Abstract
There are lots of question-based data elements from the pharmacogenomics research network (PGRN) studies. Many data elements contain temporal information. To semantically represent these elements so that they can be machine processiable is a challenging problem for the following reasons: (1) the designers of these studies usually do not have the knowledge of any computer modeling and query languages, so that the original data elements usually are represented in spreadsheets in human languages; and (2) the time aspects in these data elements can be too complex to be represented faithfully in a machine-understandable way.
In this paper, we introduce our efforts on representing these data elements using semantic web technologies. We have developed an ontology, CNTRO, for representing clinical events and their temporal relations in the web ontology language (OWL). Here we use CNTRO to represent the time aspects in the data elements. We have evaluated 720 time-related data elements from PGRN studies. We adapted and extended the knowledge representation requirements for EliXR-TIME to categorize our data elements. A CNTRO-based SPARQL query builder has been developed to customize users’ own SPARQL queries for each knowledge representation requirement. The SPARQL query builder has been evaluated with a simulated EHR triple store to ensure its functionalities.
Introduction
Pharmacogenomics deals with the influence of genetic variation on drug response in patients by correlating gene expression or single-nucleotide polymorphisms with a drug’s efficacy or toxicity1. The Pharmacogenomics Research Network (PGRN) is a collaborative partnership of research groups funded by the U.S. National Institutes of Health to investigate pharmacogenomics studies. Huge volume of pharmacogenomics data including a large portion of time related data elements is accumulated across this network. We presented the study of harmonization and semantic annotation of the pharmacogenomics data from PGRN network previously3, but we have not yet focused the associated temporal issues.
Temporal information is very important in pharmacogenomics studies. For example, investigators need to track patients’ medical history in order to find the drug effects for genetic variations during a specific time frame. Therefore, there is an urgent need to capture the temporal information not only for the purpose of representing these data elements in a standard way for interoperability and reuse, but also for querying and retrieving the corresponding patient data matches these data elements from electronic health records (EHRs).
Representing and querying time-related data from EHR, however, is not an easy task due to the following reasons: (1) the time aspects in EHR data is often embedded in clinical narratives, which is not usually machine queriable; (2) useful temporal relations of clinical events sometimes were not stated explicitly in the original documents, but rather need to inferred, and (3) the designers of pharmacogenomics studies usually do not have the knowledge of any computer modeling and query languages, so that the original data elements usually are represented in spreadsheets in human languages, which are not machine executable without further processing.
We propose a semantic web based approach for representing the time-related data elements in PGRN studies. Our previous research has been focusing on the first two challenges mentioned in the previous paragraph. We have developed the Clinical Narrative Temporal Relation Ontology (CNTRO)4 to semantically represent clinical events, their time features, and temporal relationships, so that they can be machine queriable. Based on CNTRO, we have also implemented a reasoning framework, which can automatically infer new temporal relations, durations, and time stamps from clinical data5. Our preliminary evaluation results indicate that the CNTRO-based technique can successfully represent and infer temporal information.
In this paper, we focus the third challenge. We developed an ontology-driven SPARQL query builder to automatically represent queries for the question-based temporal data elements with respect to CNTRO. We classified the pharmacogenomics temporal data elements into eight pre-defined knowledge representation categories. The automatically generated queries for the first four categories were applied to a simulated RDF triple store with patient data. The evaluation results indicated that the system can successfully represent the questions in SPARQL queries.
Method
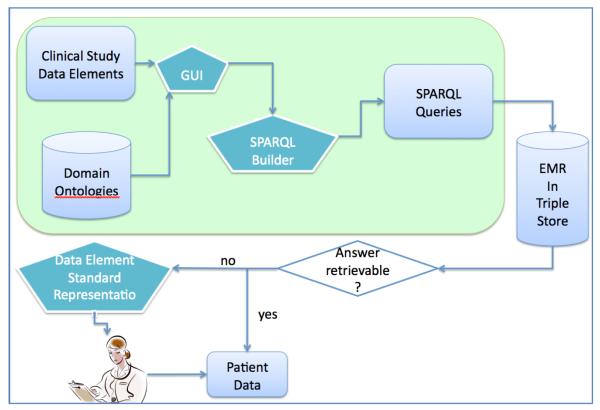
Figure 1 shows the system overview. The whole system is built on the top of the PGRN standardized pharmacogenomics data. We plan to build a Graphical User Interface (GUI), which can help end users to normalize their data elements in the PGRN standardized representations. It will take time-related data elements from the PRGN clinical studies and domain ontologies (in our case, CNTRO) and call the SPARQL query builder to compose SPARQL queries that represent the question-based data elements with respect to the ontologies. We will then run the SPARQL queries to our EHR data in RDF triple stores. If the answers of the queries can be retrieved from the triple stores, we will return the answers to the study specialists. In this paper, we focus on the steps in the big green rounded rectangle in Figure 1, where users can compose question-based data elements using our GUI and translate them to SPARQL queries.
