Abstract
Clostridium perfringens iota-toxin (Ia) mono-ADP ribosylates Arg177 of actin, leading to cytoskeletal disorganization and cell death. To fully understand the reaction mechanism of arginine-specific mono-ADP ribosyl transferase, the structure of the toxin-substrate protein complex must be characterized. Recently, we solved the crystal structure of Ia in complex with actin and the nonhydrolyzable NAD+ analog βTAD (thiazole-4-carboxamide adenine dinucleotide); however, the structures of the NAD+-bound form (NAD+-Ia-actin) and the ADP ribosylated form [Ia-ADP ribosylated (ADPR)-actin] remain unclear. Accidentally, we found that ethylene glycol as cryo-protectant inhibits ADP ribosylation and crystallized the NAD+-Ia-actin complex. Here we report high-resolution structures of NAD+-Ia-actin and Ia-ADPR-actin obtained by soaking apo-Ia-actin crystal with NAD+ under different conditions. The structures of NAD+-Ia-actin and Ia-ADPR-actin represent the pre- and postreaction states, respectively. By assigning the βTAD-Ia-actin structure to the transition state, the strain-alleviation model of ADP ribosylation, which we proposed previously, is experimentally confirmed and improved. Moreover, this reaction mechanism appears to be applicable not only to Ia but also to other ADP ribosyltransferases.
Keywords: α-actin, mono-ADP ribosylation
Mono- and poly-ADP ribosylation are ubiquitous and important posttranslational modifications in which one or more ADP ribosyl moieties from NAD+ are added to a target protein in a reaction catalyzed by an ADP ribosyl transferase (ART) (1) (Fig. S1A). Mono-ADP ribosylation was originally identified as the pathogenic mechanism of certain bacterial toxins (2), which were classified into several types based on their respective targets. As type I toxins, cholera toxin (3, 4), pertussis toxin (5), and Escherichia coli heat-labile enterotoxin (6) target the cysteine or arginine residues of heteromeric GTP-binding protein. As type II toxins, diphtheria toxin (7) and Pseudomonas exotoxin A modify elongation factor 2 diphthamide. As type III toxins, Clostridium botulinum C3 exotoxin ADP-ribosylates small GTP-binding protein asparagine (8). As type IV toxins, C. botulinum C2 (9) and Clostridium perfringens iota toxin (10) ADP-ribosylate arginine 177 of actin. Recently, a nontypical ADP-ribosylating toxin was discovered: Photorhabdus luminescens TccC3 modifies actin threonine 148 (11).
C. botulinum C2 toxin is an actin-specific ART, and C. perfringens iota-toxin (Ia) has been shown to have striking similarities in both its enzymatic component (C2I or Ia) and binding/translocation component (C2II or Ib). Thus, both C2I and Ia ADP-ribosylate G-actin Arg177, which leads to cytoskeletal disorganization and cell death (12, 13). Interestingly, Ia and C2I recognize subtle differences in the actin molecule; consequently, Ia modifies both α-actin and β-actin, whereas C2I modifies only β-actin. The structures of catalytic components or domains from various ARTs have been determined with and without NAD+ [VIP2 (14), Ia (15), C2I (16), and CDTa (17) as two-component toxin and SpvB (18) as single-component toxins]. In usual two-component toxin, the catalytic component has two similar domains whose C domains and N domains possess ADP ribosyltransferase activity and membrane-binding/translocation activity, respectively. The C domains have a highly similar area of strong electrostatic potential (15). The C-terminal domain shares a conserved β-sandwich core structure consisting of eight β-strands. Around the β-sandwich core, two helices and a loop form the NAD+-binding site. In not only type IV but also type III toxin, the similar structure of the catalytic domain was kept, but differs in the target protein and residue to be modified [C3bot (19, 20), C3stau (21), and C3lim (22)]. Furthermore, the structure of ectoART has revealed strong structural similarity with ART toxins (23). These structural and biochemical studies of ARTs have given us detailed structural information about the NAD+ binding. A particularly important point is that the nicotinamide mononucleotide (NMN) portion is highly folded into a strained conformation within all ARTs (15, 24).
