Abstract
Kindred cells can have different genomes because of dynamic changes in DNA. Single cell sequencing is needed to characterize these genomic differences but has been hindered by whole-genome amplification bias, resulting in low genome coverage. Here we report a new amplification method: Multiple Annealing and Looping Based Amplification Cycles (MALBAC) that offer high uniformity across the genome. Sequencing MALBAC amplified DNA achieves 93% genome coverage ≥1x for a single human cell at 25x mean sequencing depth. We detected digitized copy number variations (CNVs) of a single cancer cell. By sequencing three kindred cells, we were able to call individual single nucleotide variations (SNVs) with no false positives observed. We directly measured the genome-wide mutation rate of a cancer cell line and found that purine-pyrimidine exchanges occurred unusually frequently among the newly acquired SNVs.
Single molecule and single cell studies reveal behaviors that are hidden in bulk measurements (1, 2). In a human cell, the genetic information is encoded in 46 chromosomes. The variations occurring in these chromosomes, such as single nucleotide variations (SNVs) and copy number variations (CNVs) (3), are the driving forces in biological processes such as evolution and cancer. Such dynamic variations are reflected in the genomic heterogeneity among a population of cells, which demands characterization of genomes at the single cell level (4–6). Single cell genomics analysis is also necessary when the number of cells available is limited to few or one, such as prenatal testing samples (7, 8), circulating tumor cells (9), and forensic specimens (10).
Prompted by rapid progress in next generation sequencing techniques (11), there have been several reports on whole genome sequencing of single cells (12–16). These methods have relied on whole genome amplification (WGA) of an individual cell to generate enough DNA for sequencing (17–21). However, WGA methods in general are prone to amplification bias, which results in low genome coverage. PCR-based WGAintroduces sequence-dependent bias because of the exponential amplification with random primers (17, 18, 22). Multiple Displacement Amplification (MDA), which uses random priming and the strand-displacing phi29 polymerase under isothermal condition (19), has provided improvements over PCR-based methods but still exhibits considerable bias, again due to nonlinear amplification.
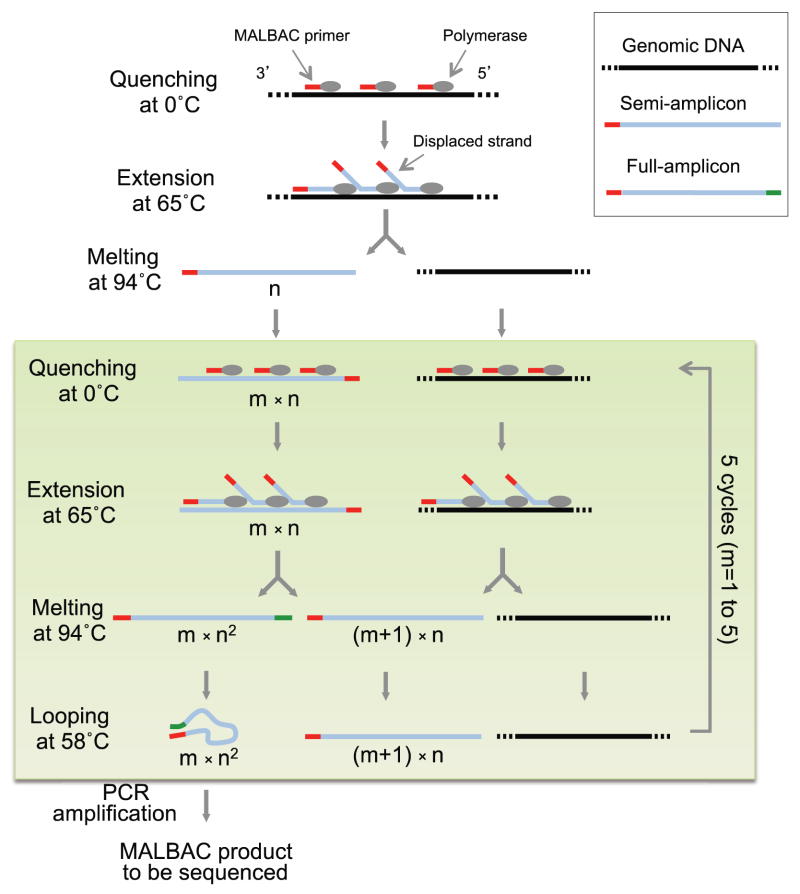
Here we report a new WGAmethod, Multiple Annealing and Looping Based Amplification Cycles (MALBAC), which introduces quasi-linear preamplification to reduce the bias associated with nonlinear amplification. Picograms of DNA fragments (~10 to 100kb) from a single human cell serve as templates for amplification with MALBAC (Fig. 1). The amplification is initiated with a pool of random primers, each having a common 27-nucleotide sequence and 8 variable nucleotides that can evenly hybridize to the templates at 0°C. At an elevated temperature of 65°C, DNA polymerases with strand displacement activity are used to generate semiamplicons with variable lengths (0.5–1.5kb), which are then melted off from the template at 94°C. Amplification of the semiamplicons give full amplicons which have complementary ends. The temperature is cycled to 58°C to allow the looping of full amplicons, which prevents further amplification and cross hybridizations. Five cycles of preamplification are followed by exponential amplification of the full amplicons by PCR in order to generate micrograms of DNA required for next generation sequencing (Fig. 1). In the PCR, oligos with the common 27-nucleotide sequence are used as the primers.
