siRNA control (NS). β-actin was used as loading control. (middle panels) Seventy-two hours after siRNA transfection with control (NS) or LMO4-directed siRNA (LMO4-KD), cells were harvested and stained with propidium iodide to analyze DNA content by FACS. The proportion of cells at each of the phases of the cell cycle (subG1, G1, S and G2/M) is depicted in the bar graph ± standard deviation. (right panels) Cells were transfected with siRNA and harvested every 24 hrs thereafter for 6 days. The growth of LMO4 knock-down cells was then compared to control cells (NS) by directly counting the number of viable cells at each time. * p<0.05 compared to NS transfected cells.
Alterations in LMO4 levels induce centrosome amplification
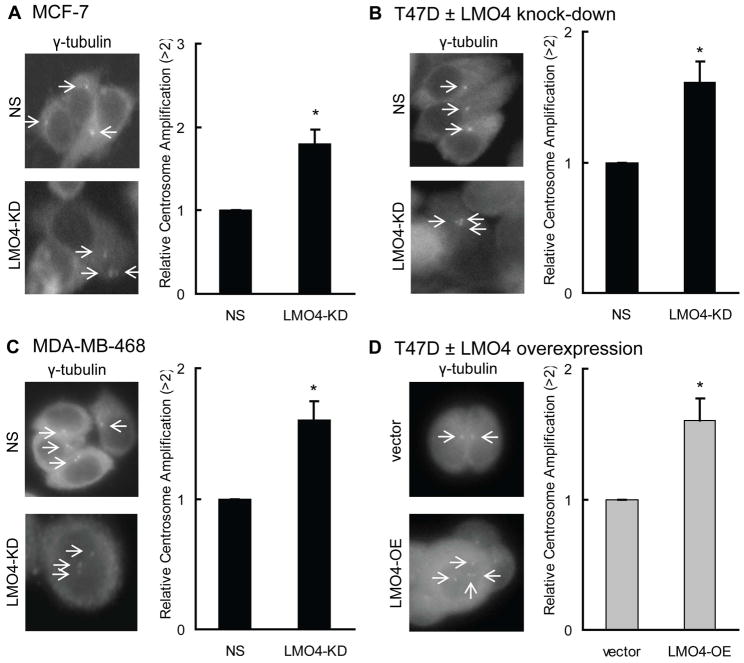
The consistent phenotypic impact of LMO4 silencing in distinct cell lines led us to postulate that the role of LMO4 in cell cycle progression may involve a common mechanism among all breast cancer subtypes. LMO4 controls expression or activity of cell cycle genes, including p21, p27 and BRCA1 (10,11,14), all of which regulate the DNA and the centrosome cycles (21,22,38), hence we hypothesized that LMO4 may control centrosome replication. To assess the impact of LMO4 silencing on centrosome number, the cell lines that were used in our analysis of cell cycle progression (with the exception of the multi-nucleated BT-549 cells) were transfected with control siRNA (NS) or LMO4-siRNA (LMO4-KD) and immunostained for the centrosome protein γ-tubulin. This analysis revealed that LMO4 knock-down leads to a 60–80% increase in cells with more than 2 centrosomes compared to non-silenced controls (Figure 4A–C). Of note, overexpression of LMO4 in the luminal breast cancer cell line T47D also caused a 60% increase in cells with abnormal centrosome numbers (Figure 4D and Supplemental Figure S3).
Figure 4.
Alterations in LMO4 levels induce centrosome amplification. (A–D) LMO4 was silenced in the luminal-like MCF7 (A) and T47D (B) as well as basal-like MDA-MB-468 (C) breast cancer cells via an LMO4 targeted siRNA. LMO4 was also overexpressed in T47D cells using retroviral infection. (D) After manipulation of LMO4 levels, cells were immunostained with an antibody to the centrosomal protein γ-tubulin. The number of centrosomes/cell was then counted and compared between cells with altered LMO4 levels [either loss of LMO4 (LMO4-KD), A–C, or LMO4 overexpression (LMO4-OE), D], and control cells [cells transfected with either non-silencing (NS) siRNA or empty expression vector (vector)]. Representative images from a total of 3 independent experiments/cell line are provided for each cell type with arrows pointing to centrosomes. Normal cells have either 1 or 2 centrosomes/cell. Bars represent the relative centrosome amplification ± standard deviation compared to the control cells in three independent experiments.