Ia is known to contain three conserved regions: an aromatic residue-R/H, an Glu-X-Glu (EXE) motif, and an STS motif. The EXE motif, which is on the ADP-ribosylating turn-turn (ARTT) loop, is particularly important for the enzyme activity and has been investigated in point mutation and crystallography studies (15). Still, the available information on the structural basis of the catalytic mechanism of ARTs remains limited and incomplete because of the limited information on the ART–substrate protein complex. To understand the mechanism underlying molecular recognition and arginine ADP ribosylation by ART, we recently reported the crystal structure of the Ia–actin complex using a nonhydrolyzable NAD+ analog, βTAD (thiazole-4-carboxamide adenine dinucleotide) (25). The structure of the complex revealed the mechanism of Ia–actin recognition and suggested a possible reaction mechanism. Here we report high-resolution structures of NAD+-Ia-actin (prereaction state) and Ia-ADP ribosylated (ADPR)-actin (postreaction state), as well as apo-Ia-actin and NAD+-Ia (mutant)-actin. Based on these structures from each reaction step, the strain and alleviation mechanism, which we proposed earlier, was experimentally confirmed and improved (25).
Results
Structures of Apo-Ia-Actin, NAD+-Ia-Actin, and Ia-ADPR-Actin.
To investigate the reaction mechanism underlying ART-catalyzed arginine ADP ribosylation, our aim was to examine structural snapshots obtained during the reaction from NAD+ to ADPR-arginine. Unfortunately, cocrystallization of the NAD+-Ia-actin complex failed because ADP ribosylation proceeded in the crystallization buffer. However, we were able to produce small apo-Ia-actin crystals, after which we refined the crystallization conditions to grow larger crystals. The structure of apo-Ia-actin was solved by molecular replacement using βTAD-Ia-actin as a model. Although the relative orientation of apo-Ia-actin differs slightly from βTAD-Ia-actin, the basic structural framework of the complex was retained, which is necessary for the ADP ribosyltransferase reaction. This suggests that the unique apo-Ia-actin complex crystal can be thought of as a reaction chamber with which to examine the ADP ribosylation reaction and the structural changes that occur upon ADP ribosylation (Fig. S1B).
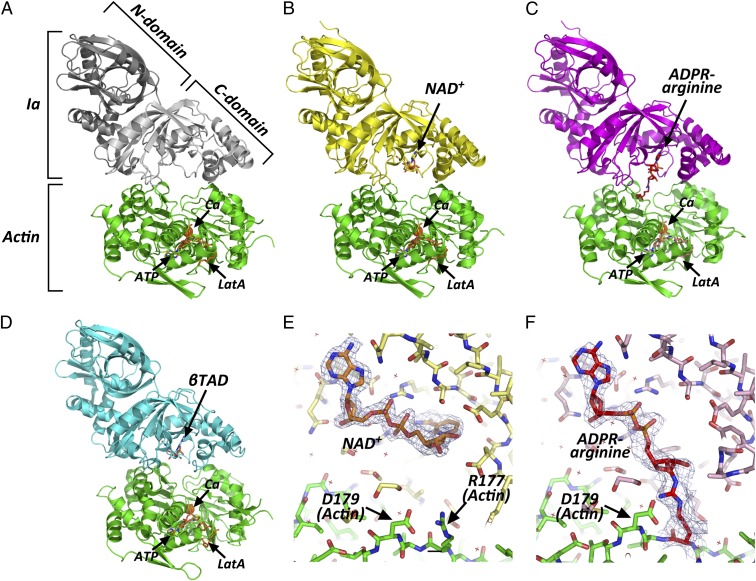
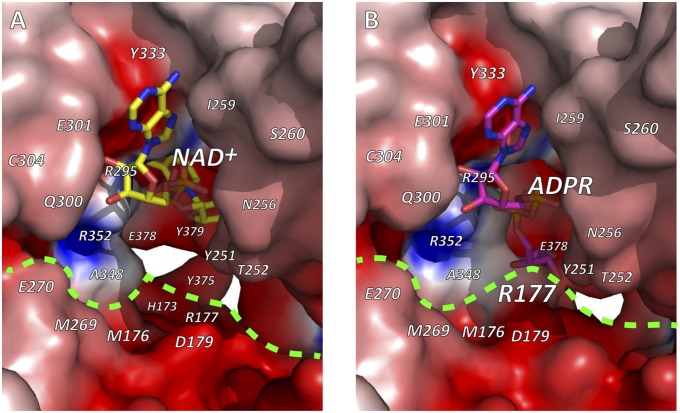
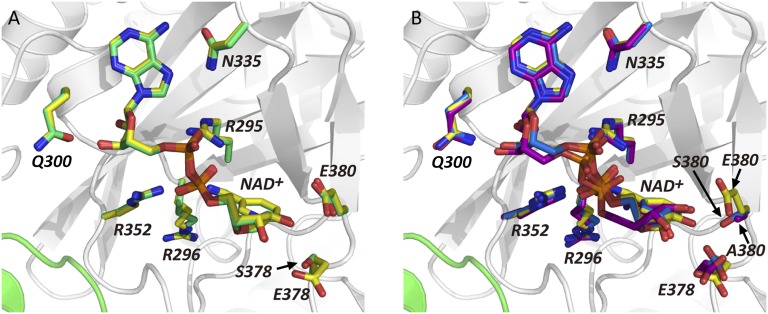