Figure 1.
MALBAC single cell whole genome amplification. A single cell is picked and lysed. First, genomic DNA of the single cell is melted into single-stranded DNA molecules at 94°C. MALBAC primers then anneal randomly to single-stranded DNA molecules at 0°C and are extended by a polymerase with displacement activity at elevated temperatures, creating semi-amplicons. In the following five temperature cycles, after the step of looping the full amplicons, single stranded amplicons and the genomic DNA are used as template to produce full amplicons and additional semi-amplicons, respectively. For full amplicons, the 3′ end is complementary to the sequence on the 5′ end. The two ends hybridize will form the looped DNA, which can efficiently prevents the full amplicon from being used as template, therefore warrant a close-to-linear amplification. After the five cycles of linear preamplification, only the full amplicons can be exponentially amplified in the following PCR using the common 27-nucleotide sequence as the primer. PCR reaction will generate microgram level of DNA material for sequencing experiments.
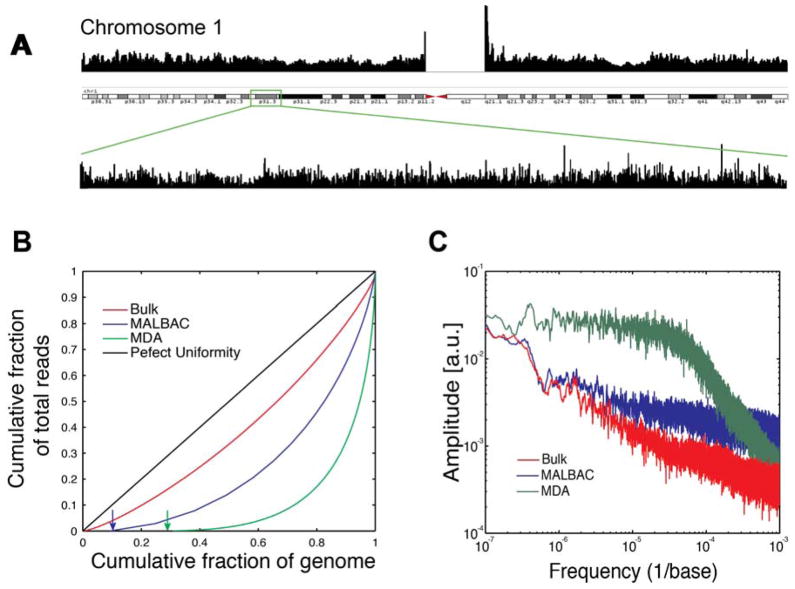
We used MALBAC to amplify the DNA of single SW480 cancer cells. With ~25x mean sequencing depth, we consistently achieved ~85% and up to 93% genome coverage at ≥1x depth on either strand (Fig. 2A). As a comparison, we performed MDA on a single cell from the same cancer cell line. At 25x mean sequencing depth, MDAcovered 72% of the genome at ≥1x coverage. While significant variations of the coverage have been reported for MDA(15, 16, 20, 23), MALBAC coverage is reproducible.
Figure 2.
