Abstract
Fetal supply with long-chain PUFA (LC-PUFA) during pregnancy is important for brain growth and visual and cognitive development and is provided by materno–fetal placental transfer. We recently showed that maternal fatty acid desaturase (FADS) genotypes modulate the amounts of LC-PUFA in maternal blood. Whether FADS genotypes influence the amounts of umbilical cord fatty acids has not been investigated until now. The aim of the present study was to investigate the influence of maternal and child FADS genotypes on the amounts of LC-PUFA in umbilical cord venous plasma as an indicator of fetal fatty acid supply during pregnancy. A total of eleven cord plasma n-6 and n-3 fatty acids were analysed for association with seventeen FADS gene cluster SNP in over 2000 mothers and children from the Avon Longitudinal Study of Parents and Children. In a multivariable analysis, the maternal genotype effect was adjusted for the child genotype and vice versa to estimate which of the two has the stronger influence on cord plasma fatty acids. Both maternal and child FADS genotypes and haplotypes influenced amounts of cord plasma LC-PUFA and fatty acid ratios. Specifically, most analysed maternal SNP were associated with cord plasma levels of the precursor n-6 PUFA, whereas the child genotypes were mainly associated with more highly desaturated n-6 LC-PUFA. This first study on FADS genotypes and cord fatty acids suggests that fetal LC-PUFA status is determined to some extent by fetal fatty acid conversion. Associations of particular haplotypes suggest specific effects of SNP rs498793 and rs968567 on fatty acid metabolism.
Keywords: FADS, Fetal fatty acid supply, Cord blood, Avon Longitudinal Study of Parents and Children (ALSPAC)
Introduction
A sufficient supply with PUFA and long-chain PUFA (LC-PUFA) of the n-6 and n-3 family is important in every stage of human life from pregnancy to adulthood for the maintenance of metabolism and health. The PUFA and LC-PUFA composition in blood and tissues has been associated with the occurrence of the metabolic syndrome and CVD, immunological and inflammatory responses and related diseases such as allergies, early visual, cognitive and motor development, as well as mental health and psychiatric disorders( 1 , 2 ). Especially LC-PUFA with twenty or more carbon atoms and three or more double bonds, such as arachidonic acid (AA; 20 : 4n-6), EPA (20 : 5n-3) and DHA (22 : 6n-3), are considered as important biologically active compounds( 3 , 4 ). LC-PUFA are indispensable components of cell membranes and modulate their integrity and fluidity( 5 ), they act as second messengers in intracellular signalling pathways or regulate transcription, and they serve as precursors for the synthesis of eicosanoids and docosanoids, which are potent regulators of inflammatory processes( 6 ).
In addition to the dietary intake of LC-PUFA from animal lipids contained in meat, eggs, fish, and in human breast milk, they can also be synthesised endogenously from the essential fatty acids linoleic acid (18 : 2n-6) and α-linolenic acid (18 : 3n-3) derived primarily from vegetable oils( 7 , 8 ). The rate-limiting enzymes in the desaturation–elongation cascade of LC-PUFA synthesis are Δ-5 desaturase and Δ-6 desaturase. A detailed overview of the complete pathway has been shown elsewhere( 9 , 10 ). Numerous genetic association studies of SNP in the desaturase-encoding genes fatty acid desaturase 1 and 2 (FADS1 and FADS2) have reported significant associations between these SNP and fatty acid levels in various tissues including serum, plasma, erythrocyte phospholipids, adipose tissue and breast milk( 11 – 22 ). Carriers of the minor alleles showed increased levels of direct substrates of the corresponding desaturase reactions (18 : 2n-6, 18 : 3n-3 and 20 : 3n-6) and reduced levels of direct and derived products of the respective desaturase reactions (for example, 18 : 3n-6, 20 : 4n-6, 22 : 4n-6, 20 : 5n-3, 22 : 6n-3), suggesting a decline in desaturase activity due to the polymorphisms.
Supply of the fetus with PUFA and LC-PUFA during pregnancy, which is considered especially important for brain growth and optimal visual and cognitive development( 23 ), is provided by placental transfer from the mother to the fetus via the umbilical cord( 24 ). The amounts of umbilical cord fatty acids are correlated with the amounts in maternal blood( 25 ) and several supplementation studies have shown that the amounts of DHA in cord blood are influenced by the mother’s diet( 26 , 27 ). The composition of LC-PUFA in human cord blood has been suggested to be important for later health outcomes of children. There is especially a debate ongoing on the importance of n-6 and n-3 fatty acids for the development of atopic diseases such as atopic dermatitis, asthma and allergic rhinitis. Several studies have demonstrated altered levels of especially n-6 fatty acids in atopic patients( 28 – 31 ). Few studies have also reported an association between fatty acid composition of umbilical cord blood and atopic disease risk or development( 32 – 34 ). In the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort, hints for associations between cord blood erythrocyte fatty acids and atopic diseases have also been reported earlier, although these associations failed to reach statistical significance after adjusting for multiple comparisons( 35 ). In addition to atopic diseases, cognitive function is another important phenotype that has been associated with cord blood fatty acid levels in some studies( 36 , 37 ). Additionally, a recent study reported associations between cord plasma fatty acid composition and risk for childhood adiposity( 38 ).
In a recent study, we showed that polymorphisms in the FADS gene cluster determine the amounts of PUFA and LC-PUFA in erythrocyte phospholipids of pregnant women( 21 ) independent of maternal diet. These results suggest that FADS genotypes may modify the fatty acid supply of the child during pregnancy; however, proof for this hypothesis has not been shown until now. The aim of the present study was therefore to investigate the influence of maternal and child FADS genotypes on the amounts of PUFA and LC-PUFA in umbilical cord venous plasma as an indicator of fetal fatty acid supply during pregnancy.
Methods
Subjects
The ALSPAC (http://www.alspac.bris.ac.uk) is a multi-purpose birth cohort study based at Bristol University, England, involving over 14 000 pregnancies in the Avon area of England in the early 1990s, the children from which have been followed through childhood( 39 ).
Analysis of umbilical cord venous plasma fatty acids
Umbilical cord plasma collected from the umbilical vein was separated and frozen within 0–8 d after birth, stored at − 80°C, thawed once to obtain a 100μl sample, shipped by airfreight on dry ice to Rockville, MD, USA and thawed a second time for analysis. Transmethylation of lipids with acetyl chloride and methanol was performed using a simplified method based upon the Lepage & Roy procedure( 40 ), using a high-throughput automated method( 41 ). Internal calibration was conducted by adding internal standards to each assay. A second standard was used to quantify the exact amount of internal standard in every batch for ongoing assay of experimental variability. Freedom Evo Instrument 200 (TECAN Trading AG) was utilised for the automatic transmethylation and extraction of fatty acids employing the customised control and automation software (EVOware v2.0, SP1, Patch3). The GC 6890 Plus LAN system (Agilent Technologies, Inc.) coupled with a fused-silica, narrow-bored DB-FFAP capillary column (Agilent 127-32H2; 15 m × 0·1 mm internal diameter × 0·1μm film thickness) was used for chromatographic separation of the fatty acid methyl esters as reported previously( 41 ). The assay was linear in the range of 1–600μg/ml plasma. The within- and between-day imprecision was 3·26 ± 1·2 %, and 2·95 ± 1·6 %, respectively, for fatty acid concentrations. From twenty-two measured fatty acids in total, eleven n-6 and n-3 fatty acids were measured and expressed as a percentage by weight of total fatty acids.
Genetic analyses
Genomic DNA was extracted from leucocytes from blood samples obtained during pregnancy for mothers and, for the children, from cord blood or from samples obtained at ALSPAC clinics from age 7 years onwards( 42 ).
SNP genotyped in the present study were chosen based on a minor allele frequency of >10 % and linkage disequilibrium (LD) information provided by HapMap (http://hapmap.ncbi.nlm.nih.gov/). SNP with r 2 >0·8 were considered to have a high degree of co-inheritance in Europeans and were therefore selected as ‘tagSNP’. Twelve ‘tagSNP’ (rs174576, rs174579, rs174448, rs2727271, rs174634, rs174449, rs968567, rs526126, rs174455, rs174602, rs498793 and rs174570) located in the genomic region spanning FADS1, FADS2 and FADS3 were selected for genotyping together with six additional SNP (rs174556, rs174561, rs3834458, rs174548, rs174574 and rs174578) that have already been associated with fatty acid levels in previous studies. A quantity of 5 ng of genomic DNA was subjected to PCR amplification followed by the genotyping procedure using iPLEXTM chemistry (Sequenom) according to the manufacturer’s protocol and a matrix-assisted laser desorption/ionisation time-of-flight-based allele detection method. The procedure has been described in detail elsewhere( 13 ).
Genotyping failure rates for maternal samples ranged from 1·1 to 4·1 %. Error rates based upon about 790 duplicate samples ranged from 0 to 0·64 %. For child DNA, failure and error rates ranged from 2·1 to 4·8 % and from 0 to 1·3 %, respectively, for 220 duplicate samples.
Statistics and confounding
All SNP except SNP rs174634 passed all quality criteria and genotypes were used for statistical analysis. Linear regression analysis was used to investigate the associations of FADS polymorphisms with PUFA levels. Due to the skewness of the data, log transformations were applied to all fatty acids and all fatty acid outcomes were standardised to have a variance of 1 to produce more comparable effect sizes. Genetic variants were modelled assuming a linear relationship between each fatty acid and the number of copies of the minor allele as has been reported in other studies( 11 – 22 ). Hardy–Weinberg equilibrium was tested using the maximum likelihood estimate of the minor allele frequency to derive expected genotype frequencies. Haplotypes were generated using PLINK version 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/). Haplotype blocks were identified using Haploview version 4.2 (Broad Institute). Data were restricted to the haplotype for each individual with a conditional probability exceeding 0·9 for the analyses of the two main blocks in high LD and 0·5 for haplotypes generated from all seventeen SNP. To estimate the independent contribution of the maternal and fetal metabolism to cord blood levels, each pair of SNP relating to the mother and offspring were entered simultaneously into separate linear regression models.
