Abstract
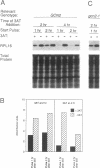
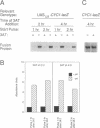
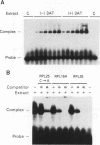
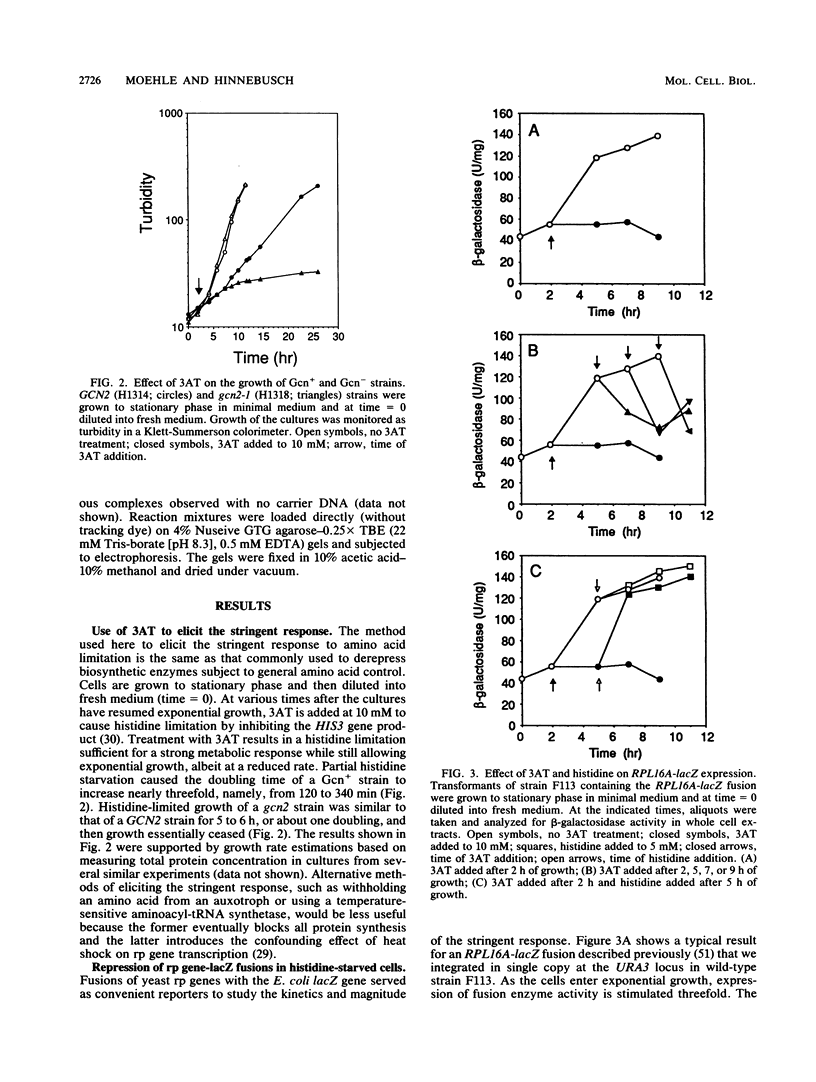
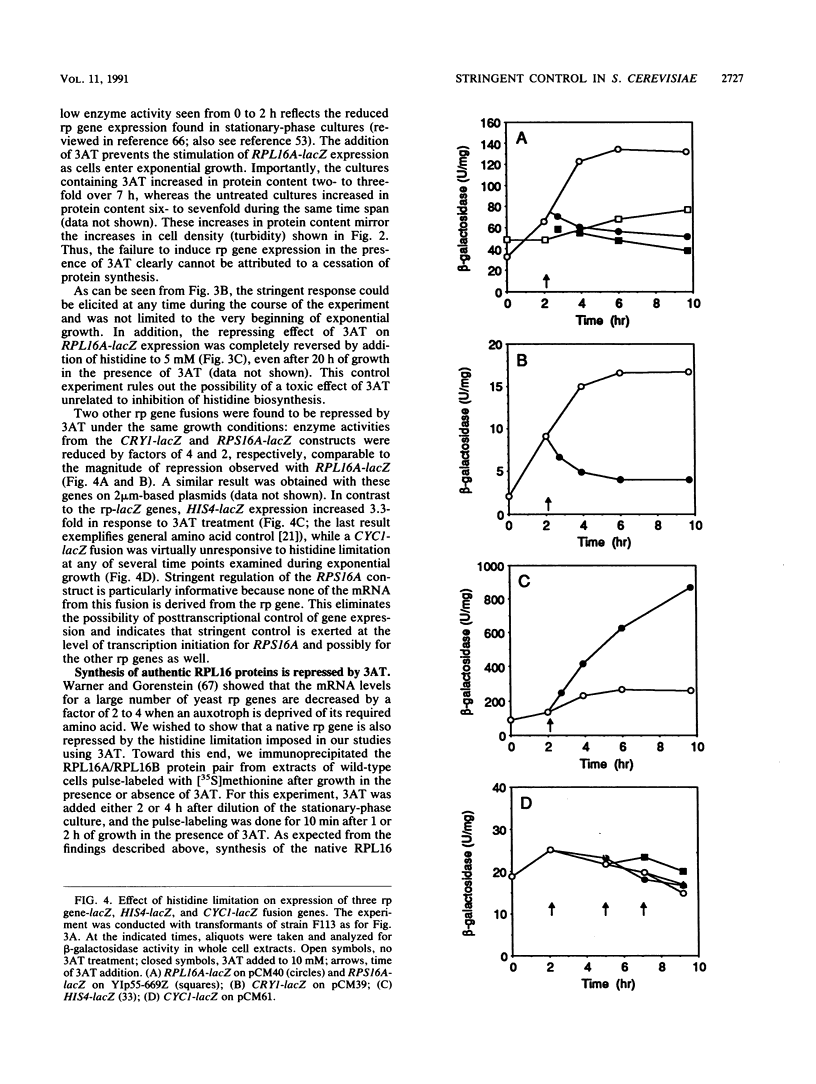
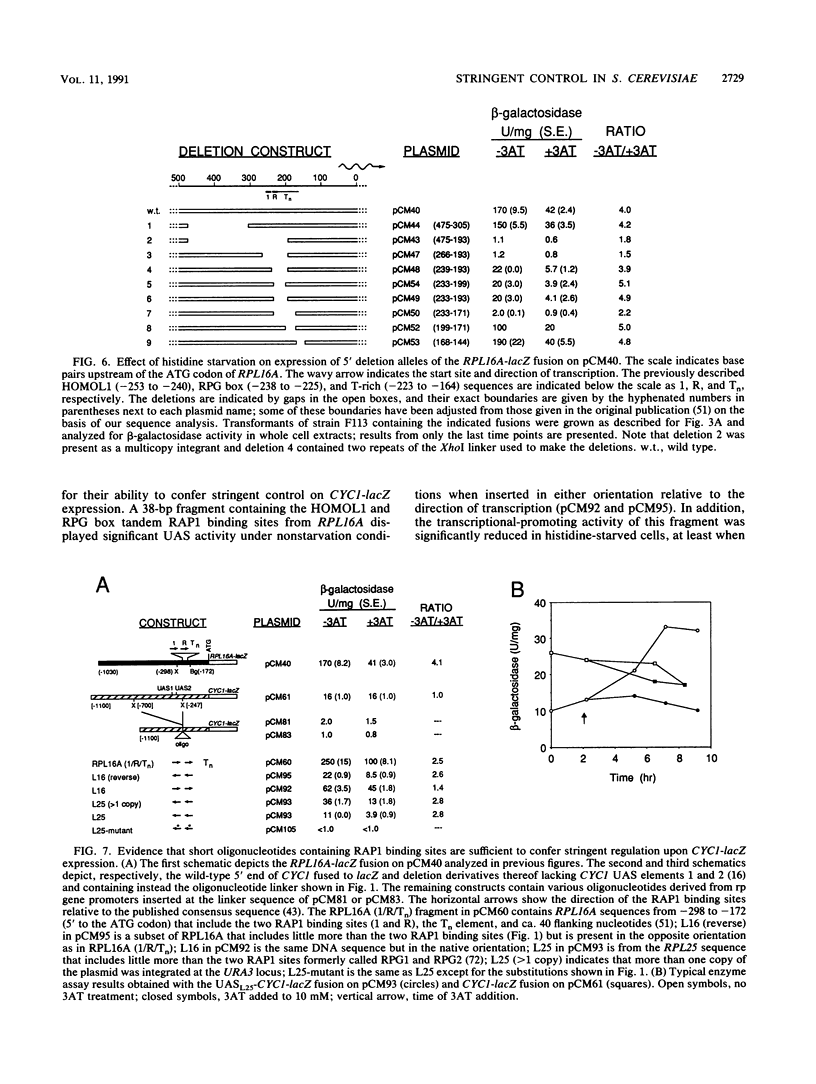
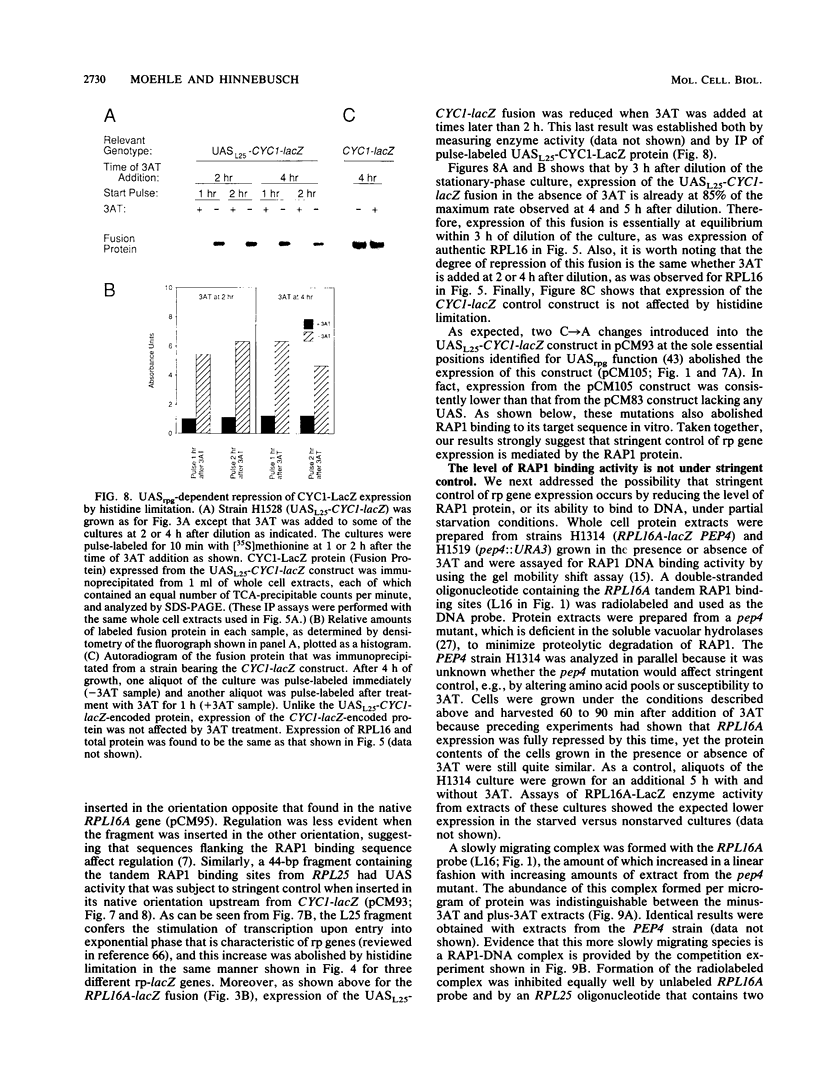
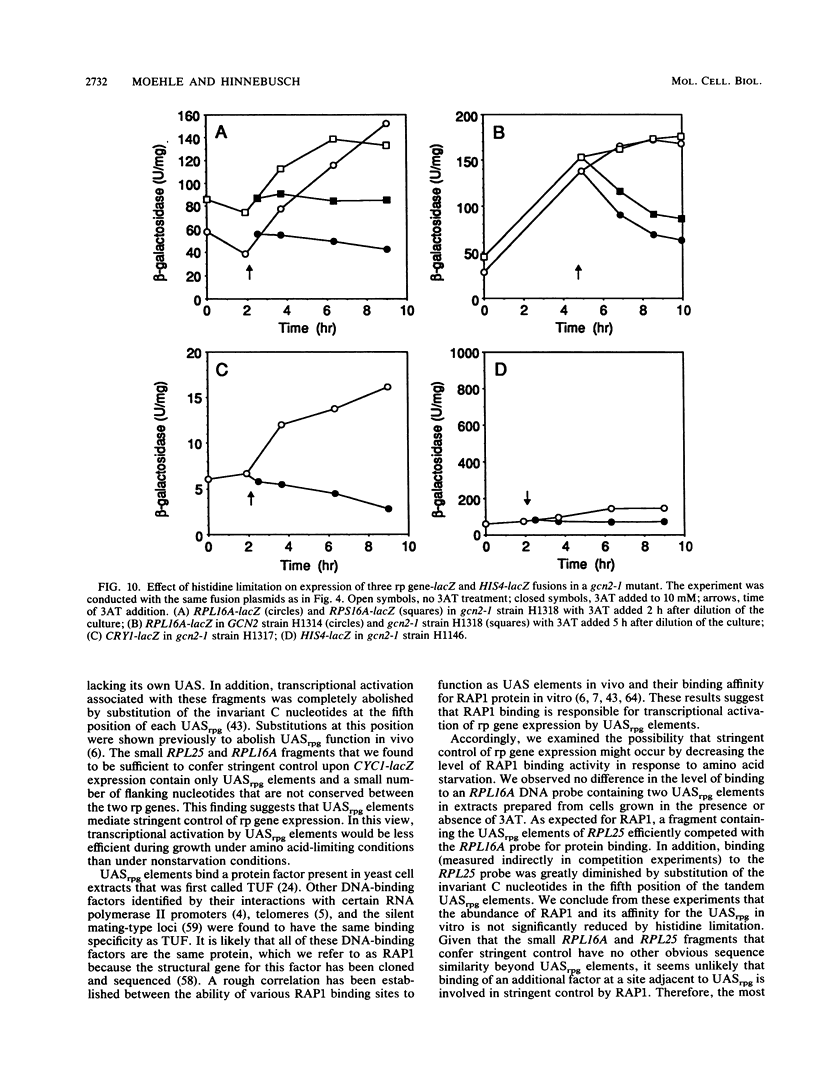
An amino acid limitation in bacteria elicits a global response, called stringent control, that leads to reduced synthesis of rRNA and ribosomal proteins and increased expression of amino acid biosynthetic operons. We have used the antimetabolite 3-amino-1,2,4-triazole to cause histidine limitation as a means to elicit the stringent response in the yeast Saccharomyces cerevisiae. Fusions of the yeast ribosomal protein genes RPL16A, CRY1, RPS16A, and RPL25 with the Escherichia coli lacZ gene were used to show that the expression of these genes is reduced by a factor of 2 to 5 during histidine-limited exponential growth and that this regulation occurs at the level of transcription. Stringent regulation of the four yeast ribosomal protein genes was shown to be associated with a nucleotide sequence, known as the UASrpg (upstream activating sequence for ribosomal protein genes), that binds the transcriptional regulatory protein RAP1. The RAP1 binding sites also appeared to mediate the greater ribosomal protein gene expression observed in cells growing exponentially than in cells in stationary phase. Although expression of the ribosomal protein genes was reduced in response to histidine limitation, the level of RAP1 DNA-binding activity in cell extracts was unaffected. Yeast strains bearing a mutation in any one of the genes GCN1 to GCN4 are defective in derepression of amino acid biosynthetic genes in 10 different pathways under conditions of histidine limitation. These