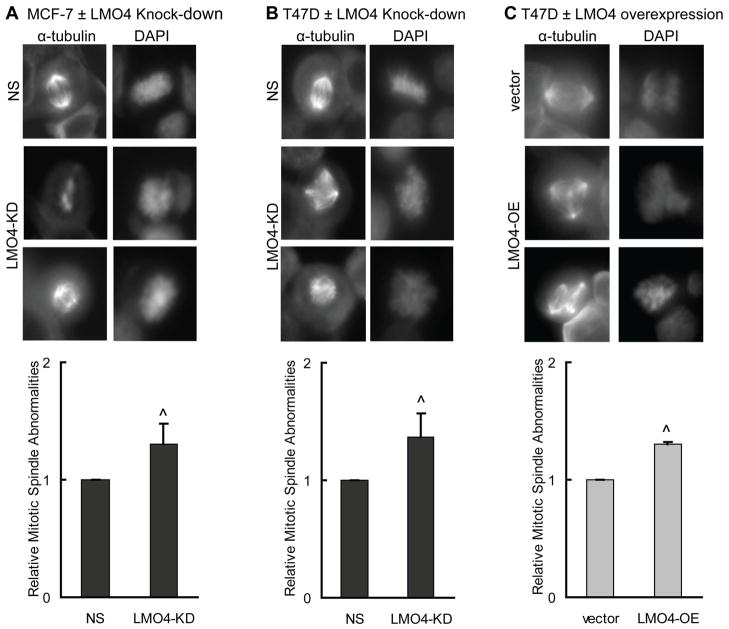
Altered LMO4 levels induce abnormal mitotic spindles
Supernumerary centrosomes often lead to formation of abnormal mitotic spindles and subsequent missegregation of chromosomes (24,39). To examine whether the aberrant centrosome number that occurs in cells with altered LMO4 levels correlates with an increase in the number of abnormal mitotic figures, breast cancer cells with silenced (LMO4-KD) or overexpressed LMO4 (LMO4-OE) were immunostained with an antibody to the mitotic spindle protein, α-tubulin, and co-stained with DAPI. LMO4 silencing leads to a 30–40% increase in the formation of abnormal mitotic spindles in MCF-7 (Figure 5A) and T47D cells (Figure 5B). Overexpression of LMO4 in T47D cells also results in a ~30% increase in abnormal mitotic figures (Figure 5C). Specifically, cells with altered LMO4 levels have increased incidence of multipolar spindles as well as asymmetrical, disorganized spindle fibers (Figure 5, α-tubulin stain). Aberrant LMO4 expression was also associated with an increase in the number of cells with unequal distribution of chromosomes (Figure 5, DAPI stain). Recapitulating the observations in Figure 4, either up- or down-regulation of LMO4 resulted in the same phenotype.
Figure 5.
Altered LMO4 levels induce abnormal mitotic spindles. (A–C) MCF-7 (A) and T47D (B) cells were transfected with either control siRNA (NS) or LMO4-specific siRNA (LMO4-KD), and synchronized using aphidicolin. T47D cells overexpressing LMO4 (LMO4-OE) or an empty vector control (vector) were also synchronized (C). After removal of aphidicolin, cells were immunostained with an antibody to α-tubulin and counterstained with DAPI for visualization of the mitotic spindle and DNA, respectively. The spindles were blindly scored for their symmetry and organization. Equal segregation of DNA was considered and percent of cells with abnormal spindles was normalized to that of control (either NS or empty vector transfected) cells. The average relative number of abnormal mitotic figures is shown with error bars depicting standard deviations. Three independent experiments were performed for each cell line.