Although genetic effects are unlikely to be confounded with sociodemographic variables( 43 ), the validity of results was explored in an additional analysis adjusting for ten potential confounders: sex of the child, multiple pregnancy (singleton, multiple), parity (primiparous, multiparous), maternal smoking at 32 weeks of gestation (yes, no), gestation, maternal age, maternal pre-pregnancy BMI, maternal diet in pregnancy (assessed using eighty-one food frequency questions) and a measure of family adversity, and storage time at 4°C. The first four confounders were considered as categorical variables while the latter six were treated as continuous variables. To avoid complications of allele frequencies and effect sizes varying with ethnicity, analyses were restricted to those mothers of white ethnic origin.
Ethics
All parents signed an informed consent for the participation in the study including approval for analysis of genetic information. Ethical approval for the study was obtained from the ALSPAC Law and Ethics Committee and the Local Research Ethics Committees. The data were deemed exempt for additional review by the Office of Human Subjects Research, National Institutes of Health.
Results
Characteristics of the sample
Cord plasma fatty acid data with matching genetic data were available for 3750 children (92·4 % of total samples and 36·5 % of total genotyped), of which 3368 were of white ethnic origin. In total, fatty acid and genotype data of 2035 mother–child pairs were available.
In the adjusted analysis including ten potential sociodemographic confounders, a total number of 2659 samples were analysed. The demographics of the sample in this adjusted analysis are shown in online Supplementary Table 1.
The composition of umbilical cord plasma PUFA in the ALSPAC cohort is shown in Table 1. Contents of individual fatty acids in our cord plasma samples were generally in the same range as in previous reports on cord plasma fatty acids, although the mean amounts of especially AA and DHA seem to be highly variable between studies( 34 , 44 – 48 ). Specifically, AA levels in the present study were slightly lower compared with most other reports, whereas DHA levels were in the medium range compared with other studies.
Table 1.
Fatty acids (wt% of total fatty acids) in cord plasma of 3368 children of white ethnic origin*
(Medians, interquartile ranges (IQR) and standard deviations)
| Fatty acid | Median | IQR | SD |
|---|---|---|---|
| Saturates | 46·38 | 41·60–49·39 | 4·92 |
| Monounsaturates | 26·82 | 24·93–28·90 | 3·14 |
| n-6 | |||
| 18:2 | 9·74 | 8·65–11·03 | 2·11 |
| 18:3 | 0·35 | 0·26–0·74 | 0·95 |
| 20:2 | 0·29 | 0·25–0·34 | 0·11 |
| 20:3 | 2·64 | 2·20–3·07 | 0·63 |
| 20:4 | 8·92 | 7·15–11·49 | 2·77 |
| 22:4 | 0·41 | 0·33–0·52 | 0·14 |
| 22:5 | 0·47 | 0·35–0·62 | 0·21 |
| n-3 | |||
| 18:3 | 0·14 | 0·11–0·18 | 0·07 |
| 20:5 | 0·19 | 0·14–0·25 | 0·10 |
| 22:5 | 0·20 | 0·15–0·27 | 0·11 |
| 22:6 | 2·43 | 1·84–3·42 | 1·18 |
The sample reflects observations where genetic data are present for at least one polymorphism. All fatty acids showed evidence of skewness (P< 0·001) although the IQR suggested that the deviations from symmetry were minor for some outcomes.
Test for Hardy–Weinberg equilibrium
Genetic variants were generally in Hardy–Weinberg equilibrium although disequilibrium existed for rs174579 in both mothers and children (P < 0·001) and rs174570 in mothers (P = 0·001) (see online Supplementary Table 2). In general, heterozygotes were under-represented in these samples although the reasons for these departures remain unclear. Possible explanations associated with genotyping problems, such as poor cluster separation, were not applicable for these data. Differential fetal survival may provide an alternative explanation( 49 ). However, overall, these departures were considered minor with the statistical significance reflecting the large sample size. It was considered important to report results for these two SNP but, if nevertheless these departures reflected genotype misclassifications, it should be noted that any associations would be biased towards the null. Hence reported results would tend to underestimate the true effect sizes.
Single SNP analysis
In a univariable model to analyse the associations of maternal and child FADS genotypes with fatty acid levels separately, most of maternal and child genotypes showed significant associations with all analysed n-6 fatty acids, except for γ-linolenic acid (18 : 3n-6). Minor alleles were associated with higher amounts of the shorter-chain fatty acids 18 : 2n-6, 20 : 2n-6 and 20 : 3n-6, and with lower amounts of the longer-chain fatty acids 20 : 4n-6, 22 : 4n-6 and 22 : 5n-6 (see online Supplementary Tables 3 and 4). One exception was the association with SNP rs498793, where the direction of effects was the opposite of all other SNP. R 2 values that reflect the genetically explained variability of fatty acid concentrations ranged from 0·13 % (18 : 3n-6) to 8·48 % (20 : 3n-6) for the seventeen maternal SNP and 0·05 % (18 : 3n-6) to 10·69 % (20 : 3n-6) for the seventeen child SNP (see Table 2).
Table 2.
Maximum R2 (%) across the seventeen genetic variants by mother–offspring for each fatty acid and ratio (log transformed) in unadjusted analyses (reflecting the genetic association) and in adjusted analyses (including the effect of confounders)
| Unadjusted |
Adjusted |
|||
|---|---|---|---|---|
| Mother | Offspring | Mother | Offspring | |
| n-6 | ||||
| 18:2 | 3·28 | 1·10 | 16·97 | 14·20 |
| 18:3 | 0·13 | 0·05 | 5·26 | 5·58 |
| 20:2 | 2·21 | 1·03 | 8·52 | 7·59 |
| 20:3 | 8·48 | 10·69 | 16·11 | 19·16 |
| 20:4 | 1·86 | 2·99 | 10·41 | 9·00 |
| 22:4 | 0·71 | 1·26 | 9·30 | 9·66 |
| 22:5 | 0·40 | 0·89 | 11·09 | 10·46 |
| 20:3/18:2 | 3·02 | 6·52 | 15·50 | 19·44 |
| 20:4/20:3 | 23·07 | 31·24 | 29·40 | 35·81 |
| 20:4/18:2 | 6·06 | 5·63 | 19·79 | 16·84 |
| n·3 | ||||
| 18:3 | 0·29 | 0·52 | 13·45 | 12·98 |
| 20:5 | 0·23 | 0·13 | 10·63 | 10·30 |
| 22:5 | 0·29 | 0·18 | 12·99 | 13·67 |
| 22:6 | 1·04 | 0·68 | 8·10 | 7·56 |
| 20:5/18:3 | 0·45 | 0·54 | 10·21 | 9·85 |
| Maximum n | 2491 | 2876 | 1983 | 2287 |
Maximum R 2 (%) across the seventeen genetic variants by mother–offspring for each fatty acid and ratio (log transformed) in unadjusted analyses (reflecting the genetic association) and in adjusted analyses (including the effect of confounders)
Significant associations were also observed between maternal and child SNP and the n-3 fatty acids α-linolenic acid (18 : 3n-3) and DHA (22 : 6n-3). Carriers of minor alleles had higher amounts of α-linolenic acid and lower amounts of the n-3 product DHA. However, the minor allele of SNP rs498793 in mothers was associated with higher levels of 22 : 6n-3 in cord blood, and children carrying the minor allele had lower levels of 18 : 3n-3, which is again contrary to the results for all other SNP (see online Supplementary Tables 3 and 4). The genetically explained variability for n-3 fatty acids ranged from 0·29 % (18 : 3n-3) to 1·04 % (22 : 6n-3) for the seventeen maternal SNP and from 0·13 % (20 : 5n-3) to 0·68 % (22 : 6n-3) for the seventeen child SNP (Table 2).
In addition to the SNP effects on single fatty acids, we investigated whether the effects would be stronger and could possibly be refined by using specific fatty acid ratios to estimate desaturase activities. In general, maternal and child genotypes showed the same effect direction on fatty acid ratios, and effect sizes were comparable as well. The minor alleles were consistently associated with higher levels of the n-6 ratio 20 : 3/18 : 2, which estimates Δ-6 desaturase activity, lower levels of the 20 : 4/20 : 3 ratio, which estimates Δ-5 desaturase activity, and also lower levels of 20 : 4/18 : 2, which is a combination of both. Minor alleles of most SNP were also associated with lower levels of the n-3 ratio 20 : 5/18 : 3. As in the analysis of single fatty acids, SNP rs498793 showed the opposite in terms of effect direction compared with all other SNP (see online Supplementary Tables 3 and 4).
The validity of these results was explored in an additional analysis adjusting for ten potential confounders. Overall, the results of the initial analysis were not changed by this adjustment. For instance, for AA, the effect sizes for the seventeen mother and offspring SNP attenuated on average by only 0·7 % (range 18 % attenuation to 20 % amplification) with similar P values (range 0·035 to 2·9 × 10− 15 in adjusted analyses compared with 0·021 to 6·1 × 10− 16 in unadjusted analyses but restricted to the adjusted sample). In the adjusted analyses, up to 29·4 % (AA:dihomo-γ-linolenic acid ratio) of variance could be explained by the maternal SNP and the included confounders, and up to 35·8 % (AA:dihomo-γ-linolenic acid ratio) of variance by the child SNP and the included confounders (Table 2).