Gcn- mutants showed wild-type regulation of ribosomal protein gene expression, which suggests that separate regulatory pathways exist in S. cerevisiae for the derepression of amino acid biosynthetic genes and the repression of ribosomal protein genes in response to amino acid starvation.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammerer G., Hunter C. P., Rothman J. H., Saari G. C., Valls L. A., Stevens T. H. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986 Jul;6(7):2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel P., Böck A. Lack of accumulation of unusual guanosine nucleotides upon amino acid starvation of two eukaryotic organisms. Biochim Biophys Acta. 1973 Sep 28;324(1):184–187. doi: 10.1016/0005-2787(73)90262-1. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chambers A., Tsang J. S., Stanway C., Kingsman A. J., Kingsman S. M. Transcriptional control of the Saccharomyces cerevisiae PGK gene by RAP1. Mol Cell Biol. 1989 Dec;9(12):5516–5524. doi: 10.1128/mcb.9.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989 Nov 24;246(4933):1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- Donovan D. M., Pearson N. J. Transcriptional regulation of ribosomal proteins during a nutritional upshift in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jul;6(7):2429–2435. doi: 10.1128/mcb.6.7.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakle K. A., Bernstein M., Emr S. D. Characterization of a component of the yeast secretion machinery: identification of the SEC18 gene product. Mol Cell Biol. 1988 Oct;8(10):4098–4109. doi: 10.1128/mcb.8.10.