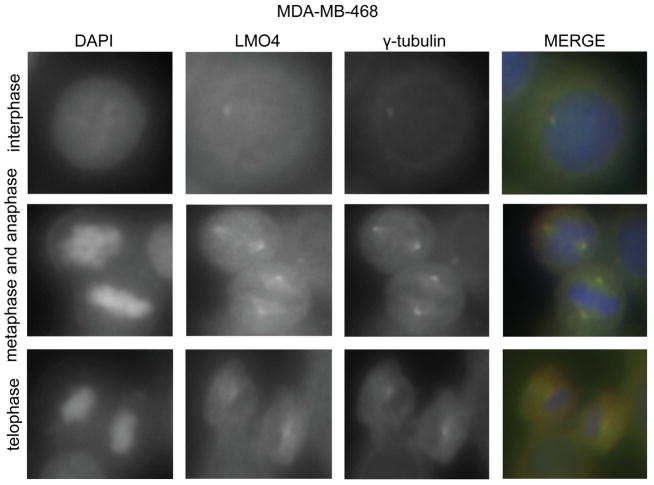
LMO4 localizes to the centrosome
Many regulators of the cell cycle, such as BRCA1 (40), co-localize or traffic to the centrosome, suggesting centrosomes act as a “command center for cellular control” (41). LMO4 regulates proteins that co-localize to the centrosome (15,42), hence we determined if LMO4 also localizes to centrosomes by co-immunostaining with an antibody to γ-tubulin. Several control experiments were performed to confirm an absence of cross-reactivity between the antibodies used in this analysis (data not shown, see Materials and Methods). LMO4 staining occurred primarily in the cytoplasm with some expression in the nucleus (Figure 6 and Supplemental Figure S4). This is consistent with other studies demonstrating that LMO4 can bind cell surface receptors and localize to the cytoplasm (43,44). We also observed LMO4 localization in centrosomal structures containing γ-tubulin in both basal-like (MDA-MB-468, Figure 6) and luminal-like (T47D, Supplemental Figure S4) cells. The co-localization of LMO4 and γ-tubulin was independent of cell cycle progression as co-staining was observed in interphase and mitotic cells (Figure 6 and Supplemental Figure S4).
Figure 6.
LMO4 localizes to the centrosome. MDA-MB-468 cells were synchronized with aphidicolin and immunostained for both LMO4 and the centrosome protein γ-tubulin. DAPI was use to stain DNA. Cells undergoing mitosis as well as cells in interphase were analyzed for co-localization of LMO4 and γ-tubulin. Images of individual stains are shown in black and white, with overlapping staining shown in yellow.
Discussion
Although mRNA array data suggests that LMO4 is a marker of aggressive breast cancers, the results presented herein reveal that LMO4 is present in all breast tumor subtypes with the highest expression in tumors with elevated mitotic indices, such as basal-like or triple negative tumors (Figures 1–2). This suggests that LMO4 is a novel indicator of a tumor’s mitotic index and that the overexpression of LMO4 observed in basal-like or ER-negative tumors is likely due to their inherently high proliferation rate (36). Our studies also revealed that LMO4 regulates G2/M progression in several breast cancer cell lines regardless of subtype (Figure 3). Furthermore, the accumulation of cells in G2/M was associated with decreased cell growth (Supplemental Figure S2), suggesting that loss of LMO4 induces a cell cycle arrest or delay in both luminal and basal-like breast cancer cells. While breast cancer subtypes are postulated to undergo distinct initiation events that drive their oncogenesis (45,46), these upstream pathways likely converge to promote growth. We propose that LMO4 is a common downstream effector that integrates signals from multiple oncogenes. This is supported by the interaction of LMO4 with proteins involved in the genesis of luminal and basal-like breast cancers such as ER (9) and BRCA1 (10), respectively and further strengthened by PI3K-mediated regulation of LMO4 expression (13), a commonly activated protein in breast cancers (47–49).
Our studies revealed that LMO4 regulates centrosome replication and mitotic spindle integrity, uncovering a novel mechanism by which LMO4 contributes to the aggressiveness of breast tumor cells. LMO4 co-localized with the centrosome protein γ-tubulin (Figure 6) suggesting that LMO4 may regulate the centrosome cycle, in part, by interacting with centrosomal proteins. Supporting this possibility, other LIM-containing proteins regulate the centrosome cycle and bind centrosomal proteins, suggesting the LIM domain may serve as a general centrosome binding domain involved in the regulation of centrosome replication. LIM Kinases 1/2 bind γ-tubulin (50) and Ajuba interacts with Aurora Kinase A, a regulator of the spindle checkpoint and the centrosome cycle (51). Alternatively, LMO4 may regulate centrosome replication by altering expression of genes involved in the centrosome cycle (i.e. Cyclin E, p21 and/or p27).