Figure 1.
System Overview
Temporal Data Elements Extraction
We have collected more than 4,000 individual data elements arising from study questionnaires from 8 PGRN groups. To extract time-related data elements, we utilized the NCBO (National Center for Biomedical Ontology) BioPortal REST services5 to semantically annotate the words/phrases decomposed from the individual data elements by semantic types6. More details about such annotation were presented in our previous paper7. For this study, we extracted the 720 data elements with temporal semantic type “Temporal Concept” assigned.
Temporal Questions Categories
The 720 time-related data elements selected from the PGRN studies have been evaluated and classified into 8 knowledge representation categories as Table 1 shows. We adapted and extended the knowledge representation requirements for EliXR-TIME8 to categorize our questions. The first 2 categories are proposed by us and the last 6 categories are from EliXR-TIME. Each of the 720 data elements we evaluated can be classified to one or more knowledge representation categories.
Table 1.
Classification of Temporal Knowledge Categories
| Temporal knowledge representation categories |
Example |
|---|---|
| Time point |
Age hypertension diagnosed; Date Complete Blood count collected |
| Time intervals (start time, end time, or duration of an event) |
Date of last antihypertensives
Beginning home blood pressure date |
| Temporal patterns | Drink at least 1 per week |
| Temporal relations | After, before, during, within, onset, until. |
| Comparison operators | irritable mood lasting at least 1 week |
| Conjunction | And, or |
| Combinatory temporal expressions that modify a single Event or Anchor |
Do you smoke more frequently during
the first hours after waking than during the rest of the day |
| Recursive or hierarchical representation of complex temporal expressions |
How many symptoms have been present
during the same TWO-WEEK period within the last hospitalization |
OWL representation
Our next step is to check if we can represent the EHR data that potentially contains the corresponding data that matches the data elements in each knowledge representation category. In our study, we use CNTRO for modeling the time aspects on the instance level4. CNTRO is an OWL ontology designed for modeling the temporal information and relations found both in structured databases and in native language-based clinical reports. It can represent clinical events, temporal expressions (such as time instants, time intervals, repeated time periods, and durations), different levels of temporal granularity (such as minute, hour, day, month, year), temporal relationships (adopted from Allen’s temporal algebra9, such as before, after, equal), and time uncertainties.
We have evaluated CNTRO using real-world clinical notes and our preliminary results indicated that CNTRO can faithfully represent most of the time-related information from clinical notes4. From this previous study, we have verified that CNTRO is able to represent (1) time points; (2) time intervals; (3) temporal patterns; and (4) temporal relations. CNTRO does not focus on the representation of comparison operators and conjunctions, but they can be handled by either downstream tools or any rule-based language such as SWRL10. We have not focused on “combinatory temporal expressions” and “recursive or hierarchical representation” categories for the CNTRO evaluation. We believe most cases in these categories can be covered by the RDF linked-data approach using which we can integrate different time aspects for the same events easily.
SPARQL Query Builder
The prerequisite of our application is that the time aspects of the data elements and corresponding patient data are both represented with respect to the CNTRO ontology. Our previous research has already indicated that the CNTRO is capable of representing the time aspect in EHR data4. If the question-based data elements can be represented in SPARQL queries using the properties and classes defined by CNTRO, we can query the EHR data directly.
SPARQL is a complex language. Without any appropriate knowledge, it is not easy for the users to compose their own SPARQL queries. To help users ask time-related questions to SPARQL, we implemented a SPARQL Query Builder, which was adapted and extended from our previous work11.
The SPARQL query builder was built on top of a Sesame triple store12. Starting with a central subject (e.g., a particular event), a user can add data properties and object properties associated with the subject through the GUI prompted drop-down boxes which are generated dynamically using the ontology definitions. The GUI only allows users to select properties that are directly related to the subject based on the domain and range definitions of the properties. Similarly, after a property has been chosen, the GUI will allow the users to choose those objects that are related to the property (i.e., those classes that are defined as the range of the property). Step by step, the SPARQL query builder provides an intuitive way to translate a user question into a graph pattern, and then encode it into a SPARQL query. The whole query building process is done dynamically based on the back end ontology. In our case, the CNTRO ontology was loaded into the SPARQL query builder. The detailed examples with SPARQL query builder are shown in “Implementation Result” section.