Characterization of amplification uniformity. (A) Histograms of reads over the entirety of Chromosome 1 of a single cell from the SW480 cancer cell line and the zoom-in of a ~8 million base region (chr1: 62,023,147–70,084,845). (B) Lorenz curves of MALBAC, MDA and bulk sample. A Lorenz curve gives the cumulated fraction of reads as a function of the cumulated fraction of genome. Perfectly uniform coverage would result in a diagonal line and a large deviation from the diagonal is indicative of a biased coverage. The green and blue arrows indicate the uncovered fractions of the genome for MALBAC and MDA respectively. All samples are sequenced at 25x depth. (C) Power spectrum of read density throughout the genome (as a function of spatial frequency). MALBAC performs similarly to bulk, while the MDA spectrum shows high amplitude at low frequency, demonstrating that regions of several megabases suffer from under- and over- amplification. This observation is consistent with the variations of read depth in Fig. S3 (SOM).
We use Lorenz curves to evaluate coverage uniformity along the genome. Here, we plotted the cumulative fraction of the total reads that cover a given cumulative fraction of genome (Fig. 2B). The diagonal line indicates a perfectly uniform distribution of reads, and deviation from the diagonal line indicates an uneven distribution of reads. We compared the Lorenz curves for bulk sequencing, MALBAC, and MDA at ~25x mean sequencing depth (Fig. 2B). It is evident that MALBAC outperforms MDAin uniformity of genome coverage. We also plotted the power spectrum of read density variations to show the spatial scale at which the variations take place. For MDA, large amplitudes at low frequencies (1/genome distance) were observed, indicating that large contiguous regions of the genome are over- or under-amplified. In contrast, MALBAC has a power spectrum similar to that of the unamplified bulk.
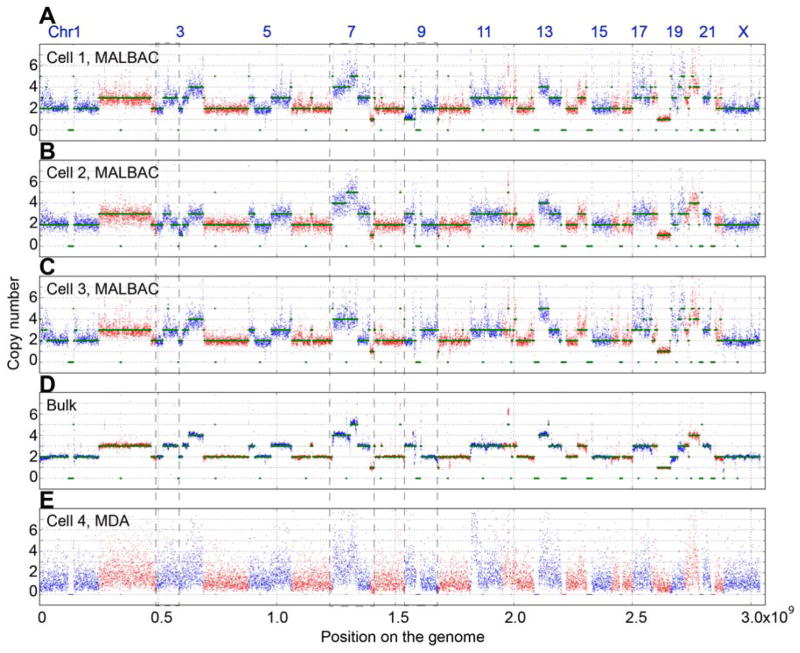
CNVs due to insertions, deletions, or multiplications of genome segments are frequently observed in almost all categories of human tumors (13, 24, 25). MALBAC’s lack of large-scale bias makes it amenable to probing CNVs in single cells. We determined the digitized CNVs across the whole genomes of three individual cells from the SW480 cancer cell line (Fig. 3A–C). CNVs of five cells are included in the SOM (Supplemental Online Material). The chromosomes exhibit distinct CNV differences among the three individual cancer cells and in the bulk result (Fig. 3D), which are difficult to resolve by MDA (Fig. 3E). For the MALBAC data, we used a hidden Markov model to quantify CNVs (SOM). We confirmed the gross features of CNVs detected by MALBAC with a previously published karyotyping study (26). For example, both MALBAC-based quantification of CNVs and spectral karyotyping show one copy of chromosome 18 and three copies of chromosome 17 in the SW480 cancer cell line. Although the majority of copy numbers are consistent between single cells, we also observe cell-to-cell variations as labeled by the dashed box in Fig. 3.
Figure 3.