Predominant effect of maternal FADS genotypes on less desaturated n-6 fatty acids
Maternal and child genotypes are related to each other due to the rules of inheritance. Therefore, in a second step, we repeated the single SNP analysis by adjusting the maternal genotype effect for the child genotype and vice versa. The purpose of these analyses was to estimate which of the two has the stronger influence on cord blood fatty acid composition, i.e. whether cord blood fatty acid composition is determined to a greater extent by fetal or maternal metabolism. By adjusting the maternal genotype effect for the child genotype, the minor alleles of most analysed maternal SNP were positively associated with the precursor n-6 PUFA 18 : 2n-6, 20 : 2n-6 and 20 : 3n-6 (P values between 0·020 and 3·2 × 10− 14 for 88 % of results) (Table 3). In terms of regression coefficients, associations with 20 : 3n-6 were the strongest, followed by 20 : 2n-6 and 18 : 2n-6. Negative and less significant associations were observed for the n-6 LC-PUFA AA (P < 0·027) for ten of the seventeen analysed SNP and adrenic acid (P < 0·033) for four SNP. In the initial unadjusted analysis, the genetically explained variances of 18 : 2n-6 and 20 : 2n-6 were higher for the maternal than for the child SNP (Table 2), which might also reflect the predominant influence of the maternal genotype on the levels of these fatty acids. Amounts of 18 : 3n-6 and 22 : 5n-6 were not associated with any of the tested maternal SNP. Effect sizes of the ratio associations were slightly attenuated when the maternal genotype was adjusted for the child genotype (see online Supplementary Table 3 and Table 4).
Table 3.
Regression coefficients from analyses of log n-6 and n-3 fatty acid levels as a percentage of total fatty acid levels on maternal fatty acid desaturase (FADS) SNP adjusted for child genotype† (Regression coefficients with their standard errors)
|
FADS1
|
Intergenic |
FADS2
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs174548 |
rs174556 |
rs174561 |
rs3834458 |
rs968567 |
rs174570 |
rs174574 |
||||||||
| Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | |
| n-6 | ||||||||||||||
| 18:2 | 0·190*** | 0·037 | 0·199*** | 0·037 | 0·201*** | 0·037 | 0·219*** | 0·036 | 0·105* | 0·045 | 0·219*** | 0·050 | 0·215*** | 0·036 |
| 18:3 | 0·046 | 0·039 | 0·044 | 0·040 | 0·050 | 0·040 | 0·063 | 0·039 | 0·032 | 0·048 | 0·004 | 0·054 | 0·058 | 0·038 |
| 20:2 | 0·209*** | 0·036 | 0·203*** | 0·037 | 0·212*** | 0·037 | 0·221*** | 0·036 | 0·146** | 0·045 | 0·206*** | 0·050 | 0·222*** | 0·035 |
| 20:3 | 0·253*** | 0·036 | 0·265*** | 0·036 | 0·275*** | 0·036 | 0·253*** | 0·035 | 0·241*** | 0·045 | 0·146** | 0·052 | 0·251*** | 0·035 |
| 20:4 | −0·093* | 0·038 | −0·095* | 0·038 | −0·091* | 0·038 | −0·126*** | 0·037 | −0·084 | 0·046 | −0·077 | 0·052 | −0·115** | 0·037 |
| 22:4 | −0·053 | 0·038 | −0·068 | 0·039 | −0·053 | 0·039 | −0·093* | 0·038 | −0·076 | 0·046 | −0·018 | 0·053 | −0·079* | 0·037 |
| 22:5 | 0·002 | 0·039 | −0·018 | 0·039 | −0·003 | 0·039 | −0·039 | 0·038 | −0·042 | 0·047 | 0·009 | 0·053 | −0·020 | 0·038 |
| 20:3/18:2 | 0·101** | 0·038 | 0·106** | 0·038 | 0·114** | 0·038 | 0·078* | 0·037 | 0·156*** | 0·046 | −0·028 | 0·053 | 0·079* | 0·037 |
| 20:4/20:3 | −0·388*** | 0·031 | −0·403*** | 0·031 | −0·408*** | 0·031 | −0·430*** | 0·030 | −0·362*** | 0·043 | −0·254*** | 0·050 | −0·414*** | 0·030 |
| 20:4/18:2 | −0·217*** | 0·038 | −0·224*** | 0·038 | −0·222*** | 0·038 | −0·268*** | 0·037 | −0·152** | 0·047 | −0·218*** | 0·052 | −0·254*** | 0·036 |
| n-3 | ||||||||||||||
| 18:3 | 0·039 | 0·037 | 0·018 | 0·038 | 0·018 | 0·037 | 0·037 | 0·036 | −0·003 | 0·045 | 0·021 | 0·051 | 0·041 | 0·036 |
| 20:5 | −0·048 | 0·037 | −0·046 | 0·037 | −0·040 | 0·038 | −0·041 | 0·036 | −0·079 | 0·045 | −0·000 | 0·051 | −0·039 | 0·036 |
| 22:5 | −0·002 | 0·038 | −0·019 | 0·038 | −0·008 | 0·038 | −0·018 | 0·037 | −0048 | 0·046 | −0010 | 0·052 | −0·019 | 0·037 |
| 22:6 | −0·086* | 0·038 | −0·097* | 0·038 | −0·086* | 0·038 | −0·116** | 0·037 | −0·095* | 0·046 | −0·075 | 0·052 | −0·108** | 0·037 |
| 20:5/18:3 | −0·070 | 0·037 | −0·053 | 0·037 | −0·047 | 0·038 | −0·063 | 0·036 | −0·065 | 0·045 | −0016 | 0·051 | −0·064 | 0·036 |
| n | 1938 | 1924 | 1943 | 1938 | 2003 | 1949 | 1943 | |||||||
|
FADS2
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2727271 |
rs174576 |
rs174578 |
rs174579 |
rs174602 |
rs498793 |
rs526126 |
||||||||
| Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | |
| n-6 | ||||||||||||||
| 18:2 | 0·218*** | 0·056 | 0·212*** | 0·036 | 0·210*** | 0·036 | 0·158*** | 0·041 | 0·136** | 0·043 | −0·020 | 0·036 | 0·047 | 0·045 |
| 18:3 | 0·053 | 0·061 | 0·048 | 0·039 | 0·056 | 0·038 | 0·029 | 0·044 | 0·048 | 0·046 | −0·049 | 0·038 | 0·066 | 0·048 |
| 20:2 | 0·191*** | 0·056 | 0·226*** | 0·036 | 0·226*** | 0·035 | 0·185*** | 0·041 | 0·130** | 0·042 | −0·036 | 0·036 | 0·047 | 0·045 |
| 20:3 | 0·185** | 0·057 | 0·247*** | 0·035 | 0·245*** | 0·035 | 0·242*** | 0·041 | 0·101* | 0·044 | −0·013 | 0·037 | 0·068 | 0·046 |
| 20:4 | −0·089 | 0·058 | −0·126*** | 0·037 | −0·115** | 0·037 | −0·093* | 0·042 | −0·059 | 0·044 | 0·096** | 0·037 | −0·116* | 0·046 |
| 22:4 | −0·042 | 0·059 | −0·092* | 0·038 | −0·079* | 0·037 | −0·079 | 0·042 | −0·051 | 0·044 | 0·048 | 0·037 | −0·071 | 0·046 |
| 22:5 | 0·018 | 0·059 | −0·051 | 0·038 | −0·035 | 0·038 | −0·041 | 0·043 | 0·014 | 0·045 | 0·033 | 0·038 | −0·047 | 0·047 |
| 20:3/18:2 | 0·011 | 0·059 | 0·077* | 0·037 | 0·078* | 0·037 | 0·115** | 0·042 | −0·007 | 0·045 | 0·003 | 0·038 | 0·030 | 0·047 |
| 20:4/20:3 | −0·311*** | 0·056 | −0·424*** | 0·030 | −0·409*** | 0·030 | −0·375*** | 0·039 | −0·184*** | 0·043 | 0·140*** | 0·037 | −0·224*** | 0·046 |
| 20:4/18:2 | −0·230*** | 0·059 | −0·264*** | 0·037 | −0·251*** | 0·036 | −0·195*** | 0·042 | −0·147** | 0·045 | 0·110** | 0·038 | −0·148** | 0·047 |
| n-3 | ||||||||||||||
| 18:3 | 0·044 | 0·057 | 0·026 | 0·037 | 0·031 | 0·036 | 0·001 | 0·041 | 0·043 | 0·043 | −0·072* | 0·036 | 0·020 | 0·045 |
| 20:5 | 0·014 | 0·057 | −0·043 | 0·037 | −0·043 | 0·036 | −0·071 | 0·041 | −0·028 | 0·043 | 0·014 | 0·036 | −0·061 | 0·045 |
| 22:5 | 0·065 | 0·058 | −0·023 | 0·037 | −0·019 | 0·037 | −0·064 | 0·042 | 0·008 | 0·043 | 0·008 | 0·037 | −0·023 | 0·046 |
| 22:6 | −0·060 | 0·058 | −0·120** | 0·037 | −0·115** | 0·036 | −0·110** | 0·042 | −0·035 | 0·044 | 0·057 | 0·037 | −0·099* | 0·046 |
| 20:5/18:3 | −0·021 | 0·057 | −0·056 | 0·037 | −0·059 | 0·036 | −0·061 | 0·041 | −0·056 | 0·043 | 0·066 | 0·036 | −0·067 | 0·045 |
| n | 1944 | 1911 | 1938 | 1931 | 1959 | 1923 | 1950 | |||||||
| Intergenic |
FADS3 | |||||
|---|---|---|---|---|---|---|
| rs174448 |
rs174449 |
rs174455 |
||||
| Coefficient | SE | Coefficient | SE | Coefficient | SE | |
| n-6 | ||||||
| 18:2 | 0·162*** | 0·035 | 0·163*** | 0·036 | 0·120*** | 0·036 |
| 18:3 | 0·083* | 0·038 | 0·081* | 0·038 | 0·077* | 0·039 |
| 20:2 | 0·155*** | 0·035 | 0·154*** | 0·035 | 0·137*** | 0·036 |
| 20:3 | 0·199*** | 0·035 | 0·201*** | 0·036 | 0·146*** | 0·037 |
| 20:4 | −0·058 | 0·036 | −0·060 | 0·037 | −0·073 | 0·037 |
| 22:4 | −0·021 | 0·037 | −0·023 | 0·037 | −0·027 | 0·038 |
| 22:5 | 0·011 | 0·037 | 0·010 | 0·038 | −0·002 | 0·038 |
| 20:3/18:2 | 0·070 | 0·037 | 0·071 | 0·037 | 0·050 | 0·038 |
| 20:4/20:3 | −0·285*** | 0·033 | −0·290*** | 0·034 | −0·248*** | 0·035 |
| 20:4/18:2 | −0·163*** | 0·036 | −0·165*** | 0·037 | −0·151*** | 0·038 |
| n-3 | ||||||
| 18:3 | 0·028 | 0·035 | 0·024 | 0·036 | 0·027 | 0·036 |
| 20:5 | −0·061 | 0·035 | −0·066 | 0·036 | −0·041 | 0·036 |
| 22:5 | −0·008 | 0·036 | −0·002 | 0·036 | 0·023 | 0·037 |
| 22:6 | −0·072* | 0·036 | −0·063 | 0·036 | −0·068 | 0·037 |
| 20:5/18:3 | −0·073* | 0·035 | −0·074* | 0·036 | −0·055 | 0·036 |
| n | 1966 | 1945 | 1956 | |||
P<0·05
P<0·01
P<0·001
Fatty acids have been standardised to have a variance of 1. Effect sizes are reported per copy of the minor allele.