Considering that overexpression of LMO4 induces centrosome amplification, it is not surprising that cells with elevated endogenous LMO4, such as basal-like cells (Figure 2) have an intrinsically higher degree of genomic instability and centrosome amplification compared with other subtypes (52). Additionally, centrosome re-duplication is associated with a prolonged G2/M phase (39), hence the G2/M arrest observed upon loss of LMO4 (Figure 3) may be induced by the abnormal centrosome count in these cells. Similarly, because most progeny derived from abnormal mitoses undergo apoptosis (39), the increase in abnormal mitotic figures may underlie the elevated apoptosis observed in some cell lines after LMO4 silencing (Supplemental Figure S2). However, the increase in cell death (2–3-fold increase, Supplemental Figure S2) was higher than that of the number of cells containing abnormal mitotic figures (1.3-to 1.4-fold) (Figure 5). This suggests that cell death is occurring prior to fixation and scoring for abnormal mitotic spindles or that additional cell death pathways may be activated in response to altered LMO4 levels.
Induction or repression of LMO4 resulted in the same abnormalities in centrosomes and mitotic spindles (Figures 4–5). Similarly, loss of LMO4 led to increased apoptosis (Supplemental Figure S2) while a previous report found that LMO4 overexpression in MCF-7 cells also increases apoptosis (6). Together, these data support a biphasic dose-response curve for LMO4 wherein LMO4 is required not only for the formation of functional protein complexes but may also act in a dominant-negative manner to inhibit complex formation when overexpressed. Because either elevated or diminished levels of LMO4 result in the same outcomes, changes in LMO4 expression may result in perturbations in the stoichiometry of protein complexes involved in centrosome replication. This dose-response pattern has been described previously for LMO4-mediated regulation of SMAD signaling (12) and may be a function of LMO4’s ability to act as a bridging factor in multimeric complexes.
In summary, LMO4 expression is a marker of breast cancers with high mitotic indices, indicating that altering LMO4 expression may contribute to the aggressiveness of tumors. LMO4 can localize to the centrosome, regulate centrosome number and mitotic spindle integrity, and control G2/M progression. Hence, elevated LMO4 expression is not only a marker of the proliferative rate of tumors but likely also modulates their degree of genomic instability.
Supplementary Material
Acknowledgments
Grant support: NIH RO1-CA090398 (RAK), CURE supplemental grant to RO1-CA090398 (RAK), NIH T32-CA059366 (EJ), NIH T32-GM007250 (MEMW), NIH F31-CA123642 (MEMW), and DoD W81XWH-09-1-0558 (MDS).
We thank William Bechtold for technical support and Gina Bernardo for help with manuscript preparation. We acknowledge the gene expression profile efforts of the International Genomic Consortium (IGC) and the Expression Project for Oncology (expO) and are also thankful for assistance from the Case Comprehensive Cancer Center core facilities (P30 CA43703).
Footnotes
Supplementary information: Supplemental materials and methods are available at the Journal of Pathology website.
Author contribution
MEMW and RAK conceived the study. MEMW, MDS, DDS, KLL and EJ were involved in acquiring data. JDM and FWAK performed pathology studies. JEV provided study material and expert guidance. All authors were involved in interpreting the data, writing the paper and had final approval of the submitted manuscript.
Conflicts of Interest: No conflicts of interest were declared.