By soaking the apo-Ia-actin crystals with NAD+ under different conditions, we obtained data sets of NAD+-Ia-actin and Ia-ADPR-actin at 1.75 and 2.2 Å resolution, respectively (Table S1). Specifically, soaking apo-Ia(WT)-actin crystals with 10 mM NAD+ in cryo-protectant containing 30% ethylene glycol for 30 min at room temperature yielded NAD+-Ia-actin, whereas soaking the complex with 10 mM NAD+ in mother liquor for 30 min at room temperature yielded Ia-ADPR-actin. Overall, the structures of apo-Ia-actin, NAD+-Ia-actin, and Ia-ADPR-actin were similar to βTAD-Ia-actin, but there were obvious differences around the NAD+-binding site (Fig. 1 A–D). With NAD+-Ia-actin, the difference map showed a clear NAD+ electron density (Fig. 1E). With Ia-ADPR-actin, the 2Fo-Fc (Fo: observed intensity and Fc: calculated intensity) maps showed obvious differences from NAD+-Ia-actin, including the presence of an ADP-ribosylated arginine density instead of NAD+ (Fig. 1F). In NAD+-Ia-actin, the NAD+ conformation is highly folded, as is seen in all ARTs. That is, the ADP moiety was gripped by Ia via Asn335, Gln300, Arg295, and Arg352, and the folded nicotinamide makes a hydrogen bond with the Arg296 main chain carbonyl and nitrogen (Fig. 2). In Ia-ADPR-actin, the ADP moiety is gripped by the same residues as in NAD+-Ia-actin, but the N-ribose was largely moved to Arg177 of actin after nicotinamide cleavage (Fig. 3). It is important to note that there are no water molecules close to NC1 of N-ribose, which suggests that actin binding prevents unfavorable reactions as NADase from proceeding by making water molecules unavailable. The electrostatic molecular surface was shown in each state of the Ia-actin complex (Fig. 4 and Fig. S2). A key feature is that the surface potential is acidic around NAD+ in NAD+-Ia-actin and Ia-ADPR-actin. It seems to stabilize two oxocarbenium ion intermediates as described later. When comparing the NAD+-Ia-actin and Ia-ADPR-actin complexes, another important difference could be seen at Arg177 of actin. Within the ADP-ribosylated actin, the side chain of Arg177 was tilted slightly and interacted covalently with ADPR (Fig. 2). By contrast, in apo-Ia-actin and NAD+-Ia-actin, Arg177 clearly interacted with Asp179 of actin by ionic bond. Using streptavidin-FITC and biotin-NAD+, we confirmed that ADP ribosylation occurs within apo-Ia-actin crystals soaked in mother liquor (Fig. 5A). On the other hand, within apo-Ia (E378S, E380A, and E380S)-actin crystals described in the next section, ADP-ribosylation assays showed no activity (Fig. 5A). In solution assays, the wild-type (WT) complex showed high activity, but ethylene glycol inhibited the ADP ribosylation (Fig. 5B). Within the NAD+-Ia-actin structure, several ethylene glycol molecules were assigned in the structure. None of the ethylene glycol molecules was around the NAD+-binding site, so how they inhibit the reaction is not clear. Nonetheless, the structure of NAD+-Ia-actin was obviously trapped in a prereaction state.