Table 4.
Regression coefficients from analyses of log n-6 and n-3 fatty acid levels as a percentage of total fatty acid levels on child fatty acid desaturase (FADS) SNP adjusted for the maternal genotype†
(Regression coefficients with their standard errors)
|
FADS1
|
Intergenic |
FADS2
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs174548 |
rs174556 |
rs174561 |
rs3834458 |
rs968567 |
rs174570 |
rs174574 |
||||||||
| Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | |
| n-6 | ||||||||||||||
| 18:2 | 0·044 | 0·038 | 0·042 | 0·038 | 0·030 | 0·038 | 0·057 | 0·037 | −0·025 | 0·045 | 0·137* | 0·053 | 0·059 | 0·037 |
| 18:3 | 0·002 | 0·041 | 0·008 | 0·041 | 0·000 | 0·041 | −0·011 | 0·040 | 0·045 | 0·048 | −0·053 | 0·057 | −0·017 | 0·040 |
| 20:2 | 0·025 | 0·038 | 0·018 | 0·038 | 0·026 | 0·038 | 0·022 | 0·037 | −0·086 | 0·045 | 0·144** | 0·053 | 0·032 | 0·037 |
| 20:3 | 0·368*** | 0·037 | 0·379*** | 0·037 | 0·362*** | 0·037 | 0·371*** | 0·036 | 0·295*** | 0·045 | 0·238*** | 0·054 | 0·365*** | 0·036 |
| 20:4 | −0·219*** | 0·039 | −0·218*** | 0·039 | −0·220*** | 0·039 | −0·198*** | 0·038 | −0·122** | 0·047 | −0·212*** | 0·055 | −0·202*** | 0·038 |
| 22:4 | −0·156*** | 0·039 | −0·144*** | 0·040 | −0·155*** | 0·039 | −0·120** | 0·039 | −0·116* | 0·047 | −0·094 | 0·056 | −0·126** | 0·039 |
| 22:5 | −0·154*** | 0·040 | −0·136*** | 0·040 | −0·143*** | 0·040 | −0·124** | 0·039 | −0·009 | 0·048 | −0·212*** | 0·056 | −0·135*** | 0·039 |
| 20:3/18:2 | 0·330*** | 0·039 | 0·343*** | 0·039 | 0·336*** | 0·039 | 0·323*** | 0·038 | 0·313*** | 0·046 | 0·128* | 0·056 | 0·315*** | 0·038 |
| 20:4/20:3 | −0·674*** | 0·032 | −0·683*** | 0·032 | −0·670*** | 0·032 | −0·649*** | 0·031 | −0·470*** | 0·043 | −0·528*** | 0·053 | −0·648*** | 0·031 |
| 20:4/18:2 | −0·250*** | 0·039 | −0·247*** | 0·039 | −0·242*** | 0·039 | −0·237*** | 0·038 | −0·107* | 0·047 | −0·302*** | 0·055 | −0·242*** | 0·038 |
| n-3 | ||||||||||||||
| 18:3 | 0·090* | 0·038 | 0·106** | 0·039 | 0·104** | 0·038 | 0·093* | 0·037 | 0·048 | 0·046 | 0·136* | 0·054 | 0·077* | 0·038 |
| 20:5 | −0·004 | 0·038 | 0·005 | 0·038 | 0·001 | 0·038 | −0·006 | 0·038 | −0·010 | 0·045 | −0·001 | 0·054 | −0·013 | 0·038 |
| 22:5 | −0·017 | 0·039 | −0·003 | 0·039 | −0015 | 0·039 | 0·012 | 0·038 | −0·062 | 0·046 | 0·107* | 0·054 | 0·014 | 0·038 |
| 22:6 | −0·094* | 0·039 | −0·081* | 0·039 | −0·092* | 0·039 | −0·069 | 0·038 | −0·053 | 0·046 | −0·052 | 0·055 | −0·075* | 0·038 |
| 20:5/18:3 | −0·070 | 0·038 | −0·075 | 0·038 | −0·076* | 0·038 | −0·074* | 0·038 | −0·044 | 0·046 | −0·102 | 0·054 | −0·068 | 0·038 |
| n | 1938 | 1924 | 1943 | 1938 | 2003 | 1949 | 1943 | |||||||
| FADS2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2727271 |
rs174576 |
rs174578 |
rs174579 |
rs174602 |
rs498793 |
rs526126 |
||||||||
| Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | Coefficient | SE | |
| n-6 | ||||||||||||||
| 18:2 | 0·070 | 0·056 | 0·050 | 0·037 | 0·055 | 0·037 | 0·020 | 0·041 | 0·059 | 0·043 | 0·001 | 0·036 | 0·035 | 0·045 |
| 18:3 | −0·081 | 0·060 | −0·007 | 0·040 | −0·012 | 0·040 | 0·052 | 0·044 | −0·028 | 0·046 | 0·037 | 0·039 | −0·010 | 0·048 |
| 20:2 | 0·147** | 0·055 | 0·031 | 0·037 | 0·026 | 0·037 | −0·069 | 0·041 | 0·084* | 0·043 | −0·058 | 0·036 | 0·046 | 0·045 |
| 20:3 | 0·279*** | 0·057 | 0·370*** | 0·036 | 0·365*** | 0·036 | 0·259*** | 0·041 | 0·158*** | 0·044 | −0·088* | 0·037 | 0·146** | 0·046 |
| 20:4 | −0·204*** | 0·058 | −0·200*** | 0·038 | −0·204*** | 0·038 | −0·129** | 0·042 | −0·092* | 0·044 | 0·082* | 0·037 | −0·059 | 0·046 |
| 22:4 | −0·072 | 0·058 | −0·126** | 0·039 | −0·133*** | 0·039 | −0·112** | 0·042 | −0·039 | 0·044 | 0·058 | 0·037 | −0·096* | 0·046 |
| 22:5 | −0·204*** | 0·058 | −0·116** | 0·040 | −0·124** | 0·039 | −0·045 | 0·043 | −0·078 | 0·045 | 0·062 | 0·038 | −0·041 | 0·047 |
| 20:3/18:2 | 0·221*** | 0·058 | 0·327*** | 0·039 | 0·318*** | 0·038 | 0·241*** | 0·042 | 0·110* | 0·045 | −0·089* | 0·038 | 0·118* | 0·047 |
| 20:4/20:3 | −0·560*** | 0·055 | −0·651*** | 0·031 | −0·650*** | 0·031 | −0·441*** | 0·039 | −0·287*** | 0·044 | 0·201*** | 0·037 | −0·231*** | 0·046 |
| 20:4/18:2 | −0·251*** | 0·058 | −0·235*** | 0·038 | −0·241*** | 0·038 | −0·143*** | 0·042 | −0·131** | 0·045 | 0·082* | 0·038 | −0·082 | 0·047 |
| n-3 | ||||||||||||||
| 18:3 | 0·160** | 0·056 | 0·087* | 0·038 | 0·081* | 0·038 | 0·040 | 0·041 | −0·026 | 0·043 | −0·020 | 0·036 | −0·018 | 0·045 |
| 20:5 | 0·007 | 0·056 | −0·021 | 0·038 | −0·016 | 0·037 | −0·020 | 0·041 | −0·031 | 0·043 | 0·031 | 0·036 | −0·020 | 0·045 |
| 22:5 | 0·056 | 0·057 | 0·003 | 0·038 | 0·000 | 0·038 | −0·023 | 0·042 | −0·045 | 0·044 | 0·032 | 0·037 | −0·090* | 0·046 |
| 22:6 | −0·047 | 0·057 | −0·077* | 0·038 | −0·075* | 0·038 | −0·065 | 0·042 | −0·069 | 0·044 | 0·048 | 0·037 | −0·057 | 0·046 |
| 20:5/18:3 | −0·113* | 0·056 | −0·082* | 0·038 | −0·074* | 0·038 | −0·047 | 0·041 | −0·007 | 0·043 | 0·041 | 0·036 | −0·004 | 0·045 |
| n | 1944 | 1911 | 1938 | 1931 | 1959 | 1923 | 1950 | |||||||
| Intergenic |
FADS3
|
|||||
|---|---|---|---|---|---|---|
| rs174448 |
rs174449 |
rs174455 |
||||
| Coefficient | SE | Coefficient | SE | Coefficient | SE | |
| n-6 | ||||||
| 18:2 | 0·044 | 0·036 | 0·048 | 0·037 | 0·061 | 0·037 |
| 18:3 | −0·002 | 0·039 | −0·005 | 0·039 | −0·014 | 0·039 |
| 20:2 | 0·016 | 0·036 | 0·018 | 0·036 | 0·042 | 0·037 |
| 20:3 | 0·210*** | 0·036 | 0·214*** | 0·037 | 0·227*** | 0·037 |
| 20:4 | −0·156*** | 0·037 | −0·156*** | 0·038 | −0·116** | 0·038 |
| 22:4 | −0·137*** | 0·037 | −0·133*** | 0·038 | −0·088* | 0·038 |
| 22:5 | −0·149*** | 0·038 | −0·146*** | 0·039 | −0·108** | 0·039 |
| 20:3/18:2 | 0·174*** | 0·037 | 0·175*** | 0·038 | 0·177*** | 0·038 |
| 20:4/20:3 | −0·425*** | 0·034 | −0·429*** | 0·035 | −0·390*** | 0·035 |
| 20:4/18:2 | −0·186*** | 0·037 | −0·188*** | 0·038 | −0·156*** | 0·038 |
| n-3 | ||||||
| 18:3 | 0·070 | 0·036 | 0·077* | 0·037 | 0·064 | 0·037 |
| 20:5 | 0·010 | 0·036 | 0·008 | 0·037 | 0·026 | 0·037 |
| 22:5 | −0·033 | 0·037 | −0·031 | 0·037 | −0·008 | 0·037 |
| 22:6 | −0·067 | 0·037 | −0·065 | 0·037 | −0·028 | 0·037 |
| 20:5/18:3 | −0·043 | 0·036 | −0·050 | 0·037 | −0·025 | 0·037 |
| n | 1966 | 1945 | 1956 | |||
P<0·05
P<0·01
P<0·001
Fatty acids have been standardised to have a variance of 1. Effect sizes are reported per copy of the minor allele.