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter H., Kavety B., Vandekerckhove J., Kiefer F., Gallwitz D. Sequence, expression and mutational analysis of BAF1, a transcriptional activator and ARS1-binding protein of the yeast Saccharomyces cerevisiae. EMBO J. 1989 Dec 20;8(13):4265–4272. doi: 10.1002/j.1460-2075.1989.tb08612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamil K. G., Nam H. G., Fried H. M. Constitutive transcription of yeast ribosomal protein gene TCM1 is promoted by uncommon cis- and trans-acting elements. Mol Cell Biol. 1988 Oct;8(10):4328–4341. doi: 10.1128/mcb.8.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Williams N. P., Wek R. C., Hinnebusch A. G. The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics. 1990 Nov;126(3):549–562. doi: 10.1093/genetics/126.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herruer M. H., Mager W. H., Woudt L. P., Nieuwint R. T., Wassenaar G. M., Groeneveld P., Planta R. J. Transcriptional control of yeast ribosomal protein synthesis during carbon-source upshift. Nucleic Acids Res. 1987 Dec 23;15(24):10133–10144. doi: 10.1093/nar/15.24.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W., Zubenko G. S., Parker R. R. PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics. 1982 Dec;102(4):665–677. doi: 10.1093/genetics/102.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kief D. R., Warner J. R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Nov;1(11):1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H., Warner J. R. Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol Cell Biol. 1983 Mar;3(3):457–465. doi: 10.1128/mcb.3.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopotowski T., Wiater A. Synergism of aminotriazole and phosphate on the inhibition of yeast imidazole glycerol phosphate dehydratase. Arch Biochem Biophys. 1965 Dec;112(3):562–566. doi: 10.1016/0003-9861(65)90096-2. [DOI] [PubMed] [Google Scholar]