Fig. 1.
Overall structures of Ia-actin complex during the reaction. (A) Apo-Ia-actin; Ia is depicted in white as the C domain and gray as the N domain. (B) NAD+-Ia-actin; Ia and NAD+ are in yellow. (C) Ia-ADPR-actin; Ia is in magenta and ADP-ribosylated R177 is in red. (D) βTAD-Ia-actin (25); Ia and βTAD are in cyan. All actins are in green. NAD+, ADPR-arginine, ATP, and latrunculinA (LatA) are represented in stick formation. Calcium (Ca) is represented as a sphere. (E and F) Electron density around NAD+ within the NAD+-Ia-actin and around ADPR-arginine within the Ia-ADPR-actin complex. NAD+, R177/ADPR-R177, and D179 of actin are labeled. The 2Fo-Fc electron density maps are drawn at 1 σ.
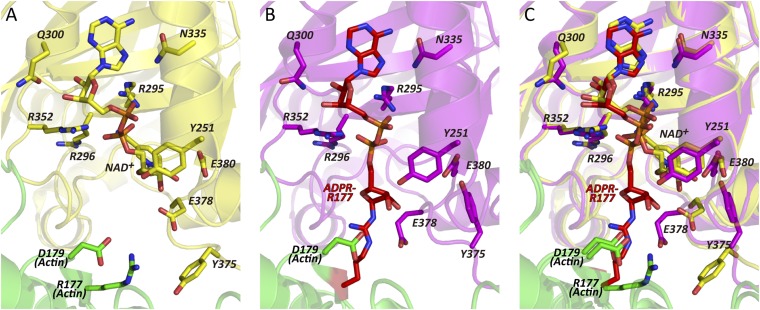
Fig. 2.
Detailed views of the area around NAD+ and ADPR-arginine. (A) Close-up view of NAD+-Ia-actin. (B) Close-up view of Ia-ADPR-actin. (C) Superposed view of NAD+-Ia-actin and Ia-ADPR-actin: all colors are the same as in Fig. 1. Side chains of R177 and D179 in actin are depicted and labeled. In Ia, Y251, R295, R296, Q300, N335, R352, Y375, E378, and E380 are labeled.
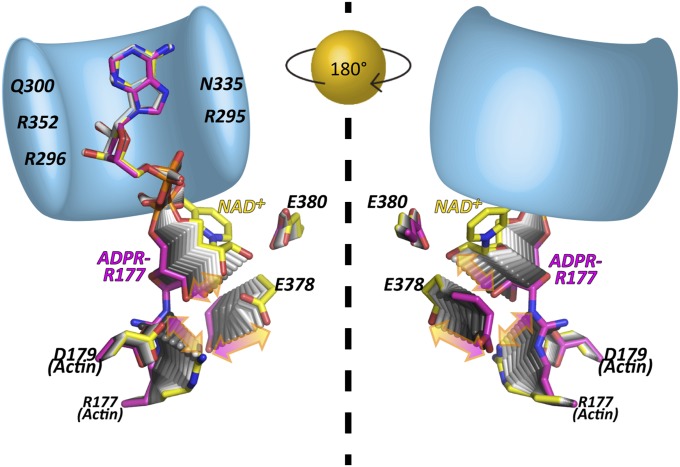
Fig. 3.