Predominant effect of child FADS genotypes on highly desaturated n-6 fatty acids
Associations of child genotypes with cord blood fatty acid levels adjusted for maternal genotype resulted in positive associations with the precursor PUFA 20 : 3n-6 for all analysed SNP (P values between 0·017 and 3·5 × 10− 24) (Table 4). Regression coefficients for this association were generally higher than in the maternal SNP analysis. In contrast to the maternal SNP results, we also found strong negative associations between minor alleles of the child SNP and the n-6 LC-PUFA 20 : 4n-6, 22 : 4n-6 and 22 : 5n-6 (P values between 0·038 and 1·7 × 10− 8 for 80 % of results), with 20 : 4n-6 exhibiting the highest regression coefficients of these three LC-PUFA. In the initial unadjusted analysis, the genetically explained variances of 20 : 3n-6, 20 : 4n-6, 22 : 4n-6 and 22 : 5n-6 were higher for the child than for the maternal SNP (Table 2), which might also reflect the predominant influence of the child genotype on the levels of these fatty acids. The PUFA 18 : 3n-6 was not significantly associated with child FADS genotypes, and 18 : 2n-6 and 20 : 2n-6 showed only sporadic associations. Again, effect sizes of the ratio associations were slightly attenuated when the offspring genotype was adjusted for the maternal genotype (see online Supplementary Tables 3 and 4).
Associations with n-3 long-chain-PUFA in the multivariable analysis
By adjusting the maternal genotype effect for the child genotype, negative associations were observed for the n-3 LC-PUFA DHA (P < 0·046) for eleven SNP. Amounts of 22 : 5n-6, 18 : 3n-3, 20 : 5n-3 and 22 : 5n-3 were not associated with any of the tested maternal SNP while 18 : 3n-6 was only associated with one SNP (Table 3). In the vice versa analysis (child genotype effect adjusted for maternal genotype), negative associations with DHA were also observed for six SNP (P < 0·0499) with a comparable regression coefficient to the maternal SNP analysis (Table 4). Additionally, in contrast to the results for the maternal genotype effect, ten SNP were associated with amounts of α-linolenic acid (18 : 3n-3) (P < 0·042), with minor allele carriers exhibiting higher levels of this fatty acid. This was also reflected in the higher genetically explained variance of 18 : 3n-3 for the seventeen child SNP compared with the maternal SNP (0·52 v. 0·29 %) (Table 2). Similar to the maternal SNP analysis, the n-3 fatty acids 20 : 5n-3 and 22 : 5n-3 were rarely associated with child FADS genotypes (6 % of results). The n-3 ratio 20 : 5/18 : 3 was hardly associated with any SNP when the maternal genotype was adjusted for the offspring genotype and vice versa (12 and 29 % of the results, respectively). Overall, the seventeen SNP had lower R 2 with all of the n-3 fatty acids than with any of the n-6 fatty acids except 18 : 3n-6 (see Table 2).
Haplotype analysis
To further investigate the genetic associations, haplotypes were generated from the seventeen SNP for mother and offspring separately. Analysis of haplotype blocks identified three regions of high LD (see online Supplementary Fig. 2). Due to the high recombination levels between the first two regions (D’ = 0·96), these regions were combined. Haplotypes were constructed for the associated ten SNP and were restricted to data with probabilities >0·9. rs174570, although within the region rs174548 to rs174579, was left unspecified due to its relatively low LD with other SNP. A total of five haplotypes (frequency >1 %) were derived for this region with about 3 % of the genetic variation associated with rare haplotypes (see online Supplementary Table 5). Of those three, the haplotype representing all the major alleles (Hap1) generally showed the strongest association with fatty acids (see online Supplementary Tables 6 and 7). Multivariate tests across the fifteen outcomes for each of the five minor haplotypes suggested that all haplotypes contributed to the explanation of outcomes (P < 0·001). To explore their associations further, equality of regression coefficients for the minor haplotypes was tested. The main differences between haplotypes related to the n-6 fatty acids with two or three unsaturated bonds and the ratios derived from these fatty acids, although differences were also observed for offspring haplotypes on 22 : 5n-6 (Hap1 column; see online Supplementary Table 10). If their effects were equal, it implied that Hap1 could serve as a proxy for these haplotypes due to their linear dependency. Even where differences between the minor haplotypes existed, the contribution was < 1 % over and above that of Hap1 (difference R 2 for Max and Hap1 columns; see online Supplementary Table 10). Subsequent models explored the relative importance of the minor haplotypes. Hap5, Hap4 and then Hap3 made decreasing contributions to the explanation of outcomes (see online Supplementary Table 10). Statistically, Hap1 could be used to reflect Hap2 and Hap.rare (and Hap3 for maternal haplotypes) although parsimony may not be the over-riding goal. These rare haplotypes may reflect distinct effects that the present study was underpowered to detect.
Although the minor haplotypes made only little contribution to the explanation of fatty acid levels, comparison between Hap3 and Hap5 might suggest a specific role of SNP rs968567 for fatty acid metabolism. Specifically, Hap3 and Hap5 differ only in the presence or absence of the rs968567 minor allele (see online Supplementary Table 5). Maternal Hap3 showed hardly any significant associations with fatty acids (which, however, may have been due to a lack of power with the haplotype frequency being 1·6 %), whereas maternal Hap5 (which contains the minor allele of rs968567) was significantly associated with various fatty acids including 22 : 6n-3 and fatty acid ratios. However, none of these differences was statistically significant. For offspring haplotypes, differences between Hap3 and Hap5 were observed for 18 : 2n-6 (P = 0·003), 20 : 5n-6 (P = 0·010) and 20 : 4/18 : 2 (P = 0·005), with genetic effects being reduced in Hap5.
Also, SNP rs498793 was atypical due to its negative LD with other SNP. To further explore this variant, haplotypes were constructed for all seventeen SNP. Due to the low LD between some SNP and the large number of SNP, a plethora of haplotypes was generated, with some individuals having many possible alternatives. A weaker criterion of haplotype assignment, probability >0·5, was chosen to increase the number of individuals in subsequent fatty acid analyses. Over 300 rare haplotypes were identified representing about 26 % of the genetic variation (see online Supplementary Table 8). Only two haplotypes had frequencies above 5 % – one involving major alleles for all SNP and the other with the minor allele for rs498793 only. This latter haplotype, reflecting the negative LD between rs498793 and other SNP, showed much stronger results than its associated SNP (compare Tables 3 and 4 with online Supplementary Table 9).
Multiple comparisons
In the univariable analyses, hypotheses relating to seventeen genetic variants by mother and offspring for the eleven fatty acids were being tested. Due to their derivation from these data, the results from fatty acid ratios and haplotypes were excluded from this analysis. Of these 374 comparisons, 132 had nominal P>0·05, 242 had P < 0·05, 203 had P < 0·01, 173 had P < 0·001 and 140 had P < 0·0001. This distribution of the observed P values differed markedly from that expected if all 374 comparisons were null. In particular, 65 % of observed P values were less than the 5 % significance level compared with 5 % (by definition) for the null scenario. Even using the overly conservative Bonferroni correction, 38 % of tests would have remained significant. A more powerful method adjusting for multiple comparisons, the false discovery rate( 50 ), suggested that the adjusted 5 % critical P value was 0·0302. Only nineteen of the 242 nominally significant P values failed to satisfy this adjusted critical value. The minor adjustment to the nominal value of 0·05 reflected the early divergence in the distributions of observed and expected P values (see online Supplementary Fig. 1).
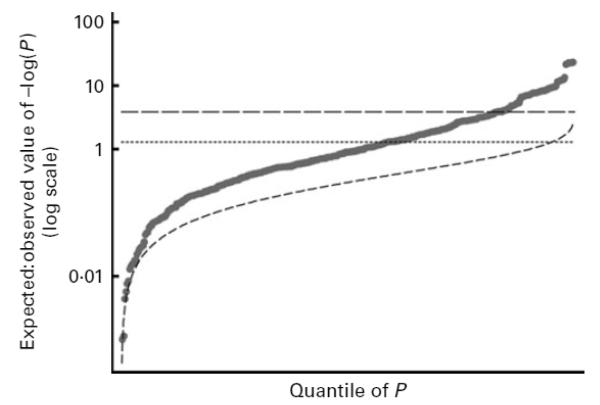
A similar pattern emerged for multivariable analyses when mother–offspring variants were jointly assessed (see Fig. 1). As expected, P values were higher in these analyses partly due to the correlation between mother–offspring genotypes introduced by inheritance. Nevertheless, 159 (41 %) results had P < 0·05, with 121 having P < 0·0163 (false discovery rate criterion), eighty with P < 0·001, and sixty-three (17 %) with P < 0·000134 (Bonferroni criterion).
Fig. 1.