References
- 1.Mousses S, Bubendorf L, Wagner U, Hostetter G, Kononen J, Cornelison R, et al. Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res. 2002;62:1256–1260. [PubMed] [Google Scholar]
- 2.Visvader JE, Venter D, Hahm K, Santamaria M, Sum EY, O’Reilly L, et al. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2001;98:14452–14457. doi: 10.1073/pnas.251547698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J, Ohuchida K, Nakata K, Mizumoto K, Cui L, Fujita H, et al. LIM only 4 is overexpressed in late stage pancreas cancer. Mol Cancer. 2008;7:93–103. doi: 10.1186/1476-4598-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sum EY, Segara D, Duscio B, Bath ML, Field AS, Sutherland RL, et al. Overexpression of LMO4 induces mammary hyperplasia, promotes cell invasion, and is a predictor of poor outcome in breast cancer. Proc Natl Acad Sci U S A. 2005;102:7659–7664. doi: 10.1073/pnas.0502990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang N, Kudryavtseva E, Ch’en IL, McCormick J, Sugihara TM, Ruiz R, et al. Expression of an engrailed-LMO4 fusion protein in mammary epithelial cells inhibits mammary gland development in mice. Oncogene. 2004;23:1507–1513. doi: 10.1038/sj.onc.1207288. [DOI] [PubMed] [Google Scholar]
- 6.Wang N, Lin KK, Lu Z, Lam KS, Newton R, Xu X, et al. The LIM-only factor LMO4 regulates expression of the BMP7 gene through an HDAC2-dependent mechanism, and controls cell proliferation and apoptosis of mammary epithelial cells. Oncogene. 2007;26:6431–6441. doi: 10.1038/sj.onc.1210465. [DOI] [PubMed] [Google Scholar]
- 7.Rabbitts TH. LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev. 1998;12:2651–2657. doi: 10.1101/gad.12.17.2651. [DOI] [PubMed] [Google Scholar]
- 8.Jurata LW, Gill GN. Structure and function of LIM domains. Curr Top Microbiol Immunol. 1998;228:75–113. doi: 10.1007/978-3-642-80481-6_4. [DOI] [PubMed] [Google Scholar]
- 9.Singh RR, Barnes CJ, Talukder AH, Fuqua SA, Kumar R. Negative regulation of estrogen receptor alpha transactivation functions by LIM domain only 4 protein. Cancer Res. 2005;65:10594–10601. doi: 10.1158/0008-5472.CAN-05-2268. [DOI] [PubMed] [Google Scholar]
- 10.Sum EY, Peng B, Yu X, Chen J, Byrne J, Lindeman GJ, et al. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J Biol Chem. 2002;277:7849–7856. doi: 10.1074/jbc.M110603200. [DOI] [PubMed] [Google Scholar]
- 11.Setogawa T, Shinozaki-Yabana S, Masuda T, Matsuura K, Akiyama T. The tumor suppressor LKB1 induces p21 expression in collaboration with LMO4, GATA-6, and Ldb1. Biochem Biophys Res Commun. 2006;343:1186–1190. doi: 10.1016/j.bbrc.2006.03.077. [DOI] [PubMed] [Google Scholar]
- 12.Lu Z, Lam KS, Wang N, Xu X, Cortes M, Andersen B. LMO4 can interact with Smad proteins and modulate transforming growth factor-beta signaling in epithelial cells. Oncogene. 2006;25:2920–2930. doi: 10.1038/sj.onc.1209318. [DOI] [PubMed] [Google Scholar]
- 13.Montanez-Wiscovich ME, Seachrist DD, Landis MD, Visvader J, Andersen B, Keri RA. LMO4 is an essential mediator of ErbB2/HER2/Neu-induced breast cancer cell cycle progression. Oncogene. 2009;28:3608–3618. doi: 10.1038/onc.2009.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Smith M, Qin Z, Zhou X, Schock SC, Chen HH. LIM domain only 4 protein promotes granulocyte colony-stimulating factor-induced signaling in neurons. Cell Mol Life Sci. 2010;67:949–957. doi: 10.1007/s00018-009-0223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- 16.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 17.Dulic V, Stein GH, Far DF, Reed SI. Nuclear accumulation of p21Cip1 at the onset of mitosis: a role at the G2/M-phase transition. Mol Cell Biol. 1998;18:546–557. doi: 10.1128/mcb.18.1.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 19.Wang RH, Yu H, Deng CX. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc Natl Acad Sci U S A. 2004;101:17108–17113. doi: 10.1073/pnas.0407585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, et al. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 21.Mantel C, Braun SE, Reid S, Henegariu O, Liu L, Hangoc G, et al. p21(cip-1/waf-1) deficiency causes deformed nuclear architecture, centriole overduplication, polyploidy, and relaxed microtubule damage checkpoints in human hematopoietic cells. Blood. 1999;93:1390–1398. [PubMed] [Google Scholar]