CNVs of single cancer cells. Digitized copy numbers across the genome are plotted for three single cells (Panel A to C) as well as the bulk sample (Panel D) from the SW480 cancer cell line. The bottom panel shows the result based on MDA amplification (Panel E). Green lines are fitted CNV numbers obtained from the hidden Markov model (SOM). The single cells are sequenced at only 0.8x depth, while the bulk and MDA are done at 25x. More single cells’ CNV analyses are included in the SOM (Fig. S4). The regions within the dashed box exhibit the CNV differences among single cells and the bulk, which cannot be resolved by MDA. The binning window is 200kb.
Attempts have been made recently to identify SNVs from a single cell by MDA (15, 16, 23). The first challenge in accurate SNV calling from a single cell is substantial human contamination from the environment and the operators, given picograms of DNA from a single human cell. The second challenge is low detection yield (high false negative), particularly where alleles drop out due to amplification bias. The third challenge is false positives associated with amplification and sequencing errors, either random or systematic (27).
To meet the first challenge, we took special precautions to decontaminate with UV radiation before each experiment was conducted in a restricted clean room. An alternative approach to reduce contamination is microfluidics (28).
With regard to the second challenge, MALBAC allowed us to call 2.2 x 106 single cell SNVs compared with 2.8 x 106 detected SNVs in bulk, yielding a 76% detection efficiency, in contrast to 41% with MDA (Table 1). This improvement resulted from improved uniformity by MALBAC (SOM, Fig S6). Listed separately in Table 1 are heterozygous and homozygous SNVs. Next we calculate the allele dropout rate. Comparison of single-cell and bulk SNVs showed that 7,288 of the SNVs genotyped as homozygous mutations by MALBAC are actually heterozygous in bulk, which corresponds to a ~1% allele dropout rate in MALBAC (SOM). In contrast, with MDAwe found 172,563 incorrect homozygous calls, corresponding to an allele dropout rate of ~65% (SOM).
Table 1.
Comparison of Single cell SNVs for bulk, MDA and MALBAC
| Heterozygous SNVs | Homozygous SNVs | Total SNVs | |
|---|---|---|---|
|
| |||
| Bulk
| |||
| SNVs | 911,958 | 1,930,204 | 2,842,162 |
|
| |||
| Single cell MDA
| |||
| SNVs | 93,140 (2,828)* | 1,238,286 (1,973) | 1,331,426 (4,801) |
| Detection efficiency | 10% | 63% | 41% |
|
| |||
| Single cell MALBAC
| |||
| SNVs | 756,812 (108,481) | 1,539,326 (6,821) | 2,296,138 (115,302) |
| Detection efficiency | 71% | 80% | 76% |
The number in the bracket indicates the number of false positives.
Compared to the bulk data, the MALBAC data contains 1.1x105 false positives (Table 1) out of 3x109 bases in the genome. This corresponds to a ~4 x10−5 false positive rate, which is due to the errors made by the polymerases in the semi-amplicons generated in first MALBAC cycle and propagated in the later amplification. Although improving the polymerase’s error rate is possible, our strategy to reduce the false positive rate was to sequence two or three kindred cells derived from the same cell. The simultaneous appearance of an SNV in the kindred cells would indicate a true SNV. The false positive rate due to uncorrelated random errors can be reduced to ~10−8 with two kindred cells and ~10−12 with three kindred cells.
However, there are false positives due to correlated errors i.e. systematic sequencing and amplification errors. We filtered out these errors by comparing two unrelated single cells that are not from the same lineage (SOM, Figure S5). After this procedure, we can call true SNVs of a single cell with no false positives observed (Table 2).
Table 2.
MALBAC calling of total SNVs and newly acquired SNVs using two and three kindred cells
| Heterozygous SNVs | Homozygous SNVs | Total SNVs | |
|---|---|---|---|
|
| |||
| Two kindred cells
| |||
| SNVs | 615,387 | 1,322,555 | 1,937,942 |
| Detection efficiency | 67% | 68% | 68% |
| Newly acquired SNVs | 145 (~100)* | 3 (~0) | 148 (~100) |
|
| |||
| Three kindred cells
| |||
| SNVs | 660,246 | 1,577,798 | 2,238,044 |
| Detection efficiency | 72% | 81% | 80% |
| Newly acquired SNVs | 30 (~0) | 5 (~0) | 35 (~0) |
The number in the bracket indicates the number of false positives.