- Kudrna R., Edlin G. Nucleotide pools and regulation of ribonucleic acid synthesis in yeast. J Bacteriol. 1975 Feb;121(2):740–742. doi: 10.1128/jb.121.2.740-742.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Hinnebusch A. G., Chen C., Fink G. R. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig J. R., 2nd, Oliver S. G., McLaughlin C. S. The regulation of RNA synthesis in yeast II: Amino acids shift-up experiments. Mol Gen Genet. 1977 Dec 30;158(2):117–122. doi: 10.1007/BF00268303. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Vaughan M. H., Warner J. R., Darnell J. E. Effects of valine deprivation on ribosome formation in HeLa cells. J Mol Biol. 1969 Oct 28;45(2):265–275. doi: 10.1016/0022-2836(69)90104-1. [DOI] [PubMed] [Google Scholar]
- McLaughlin C. S., Magee P. T., Hartwell L. H. Role of isoleucyl-transfer ribonucleic acid synthetase in ribonucleic acid synthesis and enzyme repression in yeast. J Bacteriol. 1969 Nov;100(2):579–584. doi: 10.1128/jb.100.2.579-584.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for beta-galactosidase activity. Anal Biochem. 1989 Aug 15;181(1):40–50. doi: 10.1016/0003-2697(89)90391-6. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Delforge J. Role of transfer ribonucleic acids in the regulation of several biosyntheses in Saccharomyces cerevisiae. Eur J Biochem. 1976 Aug 16;67(2):335–339. doi: 10.1111/j.1432-1033.1976.tb10696.x. [DOI] [PubMed] [Google Scholar]
- Needleman R. B., Tzagoloff A. Breakage of yeast: a method for processing multiple samples. Anal Biochem. 1975 Apr;64(2):545–549. doi: 10.1016/0003-2697(75)90466-2. [DOI] [PubMed] [Google Scholar]
- Niederberger P., Miozzari G., Hütter R. Biological role of the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jul;1(7):584–593. doi: 10.1128/mcb.1.7.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwint R. T., Mager W. H., Maurer K. C., Planta R. J. Mutational analysis of the upstream activation site of yeast ribosomal protein genes. Curr Genet. 1989 Apr;15(4):247–251. doi: 10.1007/BF00447039. [DOI] [PubMed] [Google Scholar]