Implementation Result
We implemented and evaluated the query builder based on the KR categories. In this paper, we focused on the first 4 temporal knowledge representation categories in Table 1. We have evaluated the system using a simulated EHR data triple store that covers scenarios in the first 4 categories. Here we use two examples to illustrate how the system works.
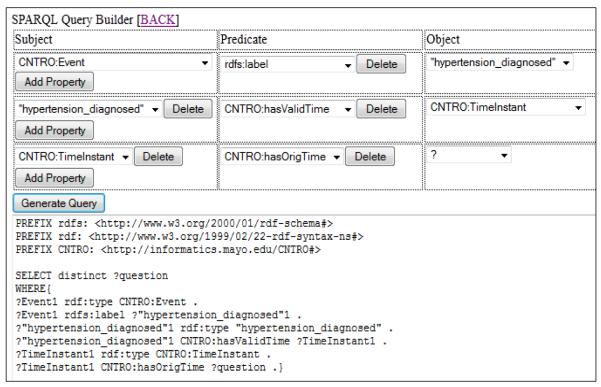
“Year hypertension diagnosed” is one PGRN temporal question-based data element belonging to the first knowledge representation category. For one particular patient, the relevant answer extracted from the EHR is “06-01-2006”. The CNTRO representation for this data element, along with the answer is shown in Figure 2. The interface of the SPARQL query builder is shown in Figure 3. As we can see in Figure 2, starting from the central subject Event, we want to find the time of the event that has the label “hypertension_diagnosed”. The query builder leads the user step by step until the original time can be retrieved. After the selection has been made, the SPARQL query will be assembled by clicking the “Generate Query” button. The query result “06-01-2006” can then be retrieved.
Figure 2.

OWL representation for the PGRN question
Figure 3.
An example of SPARQL query composed by SPARQL query builder
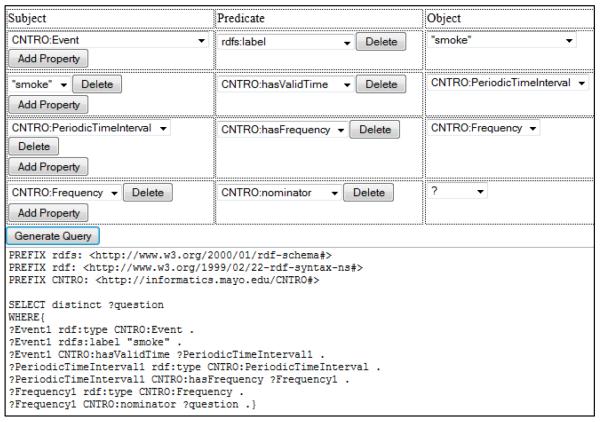
Figure 4 demonstrates how to compose a query in the third category (“Temporal Patterns”). The example we use here is “how many cigarettes did you smoke per day?” For one particular patient, we can retrieve the answer “3” from the Sesame triple store. The system can retrieve the frequency of a particular event by linking from the CNTRO PeriodicTimeInterval class. As we can see, here the central object is an Event with label “smoke”. Since we are interested in the frequency, we will need to find the PeriodicTimeInterval for the event. Then step by step, the query builder will help the users to find the nominator of the frequency.
Figure 4.
The second example of SPARQL query composed by SPARQL query builder
The query building processes for the second and fourth knowledge representation categories (“Time Interval” and “Temporal Relations) are similar to the above examples; therefore, we do not use separate examples to illustrate them. For the “Time Interval” category, we can build queries to retrieve the start time, end time, and duration of a particular event. For the “Temporal relations” category, we can build queries to retrieve the events that have a particular temporal relation with a given event, for example, “what happened during hospitalization”.
Related Work
The SPARQL query builder we introduced in this paper is an ontology driven query composer. This query builder is also fully adopted by the VIVO system, which is an ontology-based research discovery platform that hosts information about scientists, their interests, activities, and accomplishments13. Here provide further comparison of our query builder to other well-known SPARQL query building systems.
DistilBio14 is a query interface that facilitates users to ask large and complex questions in a simplified way across data sets through a graph model. SPARQL assist15 is another tool provides context-sensitive type-ahead completion during SPARQL query construction. iSPARQL16 provides a user friendly interface and functions to view predefined SPARQL queries as graph models and lets users be able to review the corresponding properties about related nodes and edges. All of these systems including ours are aiming to facilitate users to compose SPARQL queries without any knowledge of semantic web technologies. One advantage of our SPARQL query builder is that it is built on top of ontologies, so that all the predicates and their object classes and subject classes are predefined. The query builder interface can therefore dynamically load the triples step by step based on the ontology definitions. Thus, end users can easily pick up the relations from the dynamically generated pick list provided from our SPARQL query builder GUI to customize their own SPARQL queries successfully.