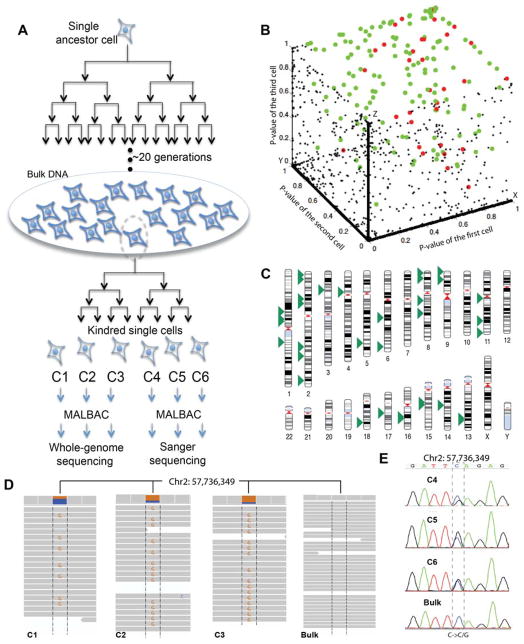
To gain insight into the mutation process in the cancer cells, we clonally expanded a single ancestor cell picked from a heterogeneous population of the SW480 cancer cell line for 20 generations (Fig. 4A). We extracted DNA from this single cell clonal expansion for bulk sequencing, which reflects the genome of the ancestor cell. We then picked a single cell from this clone. To detect SNVs acquired by the cell during expansion, we grew another four generations to obtain the kindred cells denoted C1 to C16. We individually sequenced three kindred cells, C1, C2, and C3 after MALBAC amplification. After filtering correlated and uncorrelated errors (Fig. 4B), we detected 35 unique SNVs shown in Fig. 4C.
Figure 4.
Calling newly acquired SNVs and estimation of mutation rate of a cancer cell line (SW480). (A) Experiment design. A single ancestor cell is chosen and cultured for ~20 generations. The vast majority of cells are used to extract DNA for bulk sequencing to represent the ancestor cell’s genome. A single cell from this culture is chosen for another expansion of four generations. The kindred cells are isolated for single cell whole genome amplification. Single cell sample C1, C2, and C3 are used for high-throughput sequencing. Sample C4, C5, and C6 are used for varying SNVs with Sanger sequencing. (B) 3D p-value plot of a one-sided binomial test for SNV candidates from the three kindred cells. The black dots are the false positives due to uncorrelated amplification errors; all of them are on the x-y-z axis and x-y, y-z, x-z planes. Outside of the three planes, the 166 green dots are the residual false positives due to correlated errors from homopolymers, tandem repeats, high-GC content and high density SNV regions, and the 35 red dots are the newly acquired SNVs during the 20 generations of clonal expansion (SOM). We note that the homozygous SNVs are located at the (1,1,1) position. (C) Locations of the 35 newly acquired SNVs on the chromosomes of a single cell (SOM). (D) Next-generation sequencing data of a newly acquired SNV. The SNV (C→G) exists in the high throughput data of all three kindred cells but not in the bulk data. (E) Sanger sequencing data of single cells C4, C5, and C6 confirms that this SNV is not a false positive, while the Sanger sequencing of the bulk confirms that this SNV is not a false negative of next generation sequencing of the bulk (i.e. this SNV is indeed absent in the bulk).
We randomly chose 8 out of a total of 35 unique SNVs and confirmed that they are neither false positives by Sanger sequencing C4-C6, nor false negatives by Sanger sequencing the bulk (Please refer to SOM for Sanger sequencing data). As an example, Fig. 4D and 4E shows the MALBAC and Sanger sequencing result of one such SNV.
These 35 unique SNVs are newly acquired during the 20 cell divisions. Adjusting for a detection efficiency of 72% for heterozygous SNVs, we estimate that ~49 mutations occurred in the 20 generations, yielding a mutation rate of ~2.5 nucleotides per cell generation, consistent with our estimation based on the bulk data (SOM). The mutation rate of this cancer cell line is about 10 fold higher than the mutation rate estimated based on germ line studies (29–31).
Mutations can be transitions (purine<->purine exchange i.e. A<->G or pyrimidine<->pyrimidine exchange, i.e. C<->T) or transversions (purine <-> pyrimidine exchanges, i.e. A/G<->C/T). Transitions are more common. Surprisingly, we found that the transition/transversion (tstv) ratio for the 35 newly acquired SNVs detected is only 0.30, whereas the ratio for the total SNVs of this cell line is 2.01, as expected for common human mutations (32). To further confirm that this observation is not due to single cell amplification, we sequenced the bulk DNA of the original heterogeneous culture (SOM). The tstv ratio for SNVs detected in the single cell expanded bulk but not in the original heterogeneous bulk was 0.75. Both significantly low tstv ratios indicate that transitions are not favored over transversion for newly acquired SNVs in this cancer cell line (SOM). While understanding the underlying mechanism of this phenomenon will require similar measurements in other systems, it is evident that, by allowing precise characterization of CNVs and SNVs, MALBAC can shed light on the individuality, heterogeneity, and dynamics of the genomes of single cells.