- Oliver S. G., McLaughlin C. S. The regulation of RNA synthesis in yeast. I: Starvation experiments. Mol Gen Genet. 1977 Jul 20;154(2):145–153. doi: 10.1007/BF00330830. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao C. C., Paietta J., Gallant J. A. Synthesis of guanosine tetraphosphate (magic spot I) in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1977 Jan 10;74(1):314–322. doi: 10.1016/0006-291x(77)91410-3. [DOI] [PubMed] [Google Scholar]
- Pogo A. O., Zbrzezna V. J. Cycloheximide stimulates ribosomal RNA transcription in amino acid-starved ascites tumor cells. FEBS Lett. 1978 Apr 1;88(1):135–138. doi: 10.1016/0014-5793(78)80625-5. [DOI] [PubMed] [Google Scholar]
- Rhode P. R., Sweder K. S., Oegema K. F., Campbell J. L. The gene encoding ARS-binding factor I is essential for the viability of yeast. Genes Dev. 1989 Dec;3(12A):1926–1939. doi: 10.1101/gad.3.12a.1926. [DOI] [PubMed] [Google Scholar]
- Rotenberg M. O., Moritz M., Woolford J. L., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988 Feb;2(2):160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- Rotenberg M. O., Woolford J. L., Jr Tripartite upstream promoter element essential for expression of Saccharomyces cerevisiae ribosomal protein genes. Mol Cell Biol. 1986 Feb;6(2):674–687. doi: 10.1128/mcb.6.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussou I., Thireos G., Hauge B. M. Transcriptional-translational regulatory circuit in Saccharomyces cerevisiae which involves the GCN4 transcriptional activator and the GCN2 protein kinase. Mol Cell Biol. 1988 May;8(5):2132–2139. doi: 10.1128/mcb.8.5.