Q-Q plot of 374 multivariable genetic comparisons (11 fatty acid outcomes × 17 genetic variants × mother/offspring) reported in Tables 3 and 4. The graph shows the distribution of the observed ranked P values (●; least significant to the most highly significant) and the expected distribution if all 374 comparisons were null (––). At the 5% level (----), 154 (41 %) results were significant while, using the Bonferroni adjustment (---), sixty-three (17%) were significant. The false discovery rate (FDR) criterion will always lie between these two extremes, with its precise value depending on the observed distribution. For these data, the FDR criterion was calculated as 0-0163.
In the present study it was shown that concentrations of cord venous plasma fatty acids are dependent on both the maternal and child genotype of selected polymorphisms in the FADS gene cluster. Maternal genotypes mainly influenced the concentrations of less desaturated n-6 fatty acids (18 : 2n-6, 20 : 2n-6 and 20 : 3n-6), whereas child genotypes had a predominant effect on more highly desaturated n-6 fatty acids (20 : 4n-6, 22 : 4n-6 and 22 : 5n-6). The concentrations of DHA were dependent on both the maternal and child genotype, whereas the amounts of α-linolenic acid were only associated with the child genotypes.
Effect of FADS SNP and haplotypes on cord venous plasma fatty acids and fatty acid ratios
An effect of FADS genotypes on the composition of LC-PUFA in various tissues including serum, plasma, erythrocyte phospholipids, adipose tissue and breast milk has been shown in numerous previous studies( 11 – 22 ). To our knowledge, the present study is the first to investigate the effect of maternal as well as child genotypes on cord venous plasma fatty acid composition. In our analysis, we generally observed increased levels of the direct precursors of the Δ-5 desaturase and Δ-6 desaturase reactions and lower levels of direct or derived products of the respective desaturase reactions in carriers of minor alleles, which is in accordance with the results from previous studies in other tissues.
Also the direction of SNP associations with specific ratios was generally as expected. The n-6 ratio 20 : 4/20 : 3, which is a direct estimate of the Δ-5 desaturase reaction, decreased significantly per copy of the minor allele and per copy of those haplotypes that contained minor alleles of the analysed SNP (except rs498793), which is in line with the general assumption that minor allele carriers exhibit lower Δ-5 desaturase activity. Bokor et al. ( 19 ) reported recently that the FADS2 promoter SNP rs968567 was the only SNP in their study that was associated with increased serum phospholipid Δ-6 desaturase ratios (20 : 3n-6/18 : 2n-6) in carriers of minor alleles. In the present study, the minor alleles of all analysed SNP were associated with an increase of the 20 : 3n-6/18 : 2n-6 ratio. Bokor et al. ( 19 ) concluded in their study that the increase of the 20 : 3n-6/18 : 2n-6 ratio might be a consequence of an increase in Δ-6 desaturase activity due to the minor allele of rs968567, which agrees with an earlier in vitro study on the effect of this polymorphism on FADS2 promoter activity( 51 ). In our present study, we found this effect also for all other analysed SNP in addition to rs968567. Whether the minor alleles of the investigated FADS polymorphisms really lead to an increase in Δ-6 desaturase activity, or whether the increase of the Δ-6 desaturase ratio is rather due to the massive accumulation of 20 : 3n-6 resulting from a decrease in Δ-5 desaturase activity can hardly be judged from the present data. A better estimate of Δ-6 desaturase activity would be the 18 : 3n-6/18 : 2n-6 ratio because 18 : 3n-6 is the immediate product of the Δ-6 desaturase reaction. Due to the very low concentration of 18 : 3n-6, which is very rapidly elongated to 20 : 3n-6, the precision of assessing the 18 : 3n-6/18 : 2n-6 ratio is rather limited. Further studies are therefore needed to analyse the particular effect of FADS minor alleles on desaturase activities.
In general, the contribution of FADS genetic variants to fatty acid concentrations in cord venous plasma was apparently rather low (for example, 1·86 % of the amount of AA in cord venous plasma could be explained by the maternal FADS genotype and 2·99 % by the offspring genotype). In the adjusted analysis (including the effects of all potential confounders such as maternal diet), 10·41 % of AA variability could be explained when the maternal genotype was included and 9·00 % could be explained when the offspring genotype was included, suggesting that factors such as maternal diet, age, smoking, etc. add more to the effect on fatty acid levels than the genetic background. However, the genetically explained variability of the 20 : 4n-6/20 : 3n-6 ratio, which reflects Δ-5 desaturase activity, was >20 % for maternal genotypes and >30 % for offspring genotypes and increased only slightly when confounders were included (Table 2). Usage of fatty acid concentration ratios as proxies for enzymic reaction rates has earlier already been shown to reduce variance and yield robust statistical associations with very high genetically explained variabilities of the respective ratios. In the study by Illig et al. ( 52 ) a SNP in the FADS1 gene explained more than 30 % of variability of the serum phosphatidylcholine (PC) ratio PC aa C36 : 3/PC aa C36 : 4, which is also an estimate of the Δ-5 desaturase reaction( 52 ). The present study confirms these previous findings and suggests a strong genetic effect on Δ-5 desaturase activity.
Evidence for fetal capacity of synthesising long-chain-PUFA
The fact that maternal genotypes were mainly associated with the less desaturated n-6 fatty acids and the child genotypes were mainly associated with the more highly desaturated n-6 fatty acids in our multivariable analysis suggests that the fetus might be able to synthesise n-6 LC-PUFA by itself to a certain extent. The current view of fetal fatty acid accumulation during pregnancy is that it mainly takes place through preferential placental transfer of LC-PUFA from the mother to the child( 24 ). At least for DHA, this view is supported by various tracer studies on placental fatty acid transfer, which detected increased DHA accumulation in the placenta( 53 ) as well as preferential materno–fetal transfer across the placenta after tracer administration( 54 , 55 ). For the n-6 fatty acids, data are more scarce, but suggest lower placental transfer selectivity compared with DHA( 56 ). Furthermore, it has been shown that desaturation capacity in the human fetus and newborn is apparently lower than in adult humans and other species, for example, in rat liver microsomes( 57 , 58 ), although the required desaturation enzymes for PUFA conversion are present and active in the fetal and newborn liver( 59 – 62 ). Our data suggest a significant involvement of the fetal metabolism in the synthesis of n-6 LC-PUFA, because associations with these fatty acids were only seen for the child genotypes. Actually, this result is supported by a couple of other studies on LC-PUFA synthesis in preterm infants, showing that these infants are capable of synthesising LC-PUFA from their 18C precursors at the time they should be in utero ( 63 – 65 ). Additionally, studies in fetal rat liver suggested that the rat fetus is increasingly capable of synthesising 20 : 4n-6 with advancing length of gestation, and near term relies to a lesser extent on provision of 20 : 4n-6 from the maternal organism( 66 ). Rat liver Δ-5 desaturase activity increased 3-fold up to term, whereas the activity in the maternal rat liver decreased 2–3 times. These data combined with the experimental data reported in the present paper strongly support that it is very likely that the fetus is capable of synthesising LC-PUFA at a significant rate. Whether the fetus acquires this ability in the course of gestation or whether fetal fatty acid synthesis takes place also in the first trimesters of pregnancy cannot be inferred from the present study. The observation that the content of DHA in cord plasma was dependent on both maternal and child genotypes in the present study is in concordance with the results of previous studies: on the one hand there is preferential transfer of DHA from the mother to the fetus during pregnancy( 57 – 59 ), and on the other hand the fetus is capable of synthesising additional DHA by itself( 63 – 65 ), which might be necessary to satisfy the high fetal demand of DHA during pregnancy.
Haplotype analyses might suggest specific SNP effects
In general, the associated SNP showed consistent effects on fatty acid levels. The SNP rs498793, however, which is in negative LD with the other SNP reflecting an excess of the trans compared with the cis configurations of minor alleles in double heterozygotes (D’ < −0·35; P < 6·4 × 10− 45), showed opposite effects as already reported by Bokor et al. ( 19 ). For example, AA levels were elevated in carriers of the minor allele of this SNP, whereas they were reduced in minor allele carriers of all other SNP. We also obtained similar results in our previous study on maternal FADS genotypes and maternal erythrocyte fatty acid levels( 21 ). The isolated effect of this SNP, which is located in an intronic FADS2 region, might be interesting for future functional investigations. The presence of the minor allele of this SNP on the second most common haplotype (Hap3; frequency 17 %), which produced larger effect sizes than the associated SNP, supports the special role of this SNP.
Additionally, regression coefficients of haplotypes that were generated from a region of high LD (rs174548 to rs174579) might suggest a specific effect of SNP rs968567. Hap3 and Hap5 differ only in the presence or absence of the rs968567 minor allele (see online Supplementary Table 5) and clear differences on fatty acid levels were detected between these two haplotypes. Although Hap3 occurred only at a frequency of 1·6 % (which might explain some of the non-significant results compared with Hap5), these results might also suggest a specific effect of the minor allele for SNP rs968567 on fatty acid metabolism. This is supported by our previous in vitro study on functional effects of this SNP on FADS2 promoter activity and transcription factor binding( 51 ). Further studies are needed to decipher the exact effect of this SNP on desaturase activity.
Possible explanations for weak or lack of associations with specific fatty acids
It merits mentioning that cord blood 18 : 2n-6 amounts were nearly seventy times greater than the 18 : 3n-3 amount (9·74 v. 0·14 %). This high abundance may have made an impact on the conversion of 18 : 3n-3 to 20 : 5n-3 and 22 : 6n-3, which possibly explains the weaker associations observed for these fatty acids. Another reason for weaker associations with n-3 fatty acids might be that supply with these fatty acids is more dependent on nutritional influences than the n-6 fatty acids, which are more dependent on endogenous synthesis.