Transient structures of the prereaction state [NAD+-Ia-actin (yellow)] and the postreaction state [Ia-ADPR-actin (magenta)]. Schematic presentation of Ia (bright blue: R295, R296, Q300, N335, and R352 are labeled) gripping the ADP moiety of NAD+ or ADPR. The Right panel is a back view of the Left panel.
Fig. 4.
Electrostatic molecular surfaces of the active site of Ia. (A) NAD+-Ia-actin. (B) Ia-ADPR-actin. NAD+ and ADPR are shown in stick representation. Dashed bright green line denotes interface between Ia and actin. Electrostatic molecular surfaces were calculated by GRASP (26) and drawn using PyMOL.
Fig. 5.

Detection of ADP-ribosylated actin. (A) ADP ribosylation assay in apo-Ia (WT or mutant)-actin crystals. ADP ribosylation occurs in the WT complex but not in the mutant complex. (B) Dose-dependent inhibition of ADP ribosylation by ethylene glycol.
Structures of NAD+-Ia (E378S, E380A, and E380S)-Actin.
Ia has strong ART activity, but if actin is absent, it has weak NAD+ glycohydrolase (NADase) activity (15, 27). The latter reaction is the ADP ribosylation of water molecules and is indicative of the NAD+ cleavage rate in the absence of actin. The EXE (Glu378 and Glu380) motif is important for the enzyme activity (Fig. 5A and Fig. S3A), and E380A and E380S mutations eliminated both NADase and ART activity. On the other hand, E378S blocked ART activity, but the enzyme retained about half its NADase activity, compared with WT. Using purified EXE motif mutants (E378S, E380A, and E380S), we solved the structures of the NAD+-Ia (mutants)-actin complexes. The overall crystal structure of NAD+-Ia (mutant)-actin and the electron density of NAD+ were similar to NAD+-Ia (WT)-actin (Table S2, Fig. 6, and Fig. S3 B–E). Not only the artificial structure of NAD+-Ia (WT)-actin trapped in ethylene glycol but also structures of NAD+-Ia (mutant)-actin revealed the nature of NAD+ binding in the complex. It is noteworthy, however, that within the E380A structure, the position of the N-ribose in NAD+ differs slightly from the others. This may explain how Glu380 strongly interacts with the 2′OH of N-ribose and plays an important role in nicotinamide cleavage activity.
Fig. 6.
Detailed views of the area around NAD+ of Ia mutants, E378S, E380A, and E380S. (A) Comparison of WT vs. E378S mutant; wild type of Ia is in yellow and E378S mutant is in green. (B) Comparison of WT vs. two E380 mutants. Wild type is in yellow, E380A mutant is in purple, and E380S mutant is in blue. All actins are in green. Y251, R295, R296, Q300, N335, R352, Y375, E378, and E380 are labeled in Ia.
Plasticity of the ARTT Loop Within the Complex upon ADP Ribosylation.