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge K. W., Zakian V. A. Properties of the transcriptional enhancer in Saccharomyces cerevisiae telomeres. Nucleic Acids Res. 1990 Apr 11;18(7):1783–1787. doi: 10.1093/nar/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo G. M., Tornow J. Efficient transcription of the glycolytic gene ADH1 and three translational component genes requires the GCR1 product, which can act through TUF/GRF/RAP binding sites. Mol Cell Biol. 1990 Feb;10(2):859–862. doi: 10.1128/mcb.10.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarubbi E., Rudd K. E., Cashel M. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet. 1988 Aug;213(2-3):214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- Schürch A., Miozzari J., Hütter R. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J Bacteriol. 1974 Mar;117(3):1131–1140. doi: 10.1128/jb.117.3.1131-1140.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman R. W., Sripati C. E., Warner J. R. Noncoordinated transcription in the absence of protein synthesis in yeast. J Biol Chem. 1977 Feb 25;252(4):1344–1349. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stateva L. I., Venkov P. V. Genetic analysis of Saccharomyces cerevisiae SY 15 relaxed mutant. Mol Gen Genet. 1984;195(1-2):234–237. doi: 10.1007/BF00332752. [DOI] [PubMed] [Google Scholar]
- Teem J. L., Abovich N., Kaufer N. F., Schwindinger W. F., Warner J. R., Levy A., Woolford J., Leer R. J., van Raamsdonk-Duin M. M., Mager W. H. A comparison of yeast ribosomal protein gene DNA sequences. Nucleic Acids Res. 1984 Nov 26;12(22):8295–8312. doi: 10.1093/nar/12.22.8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais M. L., Huet J., Buhler J. M., Sentenac A. Contacts between the factor TUF and RPG sequences. J Biol Chem. 1990 Aug 25;265(24):14669–14674. [PubMed] [Google Scholar]