The lack of associations between maternal genotypes and some fatty acids, particularly the more highly desaturated n-6 fatty acids, in cord blood multivariable analyses, compared with strong associations being observed in the maternal circulation( 21 ), may have different possible explanations. Fetal demand might be so high that maternal supply becomes exhausted, with associations consequentially only reflecting the fetal metabolism. Where maternal associations are strong, particularly for the less desaturated n-6 fatty acids, this may suggest an abundant supply from the mother. It is perhaps to be expected that the fetal metabolism will only operate to overcome any deficiency in the supply so that offspring genetic associations will tend to occur where there is less evidence of maternal genetic associations and vice versa. It is interesting that the fatty acids 20 : 3n-6 and 20 : 4n-6 reflected a more equal contribution from maternal supply and fetal metabolism than fatty acids at either extreme of the metabolic pathway.
An alternative explanation for the lack of maternal associations may involve the placental transfer not occurring at a fixed rate but related to maternal levels or materno–fetal gradients. Such compensatory mechanisms might mask the effects of genetic variation in the maternal circulation. Whether placental transfer mechanisms would not only preferentially transport specific fatty acids to the fetus but might also be able to increase transfer rates at low maternal levels of specific fatty acids is currently unknown and requires further investigation.
Study limitation
One of the limitations of the present study might be that we determined fatty acid amounts in umbilical cord vein plasma, which represents the part of the maternal–fetal circulation system that comes from the maternal circulation and goes to the child via the placenta and umbilical cord. In contrast, arterial blood flows from the child back to the placenta, and might therefore represent the fetal status better than the venous blood. Indeed there is evidence to support this view, for example, AA can flow from the fetal circulation back to the mother( 25 ). Such effects will weaken the fetal genetic signal in venous blood compared with arterial blood. However, it would seem unlikely that such processes could totally eliminate the fetal signal; rather, there might be an underestimation of the effect of the fetal metabolism. However, it has been shown in a couple of studies that the fatty acid composition does not differ significantly between arterial and venous cord blood( 67 , 68 ). These studies would suggest that these effects are relatively minor when one analyses composition as a percentage of total fatty acids rather than concentrations. Therefore, our overall conclusion that fetal metabolism is of influence should be correct.
Conclusion
The present study is the first to analyse the influence of maternal and offspring FADS polymorphisms on venous cord plasma fatty acid levels in a large birth cohort. We conclude that FADS genotypes significantly contribute to determining cord blood n-6 and n-3 LC-PUFA composition. Fetal metabolism seems important for neonatal supply of n-6 LC-PUFA but to a lower extent for neonatal supply of n-3 LC-PUFA. Two SNP in the FADS2 gene might have specific effects on fatty acid levels as suggested by haplotype analysis.
Supplementary Material
Abbreviations
- AA
arachidonic acid
- ALSPAC
Avon Longitudinal Study of Parents and Children
- FADS
fatty acid desaturase
- LC-PUFA
long-chain PUFA
- LD
linkage disequilibrium
References
- 1.Lattka E, Illig T, Heinrich J, et al. Do FADS genotypes enhance our knowledge about fatty acid related phenotypes? Clin Nutr. 2010;29:277–287. doi: 10.1016/j.clnu.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Lattka E, Illig T, Heinrich J, et al. FADS gene cluster polymorphisms: important modulators of fatty acid levels and their impact on atopic diseases. J Nutrigenet Nutrigenomics. 2009;2:119–128. doi: 10.1159/000235559. [DOI] [PubMed] [Google Scholar]
- 3.Chapkin RS, Kim W, Lupton JR, et al. Dietary docosahexaenoic and eicosapentaenoic acid: emerging mediators of inflammation. Prostaglandins Leukot Essent Fatty Acids. 2009;81:187–191. doi: 10.1016/j.plefa.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavie CJ, Milani RV, Mehra MR, et al. Omega-3 polyunsaturated fatty acids and cardiovascular diseases. J Am Coll Cardiol. 2009;54:585–594. doi: 10.1016/j.jacc.2009.02.084. [DOI] [PubMed] [Google Scholar]
- 5.Salem N, Jr, Niebylski CD. The nervous system has an absolute molecular species requirement for proper function. Mol Membr Biol. 1995;12:131–134. doi: 10.3109/09687689509038508. [DOI] [PubMed] [Google Scholar]
- 6.Serhan CN, Yacoubian S, Yang R. Anti-inflammatory and proresolving lipid mediators. Annu Rev Pathol. 2008;3:279–312. doi: 10.1146/annurev.pathmechdis.3.121806.151409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sprecher H. Biochemistry of essential fatty acids. Prog Lipid Res. 1981;20:13–22. doi: 10.1016/0163-7827(81)90009-6. [DOI] [PubMed] [Google Scholar]
- 8.Sprecher H, Luthria DL, Mohammed BS, et al. Reevaluation of the pathways for the biosynthesis of polyunsaturated fatty acids. J Lipid Res. 1995;36:2471–2477. [PubMed] [Google Scholar]
- 9.Nakamura MT, Nara TY. Structure, function, and dietary regulation of D6, D5, and D9 desaturases. Annu Rev Nutr. 2004;24:345–376. doi: 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- 10.Lattka E, Illig T, Koletzko B, et al. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol. 2010;21:64–69. doi: 10.1097/MOL.0b013e3283327ca8. [DOI] [PubMed] [Google Scholar]
- 11.Schaeffer L, Gohlke H, Müller M, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 12.Malerba G, Schaeffer L, Xumerle L, et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43:289–299. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- 13.Rzehak P, Heinrich J, Klopp N, et al. Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. Br J Nutr. 2009;101:20–26. doi: 10.1017/S0007114508992564. [DOI] [PubMed] [Google Scholar]
- 14.Baylin A, Ruiz-Narvaez E, Kraft P, et al. a-Linolenic acid, D6-desaturase gene polymorphism, and the risk of nonfatal myocardial infarction. Am J Clin Nutr. 2007;85:554–560. doi: 10.1093/ajcn/85.2.554. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka T, Shen J, Abecasis GR, et al. Genome-wideassociation study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5:e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie L, Innis SM. Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n-6) and (n-3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. J Nutr. 2008;138:2222–2228. doi: 10.3945/jn.108.096156. [DOI] [PubMed] [Google Scholar]
- 17.Molto-Puigmarti C, Plat J, Mensink RP, et al. FADS1 FADS2 gene variants modify the association between fish intake and the docosahexaenoic acid proportions in human milk. Am J Clin Nutr. 2010;91:1368–1376. doi: 10.3945/ajcn.2009.28789. [DOI] [PubMed] [Google Scholar]
- 18.Mathias RA, Vergara C, Gao L, et al. FADS genetic variants and v-6 polyunsaturated fatty acid metabolism in a homogenous island population. J Lipid Res. 2010;51:2766–2774. doi: 10.1194/jlr.M008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bokor S, Dumont J, Spinneker A, et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010;51:2325–2333. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rzehak P, Thijs C, Standl M, et al. Variants of the FADS1 FADS2 gene cluster, blood levels of polyunsaturated fatty acids and eczema in children within the first 2 years of life. PLoS One. 2010;5:e13261. doi: 10.1371/journal.pone.0013261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koletzko B, Lattka E, Zeilinger S, et al. Genetic variants of the fatty acid desaturase gene cluster predict amounts of red blood cell docosahexaenoic and other polyunsaturated fatty acids in pregnant women: findings from the Avon Longitudinal Study of Parents and Children. Am J Clin Nutr. 2011;93:211–219. doi: 10.3945/ajcn.110.006189. [DOI] [PubMed] [Google Scholar]
- 22.Lattka E, Rzehak P, Szabo E, et al. Genetic variants in the FADS gene cluster are associated with arachidonic acid concentrations of human breast milk at 1·5 and 6 mo postpartum and influence the course of milk dodecanoic, tetracosenoic, and trans-9-octadecenoic acid concentrations over the duration of lactation. Am J Clin Nutr. 2011;93:382–391. doi: 10.3945/ajcn.110.004515. [DOI] [PubMed] [Google Scholar]
- 23.Koletzko B, Lien E, Agostoni C, et al. The roles of long-chain polyunsaturated fatty acids in pregnancy, lactation and infancy: review of current knowledge and consensus recommendations. J Perinat Med. 2008;36:5–14. doi: 10.1515/JPM.2008.001. [DOI] [PubMed] [Google Scholar]
- 24.Hanebutt FL, Demmelmair H, Schiessl B, et al. Longchain polyunsaturated fatty acid (LC-PUFA) transfer across the placenta. Clin Nutr. 2008;27:685–693. doi: 10.1016/j.clnu.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Hendrickse W, Stammers JP, Hull D. The transfer of free fatty acids across the human placenta. Br J Obstet Gynaecol. 1985;92:945–952. doi: 10.1111/j.1471-0528.1985.tb03075.x. [DOI] [PubMed] [Google Scholar]
- 26.Krauss-Etschmann S, Shadid R, Campoy C, et al. Effects of fish-oil and folate supplementation of pregnant women on maternal and fetal plasma concentrations of docosahexaenoic acid and eicosapentaenoic acid: a European randomized multicenter trial. Am J Clin Nutr. 2007;85:1392–1400. doi: 10.1093/ajcn/85.5.1392. [DOI] [PubMed] [Google Scholar]