Conclusions and Future Work
This paper introduces our preliminary work on representing time-related question-based data elements from PGRN using semantic web technologies. We have successfully categorized the pharmacogenomics data elements from PGRN network into 8 knowledge representation categories and utilized our SPARQL query builder to compose SPARQL queries and retrieve the corresponding patient data from a simulated EHR triple store.
Several directions remain to be explored in the future. With the current SPARQL query builder, users must select one central subject (e.g., a particular event) to start with. When a data element involves multiple temporal events, or multiple temporal features about a single event, however, the data element has to be decomposed to multiple sub-queries. For example, “how many abnormal blood pressure measurements within two months after taking anti-hypertensive drug” contains three sub-queries: “when was the anti-hypertension drug taken”; “when were the abnormal blood pressure measurements taken”; “the duration in between”. Our system can successfully build queries for these questions. Our next step is to implement a downstream pipeline which can combine the answers retrieved and eventually answer the questions asked. This functionality will also facilitate building queries from the last four knowledge representation categories. In addition, the current SPARQL query builder is only designed for questions about subjects and objects. Another future direction is to improve the GUI, so that the end users can retrieve information about predicates. Finally, we would like to connect our system to the Strategic Health IT Advanced Research Projects, Area 4 (SHARPn.org), so that we can retrieve answers from real patient data.
Acknowledgement
This work was partially supported by the NIH/NIGMS (U19 GM61388; the Pharmacogenomic Research Network, PGRN) and the Strategic Health IT Advanced Research Projects (SHARP) Program (90TR002) administered by the Office of the National Coordinator for Health Information Technology. The contents of the manuscript are solely the responsibility of the authors.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Reference
- 1.Wang L. Pharmacogenomics: a systems approach. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2010;2(1):3–22. doi: 10.1002/wsbm.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.PGRN [cited; Available from: http://pgrn.org/display/pgrnwebsite/PGRN+Home.
- 3.Zhu QFR, Durski MJ, Pathak J, Chute CG. Standardization of Data Dictionaries: A Case Study from the Pharmacogenomics Research Network. AMIA Summit on Clinical Research Informatics; 2012. [Google Scholar]
- 4.Tao C, Wei WQ, Solbrig HR, Savova G, Chute CG. CNTRO: A Semantic Web Ontology for Temporal Relation Inferencing in Clinical Narratives. AMIA Annu Symp Proc. 2010;2010:787–91. [PMC free article] [PubMed] [Google Scholar]
- 5.Wei WQ, Tao C, Jiang G, Chute CG. A high throughput semantic concept frequency based approach for patient identification: a case study using type 2 diabetes mellitus clinical notes. AMIA Annu Symp Proc. 2010;2010:857–61. [PMC free article] [PubMed] [Google Scholar]
- 6.UMLS Semantic Types. http://wwwnlmnihgov/research/umls/META3_current_semantic_typeshtml.
- 7.Zhu QFR, Lian Z, Bauer HS, Durski MJ, Tao C, Pathak J, Chute CG. Harmonization and semantic annotation of data dictionaries from the Pharmacogenomics Research Network: a case study. Submitted to AMIA 2012 Annual Symposium; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boland MR, Tu SW, Carini MA, Sim I, Weng C. EliXR-TIME: A Temporal Knowledge Representation for Clinical Research Eligibility Criteria. AMIA Summits Transl Sci Proc. 2012 [PMC free article] [PubMed] [Google Scholar]
- 9.Allen JF. Maintaining Knowledge About Temporal Intervals. Commun Acm. 1983;26(11):832–43. [Google Scholar]
- 10. [02/23/2012];A Semantic Web Rule Language Combining OWL and RuleML. Available from: http://www.w3.org/Submission/SWRL/
- 11.Zhu Q, Sun Y, Challa S, Ding Y, Lajiness MS, Wild DJ. Semantic inference using chemogenomics data for drug discovery. BMC Bioinformatics. 2011;12:256. doi: 10.1186/1471-2105-12-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sesame. http://wwwopenrdforg/indexjsp.
- 13.VIVO. [cited; Available from: http://www.vivoweb.org.
- 14.DistilBio. [cited; Available from: http://distilbio.com/
- 15.SPARQL Assistant [cited; Available from: http://sadiframework.org/content/tag/sparql-assist/SPARQL Assist.
- 16.iSPARQL [cited; Available from: http://dbpedia.org/isparql/
- 17.SPARQL Assist [cited; Available from: http://sadiframework.org/content/tag/sparql-assist/