- 22.Sugihara E, Kanai M, Saito S, Nitta T, Toyoshima H, Nakayama K, et al. Suppression of centrosome amplification after DNA damage depends on p27 accumulation. Cancer Res. 2006;66:4020–4029. doi: 10.1158/0008-5472.CAN-05-3250. [DOI] [PubMed] [Google Scholar]
- 23.D’Assoro AB, Barrett SL, Folk C, Negron VC, Boeneman K, Busby R, et al. Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res Treat. 2002;75:25–34. doi: 10.1023/a:1016550619925. [DOI] [PubMed] [Google Scholar]
- 24.Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sum EY, O’Reilly LA, Jonas N, Lindeman GJ, Visvader JE. The LIM domain protein Lmo4 is highly expressed in proliferating mouse epithelial tissues. J Histochem Cytochem. 2005;53:475–486. doi: 10.1369/jhc.4A6553.2005. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 29.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 30.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 31.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 32.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 34.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, Bartelink H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65–R79. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhargava R, Striebel J, Beriwal S, Flickinger JC, Onisko A, Ahrendt G, et al. Prevalence, morphologic features and proliferation indices of breast carcinoma molecular classes using immunohistochemical surrogate markers. Int J Clin Exp Pathol. 2009;2:444–455. [PMC free article] [PubMed] [Google Scholar]
- 37.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko MJ, Murata K, Hwang DS, Parvin JD. Inhibition of BRCA1 in breast cell lines causes the centrosome duplication cycle to be disconnected from the cell cycle. Oncogene. 2006;25:298–303. doi: 10.1038/sj.onc.1209028. [DOI] [PubMed] [Google Scholar]
- 39.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu LC, Doan TP, White RL. Identification of a gamma-tubulin-binding domain in BRCA1. Cancer Res. 2001;61:7713–7718. [PubMed] [Google Scholar]
- 41.Doxsey SJ. Centrosomes as command centres for cellular control. Nat Cell Biol. 2001;3:E105–E108. doi: 10.1038/35074618. [DOI] [PubMed] [Google Scholar]
- 42.Lotti LV, Ottini L, D’Amico C, Gradini R, Cama A, Belleudi F, et al. Subcellular localization of the BRCA1 gene product in mitotic cells. Genes Chromosomes Cancer. 2002;35:193–203. doi: 10.1002/gcc.10105. [DOI] [PubMed] [Google Scholar]
- 43.Novotny-Diermayr V, Lin B, Gu L, Cao X. Modulation of the interleukin-6 receptor subunit glycoprotein 130 complex and its signaling by LMO4 interaction. J Biol Chem. 2005;280:12747–12757. doi: 10.1074/jbc.M500175200. [DOI] [PubMed] [Google Scholar]
- 44.Schaffar G, Taniguchi J, Brodbeck T, Meyer AH, Schmidt M, Yamashita T, et al. LIM-only protein 4 interacts directly with the repulsive guidance molecule A receptor Neogenin. J Neurochem. 2008;107:418–431. doi: 10.1111/j.1471-4159.2008.05621.x. [DOI] [PubMed] [Google Scholar]
- 45.Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, et al. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci U S A. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007;117:3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokunaga E, Kimura Y, Mashino K, Oki E, Kataoka A, Ohno S, et al. Activation of PI3K/Akt signaling and hormone resistance in breast cancer. Breast Cancer. 2006;13:137–144. doi: 10.2325/jbcs.13.137. [DOI] [PubMed] [Google Scholar]
- 48.Dillon RL, White DE, Muller WJ. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene. 2007;26:1338–1345. doi: 10.1038/sj.onc.1210202. [DOI] [PubMed] [Google Scholar]
- 49.Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 50.Chakrabarti R, Jones JL, Oelschlager DK, Tapia T, Tousson A, Grizzle WE. Phosphorylated LIM kinases colocalize with gamma-tubulin in centrosomes during early stages of mitosis. Cell Cycle. 2007;6:2944–2952. doi: 10.4161/cc.6.23.4957. [DOI] [PubMed] [Google Scholar]
- 51.Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, et al. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 52.Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.