Successive structural analyses showed that the ARTT loop is the most flexible part of the Ia protein during the ADP-ribosylation reaction and that Tyr375 and Glu378 are present in at least two different states: the apo- and postreaction (ADP-ribosylated) state and the prereaction state (NAD+-bound) (Fig. 2). Tyr375 is oriented inward, toward the toxin, in the apo- and postreaction state, but it is oriented toward the actin in the prereaction state. On the other hand, Glu378 is in close proximity to the 2′OH and 3′OH of the N-ribose in NAD+-Ia-actin (NAD+-binding state), but it is oriented toward actin Arg177 in the apo- and postreaction state. This plasticity of Glu378 is totally different from Glu380, which remains in the same position throughout the reaction. Within the motif, Tyr375 is at the first turn of the ARTT loop, and it was suggested that Tyr375 plays an important role in the recognition of the target protein (19). In the prereaction state, the observed interaction between Tyr375 and actin could serve to stabilize the NAD+-Ia-actin triple complex, but the weak electron density of Tyr375 suggests that the interaction is not very strong. In the second turn of the ARTT loop, Glu378 may be important for recognition of actin Arg177 residue. Experimentally, Glu378 was shown to be essential for both the NADase and ART activities (15). Upon ADP ribosylation, the observed movement of Glu378 (from the apo-reaction state to the prereaction state and from the prereaction to the postreaction state) appears to correlate with the rotation of the N-ribose and explains why Glu378 has dual activity: in the prereaction state, Glu378 acts with Glu380 to induce nicotinamide cleavage and is also important for keeping the oxocarbenium cation from reacting with unfavorable water during the transition from the prereaction to the postreaction state. Another ART, C. botulinum C3, alters cytoskeletal signaling by selectively modifying the low-molecular-mass GTP-binding protein RhoA at Asn41. Although Ia and C3 ADP ribosylate different substrates, they have a strong structural similarity, which suggests that they have a similar substrate amino acid recognition mechanism (14, 19), especially with respect to the ARTT loop. The importance of the ARTT loop in C3 and its plasticity has been discussed elsewhere (28), but the present study sheds light on the role of ARTT plasticity in ADP ribosylation within the structure of the ART-substrate protein complex.
Structural Comparison with βTAD-Ia-Actin.
Although the crystallization conditions and the protein packing of βTAD-Ia-actin were basically the same as those of apo-Ia-actin, NAD+-Ia-actin, and Ia-ADPR-actin, large movements of three helices (H7, H8, and H9) were seen in the βTAD-Ia-actin structure that were not seen in the other structures (Fig. S4). As will be described in the following section, this movement appears to increase the strain on the NMN moiety and to induce a shift toward formation of an oxocarbenium cation. For that reason, we deem βTAD-Ia-actin to be in the transition state SN1. With the exception of Arg177, the structure of the substrate actin molecule was unchanged in all of the structures, including βTAD-Ia-actin.
Reaction Mechanism Inferred from the Structures.
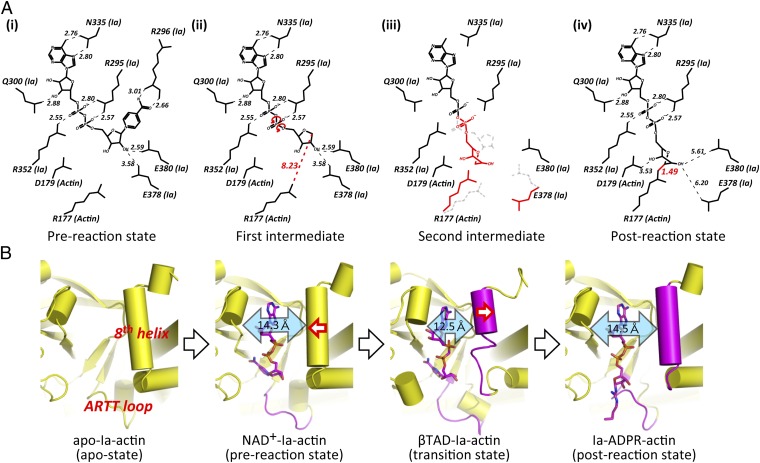
The mechanism by which an SN1 reaction leads to ADP ribosylation of Arg177 was suggested by studies of the Ia structure and its site-directed mutagenesis (15). The positively charged Arg295 and Arg352 (Ia) interact electrostatically with NAD+ phosphate and contribute to the highly folded and strained conformation of the NMN ring-like conformation. This specific structure of NAD+ is conserved among all ART family members. The specific conformation appears to induce an equilibrium shift toward formation of an oxocarbenium cation. After cleavage of the nicotinamide, the oxocarbenium cation may be stabilized via Tyr251 through a cation-pi interaction (29). In the present NAD+-Ia-actin structure, the distance between the nucleophile (Arg177) and the electrophile (NC1 of N-ribose) is 8.2 Å, making it necessary to reduce the distance between Arg177 and NC1 of N-ribose. In