Supplementary Material
Summary.
A new whole genome amplification method with significantly reduced bias allows simultaneous accurate detection of point mutations and copy number variations in single mammalian cells and the direct measurement of mutation rates.
Acknowledgments
This work was supported by the United States National Institutes of Health National Human Genome Research Institute Grant (HG005097-1 and HG005613-01) and in part by Bill & Melinda Gates Foundation OPP42867 to XSX. ARC was supported by the NIH Molecular Biophysics Training Grant (NIH/NIGMS T32 GM008313). We thank Paul Choi for his involvement on the early stage of the project and Jenny Lu and Lin Song for their assistance on the experiments. We thank Jun Yong for his help on single cell expansion and isolation and Zhang Yun at Biodynamic and Optical Imaging Center (BIOPIC) at Peking University for assistance on sequencing. The sequencing data is deposited at NCBI with accession number SRA060929.
Footnotes
Competing Financial Interests
CZ, SL and XSX are authors on a patent applied for by Harvard University that covers the MALBAC technology
References
- 1.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002 Aug 16;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 2.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011 Jul 21;475:308. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nature reviews Molecular cell biology. 2010 Mar;11:220. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998 Dec 17;396:643. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 5.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010 Oct 28;467:1114. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010 Oct 28;467:1109. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo YM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Science translational medicine. 2010 Dec 8;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 8.Kitzman JO, et al. Noninvasive whole-genome sequencing of a human fetus. Science translational medicine. 2012 Jun 6;4:137ra76. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007 Dec 20;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson EK, Ballantyne J. Whole genome amplification strategy for forensic genetic analysis using single or few cell equivalents of genomic DNA. Analytical biochemistry. 2005 Nov 15;346:246. doi: 10.1016/j.ab.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Metzker ML. Sequencing technologies - the next generation. Nature reviews Genetics. 2010 Jan;11:31. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 12.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nature biotechnology. 2011 Jan;29:51. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011 Apr 7;472:90. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gundry M, Li WG, Maqbool SB, Vijg J. Direct, genome-wide assessment of DNA mutations in single cells. Nucleic acids research. 2012 Mar;40:2032. doi: 10.1093/nar/gkr949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012 Mar 2;148:873. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Fan HC, Behr B, Quake SR. Genome-wide Single-Cell Analysis of Recombination Activity and De Novo Mutation Rates in Human Sperm. Cell. 2012 Jul 20;150:402. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, et al. Whole genome amplification from a single cell: implications for genetic analysis. Proceedings of the National Academy of Sciences of the United States of America. 1992 Jul 1;89:5847. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Telenius H, et al. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992 Jul;13:718. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 19.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang K, et al. Sequencing genomes from single cells by polymerase cloning. Nature biotechnology. 2006 Jun;24:680. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 21.Lao K, Xu NL, Straus NA. Whole genome amplification using single-primer PCR. Biotechnology journal. 2008 Mar;3:378. doi: 10.1002/biot.200700253. [DOI] [PubMed] [Google Scholar]
- 22.Dietmaier W, et al. Multiple mutation analyses in single tumor cells with improved whole genome amplification. The American journal of pathology. 1999 Jan;154:83. doi: 10.1016/S0002-9440(10)65254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012 Mar 2;148:886. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010 Feb 18;463:899. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011 Jan 7;144:27. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rochette PJ, Bastien N, Lavoie J, Guerin SL, Drouin R. SW480, a p53 double-mutant cell line retains proficiency for some p53 functions. J Mol Biol. 2005 Sep 9;352:44. doi: 10.1016/j.jmb.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 27.Macarthur D. Methods: Face up to false positives. Nature. 2012 Jul 26;487:427. doi: 10.1038/487427a. [DOI] [PubMed] [Google Scholar]
- 28.Blainey PC, Quake SR. Digital MDA for enumeration of total nucleic acid contamination. Nucleic acids research. 2011 Mar;39:e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998 Apr;148:1667. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010 Apr 30;328:636. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conrad DF, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011 Jul;43:712. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010 Oct 28;467:1061. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.