- 27.Dunstan JA, Mori TA, Barden A, et al. Effects of n-3 polyunsaturated fatty acid supplementation in pregnancy on maternal and fetal erythrocyte fatty acid composition. Eur J Clin Nutr. 2004;58:429–437. doi: 10.1038/sj.ejcn.1601825. [DOI] [PubMed] [Google Scholar]
- 28.Manku MS, Horrobin DF, Morse N, et al. Reduced levels of prostaglandin precursors in the blood of atopic patients: defective delta-6-desaturase function as a biochemical basis for atopy. Prostaglandins Leukot Med. 1982;9:615–628. doi: 10.1016/0262-1746(82)90019-1. [DOI] [PubMed] [Google Scholar]
- 29.Wright S. Essential fatty acids and atopic dermatitis. Pediatr Allergy Immunol. 1991;71:224–228. [Google Scholar]
- 30.Leichsenring M, Kochsiek U, Paul K. (n-6)-Fatty acids in plasma lipids of children with atopic bronchial asthma. Pediatr Allergy Immunol. 1995;6:209–212. doi: 10.1111/j.1399-3038.1995.tb00287.x. [DOI] [PubMed] [Google Scholar]
- 31.Rocklin RE, Thistle L, Gallant L, et al. Altered arachidonic acid content in polymorphonuclear and mononuclear cells from patients with allergic rhinitis and/or asthma. Lipids. 1986;21:17–20. doi: 10.1007/BF02534296. [DOI] [PubMed] [Google Scholar]
- 32.Galli E, Picardo M, Chini L, et al. Analysis of polyunsaturated fatty acids in newborn sera: a screening tool for atopic disease? Br J Dermatol. 1994;130:752–756. doi: 10.1111/j.1365-2133.1994.tb03413.x. [DOI] [PubMed] [Google Scholar]
- 33.Yu G, Kjellman NI, Bjorksten B. Phospholipid fatty acids in cord blood: family history and development of allergy. Acta Paediatr. 1996;85:679–683. doi: 10.1111/j.1651-2227.1996.tb14124.x. [DOI] [PubMed] [Google Scholar]
- 34.Beck M, Zelczak G, Lentze MJ. Abnormal fatty acid composition in umbilical cord blood of infants at high risk of atopic disease. Acta Paediatr. 2000;89:279–284. [PubMed] [Google Scholar]
- 35.Newson RB, Shaheen SO, Henderson AJ, et al. Umbilical cord and maternal blood red cell fatty acids and early childhood wheezing and eczema. J Allergy Clin Immunol. 2004;114:531–537. doi: 10.1016/j.jaci.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Dunstan JA, Simmer K, Dixon G, et al. Cognitive assessment of children at age 2(1/2) years after maternal fish oil supplementation in pregnancy: a randomised controlled trial. Arch Dis Child Fetal Neonatal Ed. 2008;93:F45–F50. doi: 10.1136/adc.2006.099085. [DOI] [PubMed] [Google Scholar]
- 37.Escolano-Margarit MV, Ramos R, Beyer J, et al. Prenatal DHA status and neurological outcome in children at age 5·5 years are positively associated. J Nutr. 2011;141:1216–1223. doi: 10.3945/jn.110.129635. [DOI] [PubMed] [Google Scholar]
- 38.Donahue SM, Rifas-Shiman SL, Gold DR, et al. Prenatal fatty acid status and child adiposity at age 3 y: results from a US pregnancy cohort. Am J Clin Nutr. 2011;93:780–788. doi: 10.3945/ajcn.110.005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Golding J, Pembrey M, Jones R. ALSPAC – the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 40.Lepage G, Roy CC. Direct transesterification of all classes of lipids in a one-step reaction. J Lipid Res. 1986;27:114–120. [PubMed] [Google Scholar]
- 41.Masood MA, Salem N., Jr High-throughput analysis of plasma fatty acid methyl esters employing robotic transesterification and fast gas chromatography. Lipids. 2008;43:171–180. doi: 10.1007/s11745-007-3130-9. [DOI] [PubMed] [Google Scholar]
- 42.Jones RW, Ring S, Tyfield L, et al. A new human genetic resource: a DNA bank established as part of the Avon Longitudinal Study of Pregnancy and Childhood (ALSPAC) Eur J Hum Genet. 2000;8:653–660. doi: 10.1038/sj.ejhg.5200502. [DOI] [PubMed] [Google Scholar]
- 43.Davey SG, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 44.Weiler H, Fitzpatrick-Wong S, Schellenberg J, et al. Maternal and cord blood long-chain polyunsaturated fatty acids are predictive of bone mass at birth in healthy termborn infants. Pediatr Res. 2005;58:1254–1258. doi: 10.1203/01.pdr.0000185129.73971.74. [DOI] [PubMed] [Google Scholar]
- 45.Sabel KG, Lundqvist-Persson C, Bona E, et al. Fatty acid patterns early after premature birth, simultaneously analysed in mothers’ food, breast milk and serum phospholipids of mothers and infants. Lipids Health Dis. 2009;8:20. doi: 10.1186/1476-511X-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kilari AS, Mehendale SS, Dangat KD, et al. Long chain polyunsaturated fatty acids in mothers of preterm babies. J Perinat Med. 2010;38:659–664. doi: 10.1515/jpm.2010.112. [DOI] [PubMed] [Google Scholar]
- 47.Berghaus TM, Demmelmair H, Koletzko B. Essential fatty acids and their long-chain polyunsaturated metabolites in maternal and cord plasma triglycerides during late gestation. Biol Neonate. 2000;77:96–100. doi: 10.1159/000014201. [DOI] [PubMed] [Google Scholar]
- 48.Ruyle M, Connor WE, Anderson GJ, et al. Placental transfer of essential fatty acids in humans: venous-arterial difference for docosahexaenoic acid in fetal umbilical erythrocytes. Proc Natl Acad Sci U S A. 1990;87:7902–7906. doi: 10.1073/pnas.87.20.7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pembrey M. The Avon Longitudinal Study of Parents and Children (ALSPAC): a resource for genetic epidemiology. Eur J Endocrinol. 2004;151(Suppl. 3):U125–U129. doi: 10.1530/eje.0.151u125. [DOI] [PubMed] [Google Scholar]
- 50.Benjamini Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J Royal Statist Soc Series B – Methodological. 1995;57:289–300. [Google Scholar]
- 51.Lattka E, Eggers S, Moeller G, et al. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J Lipid Res. 2010;51:182–191. doi: 10.1194/jlr.M900289-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Illig T, Gieger C, Zhai G, et al. A genomewide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larque E, Demmelmair H, Berger B, et al. In vivo investigation of the placental transfer of (13)C-labeled fatty acids in humans. J Lipid Res. 2003;44:49–55. doi: 10.1194/jlr.m200067-jlr200. [DOI] [PubMed] [Google Scholar]
- 54.Haggarty P, Page K, Abramovich DR, et al. Long-chain polyunsaturated fatty acid transport across the perfused human placenta. Placenta. 1997;18:635–642. doi: 10.1016/s0143-4004(97)90004-7. [DOI] [PubMed] [Google Scholar]
- 55.Gil-Sanchez A, Larque E, Demmelmair H, et al. Maternal-fetal in vivo transfer of [13C]docosahexaenoic and other fatty acids across the human placenta 12 h aftermaternal oral intake. Am J Clin Nutr. 2010;92:115–122. doi: 10.3945/ajcn.2010.29589. [DOI] [PubMed] [Google Scholar]
- 56.Haggarty P, Ashton J, Joynson M, et al. Effect of maternal polyunsaturated fatty acid concentration on transport by the human placenta. Biol Neonate. 1999;75:350–359. doi: 10.1159/000014115. [DOI] [PubMed] [Google Scholar]
- 57.Hrelia S, Celadon M, Rossi CA, et al. Delta-6-desaturation of linoleic and alpha-linolenic acids in aged rats: a kinetic analysis. Biochem Int. 1990;22:659–667. [PubMed] [Google Scholar]
- 58.Yamaoka K, Okayasu T, Ishibashi T. Product identification and determination of optimal condition of 8, 11, 14-eicosatrienoic acid desaturation in rat liver microsomes. Hokkaido Igaku Zasshi. 1986;61:755–765. [PubMed] [Google Scholar]
- 59.Chambaz J, Ravel D, Manier MC, et al. Essential fatty acids interconversion in the human fetal liver. Biol Neonate. 1985;47:136–140. doi: 10.1159/000242104. [DOI] [PubMed] [Google Scholar]
- 60.Poisson JP, Dupuy RP, Sarda P, et al. Evidence that liver microsomes of human neonates desaturate essential fatty acids. Biochim Biophys Acta. 1993;1167:109–113. doi: 10.1016/0005-2760(93)90149-4. [DOI] [PubMed] [Google Scholar]
- 61.Rodriguez A, Sarda P, Nessmann C, et al. Fatty acid desaturase activities and polyunsaturated fatty acid composition in human liver between the seventeenth and thirtysixth gestational weeks. Am J Obstet Gynecol. 1998;179:1063–1070. doi: 10.1016/s0002-9378(98)70216-9. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez A, Sarda P, Nessmann C, et al. D6- and D5-desaturase activities in the human fetal liver: kinetic aspects. J Lipid Res. 1998;39:1825–1832. [PubMed] [Google Scholar]
- 63.Sauerwald TU, Hachey DL, Jensen CL, et al. Intermediates in endogenous synthesis of C22:6 omega 3 and C20:4 omega 6 by term and preterm infants. Pediatr Res. 1997;41:183–187. doi: 10.1203/00006450-199702000-00005. [DOI] [PubMed] [Google Scholar]
- 64.Carnielli VP, Wattimena DJ, Luijendijk IH, et al. The very low birth weight premature infant is capable of synthesizing arachidonic and docosahexaenoic acids from linoleic and linolenic acids. Pediatr Res. 1996;40:169–174. doi: 10.1203/00006450-199607000-00029. [DOI] [PubMed] [Google Scholar]
- 65.Carnielli VP, Simonato M, Verlato G, et al. Synthesis of long-chain polyunsaturated fatty acids in preterm newborns fed formula with long-chain polyunsaturated fatty acids. Am J Clin Nutr. 2007;86:1323–1330. doi: 10.1093/ajcn/86.5.1323. [DOI] [PubMed] [Google Scholar]
- 66.Ravel D, Chambaz J, Pepin D, et al. Essential fatty acid interconversion during gestation in the rat. Biochim Biophys Acta. 1985;833:161–164. doi: 10.1016/0005-2760(85)90264-4. [DOI] [PubMed] [Google Scholar]
- 67.Ortega-Senovilla H, Alvino G, Taricco E, et al. Gestational diabetes mellitus upsets the proportion of fatty acids in umbilical arterial but not venous plasma. Diabetes Care. 2009;32:120–122. doi: 10.2337/dc08-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pankiewicz E, Cretti A, Ronin-Walknowska E, et al. Maternal adipose tissue, maternal and cord blood essential fatty acids and their long-chain polyunsaturated derivatives composition after elective caesarean section. Early Hum Dev. 2007;83:459–464. doi: 10.1016/j.earlhumdev.2006.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.