an earlier paper, we proposed a strain-alleviation model in which a second oxocarbenium ion acts as an intermediary (25). In that model, “strain” referred to the highly folded and strained conformation of NMN, whereas “alleviation” referred to the rotation of the N-ribose after scission of the nicotinamide. That is, SN1 cleavage produced the first oxocarbenium cation, after which rotation occurred via NP-NO5 of ADP ribose to produce a second cationic intermediate, which enabled NC1 of N-ribose to approach the guanidyl nitrogen of Arg177. In the present pre- and postreaction state structural study, the strain-alleviation model was confirmed experimentally and was improved. The key features of the improved model are as follows (Fig. 7A) (1). The ADP moiety is gripped by Ia, and then the ADP ribosylation reaction occurs. The grip is necessary for the subsequent rotation (2). To reduce the distance between the nucleophile (Arg177) and the electrophile (NC1 of N-ribose), after the first oxocarbenium ion intermediate is produced (SN1 reaction), the rotation of N-ribose occurs mainly via rotation of both O3-NP and NP-NO5 (3). The present structure confirms that the N-ribose 3′OH is in close proximity to Asp179 (3.5 Å) within Ia-ADPR-actin. We envision that actin Asp179 plays a stabilizing role by making contact with the N-ribose (4). Arg177 tilts slightly to react with N-ribose.
Fig. 7.
Schematic illustrating the mechanism of ADP ribosylation. (A) SN1 mechanism in Ia: (i) NAD+-Ia-Actin as the prereaction state; (ii) nicotinamide cleavage occurs via an SN1 reaction induced by an NMN ring-like structure and the first oxocarbenium cation intermediate is formed with a strained conformation; (iii) the second cationic intermediate is induced through alleviation of the strained conformation mainly by O3-NP and NP-NO5 rotation, and then NC1 of N-ribose nucleophilically attacks Arg177 of actin; (iv) Ia-ADPR-actin as the postreaction state. (B) Successive structures during ADP ribosylation and the structure of each reaction: step 1 (apo-Ia-actin), step 2 [NAD+-Ia-actin (prereaction state)], step 3 [βTAD-Ia-actin (transition state)], and step 4 [Ia-ADPR-actin (postreaction state)].
The successive structures of each reaction step are shown in Fig. 7B: the apo-state, the prereaction state, βTAD-Ia-actin as a transition state in the SN1 reaction, and the postreaction state. We speculate that the observed movement of the helices in βTAD-Ia-actin increases the strain on the nicotinamide of NAD+ so that it proceeds to the first SN1 cleavage efficiently. In summary, a simple strain-alleviation model can explain arginine ADP ribosylation occurring via two oxocarbenium ion intermediates.
Discussion
We report the structure of the NAD+-Ia-actin and Ia-ADPR-actin complexes, providing insight into the reaction mechanism of ADP ribosylation. Arginine ADP ribosylation is a modification catalyzed by both bacterial toxins and human membrane-associated ARTs. The trapped ADPR structure will provide important information about the reaction mechanism underlying ADP ribosylation of arginine. Margarit et al. reported the crystal structures of SpvB-modified ADPR-actin (18). However, their ADPR-actin structure did not include SpvB, so the modified residue was not visible due to the plasticity of the complex. Our structure of ADPR-arginine was trapped because Ia gripped the ADP moiety of ADP-ribosylated arginine. As mentioned above, we were unable to obtain NAD+ cocrystals because Ia and actin separate after ADP ribosylation. However, NAD+ soaking worked well because Ia-ADP ribosylated actin was retained within the crystal packing. Given that the structure of SpvB-modified ADPR-actin does not change from unmodified G-actin, the mechanism of inhibition of actin polymerization is thought to occur primarily through steric disruption of intrafilament contacts by the ADP-ribosylated Arg177 (18, 30). Within the crystal structure of Ia-ADPR-actin, the ADP ribosyl moiety was observed, but in solution without toxin, the flexible ADP ribosyl moiety was situated near Arg177, where it disrupted actin-actin contacts and thereby inhibited actin polymerization.