- Waltschewa L., Georgiev O., Venkov P. Relaxed mutant of Saccharomyces cerevisiae: proper maturation of ribosomal RNA in absence of protein synthesis. Cell. 1983 May;33(1):221–230. doi: 10.1016/0092-8674(83)90351-3. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Gorenstein C. Yeast has a true stringent response. Nature. 1978 Sep 28;275(5678):338–339. doi: 10.1038/275338a0. [DOI] [PubMed] [Google Scholar]
- Warner J. R. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989 Jun;53(2):256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Jackson B. M., Hinnebusch A. G. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Ramirez M., Jackson B. M., Hinnebusch A. G. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol. 1990 Jun;10(6):2820–2831. doi: 10.1128/mcb.10.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N. P., Hinnebusch A. G., Donahue T. F. Mutations in the structural genes for eukaryotic initiation factors 2 alpha and 2 beta of Saccharomyces cerevisiae disrupt translational control of GCN4 mRNA. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7515–7519. doi: 10.1073/pnas.86.19.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind M., Kolb J. M., Dodd J., Yamagishi M., Mémet S., Buhler J. M., Nomura M. Conditional expression of RPA190, the gene encoding the largest subunit of yeast RNA polymerase I: effects of decreased rRNA synthesis on ribosomal protein synthesis. Mol Cell Biol. 1990 May;10(5):2049–2059. doi: 10.1128/mcb.10.5.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt L. P., Smit A. B., Mager W. H., Planta R. J. Conserved sequence elements upstream of the gene encoding yeast ribosomal protein L25 are involved in transcription activation. EMBO J. 1986 May;5(5):1037–1040. doi: 10.1002/j.1460-2075.1986.tb04319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]