As described before (24), the NMN moiety binds in a similar compact fashion in all ART. On the other hand, the AMP moiety shows variation. Remarkably, the adenine ribose is oriented differently in type II diphtheria toxin group. Recently, the conformational analysis of NAD+ was reported (31). The conformation is different in NAD+ bound to redox enzyme and nonredox enzyme. A specific feature of ARTs is a small ChiN value that is never seen in redox NAD. Actually, the ChiN torsion angle of NAD+-Ia-actin is 3.1 ° (ChiA = −115°, GammaA = 53.2°, and ZetaA = −158.0°). Although NAD+ had been postulated to strain the N-glycosidic bond, NAD+ with different ChiN values was found to exhibit similar C1D–N1N bond cleavage barriers in water. As a next step, however, we need to evaluate the bond barriers, including supporting amino acids near NMN not in water. Two glutamates (378 and 380 in Ia) are especially important for the catalysis. The bond cleavage is determined not only by the physical property of the specific conformation of NAD+ but also by a chemical factor. Also, our strain and alleviation model is an important description of the trigger of conformational change after cleavage of nicotinamide. There should be an equilibrium between NAD+ and oxocarbenium cation/nicotinamide. If there is still strain in the first oxocarbenium cation intermediate, it creates the second conformation of the oxocarbenium cation intermediate by alleviation and finally interacts with arginine. It seems that the strain strongly supports the forward reaction not to go backward. Finally, we summarized the residue–residue interactions between Ia and actin and the properties of the key residues (Tables S3–S7).
Another available structure of an ART-substrate complex is that of exotoxin A-elongation factor 2 (ETA-EF2). With ETA, the modified residue is not arginine but diphthamide (a modified histidine) (32, 33). Jørgensen et al. reported that the diphthamide N3 nucleophilic atom remains about 10 Å from the electrophilic C1 center of the N-ribose within the complex; consequently, one additional step is necessary for consummation of the nucleophilic substitution reaction (32). They proposed a model in which the EF2 loop, which includes the diphthamide residue, migrates toward the N-ribose. If this does occur, a similar strain-alleviation model may also explain the ADP-ribosylation reaction: the L1 and L3 loops of ETA grip the ADP until the N-ribose rotates to react with the diphthamide N3 after scission of the nicotinamide. They also showed that a substrate acidic residue is important and that Asp696 forms a hydrogen bond with the N-ribose 2′OH within ADPR-eEF2, and they suggested that an Asp696 analog likely interacts with the N-ribose hydroxyl group during or after the arginine ADP ribosylation of Gα’s and actin (32). Given the βTAD-Ia-actin structure, we previously suggested that actin Asp179 serves that function and forms a hydrogen bond with N-ribose (25). In the present study, we confirmed this to be true within the Ia-ADPR-actin structure. Our strain and alleviation model is compared with the old model as mentioned in previous report (25) (Fig. S5).
ADP ribosyl transferases have been identified not only in bacteria but also in eukaryotic cells. Recently, a classification was proposed based on the key sequence similarity among all ARTs, including nonbacterial ARTs (34, 35). Within this scheme, ARTs are classified into two large groups: ARTC (R-S-E group: C2I, Ia and C3) and ARTD (H-Y-E group: diphtheria toxin group) (34). Mammalian and avian ARTs, which are known to regulate a variety of important cell functions, including immune responses, cell adhesion, cell signaling, and metabolism (36), belong to the ARTC group. Poly-ADP ribosyl transferases act as sensors of DNA strand breaks and catalyze poly-ADP ribosylation of themselves and various target proteins (37). Poly-ADP ribosyl transferases belong to the ARTD group. Although the target proteins and the residues modified by these ARTs differ, their basic core structural similarity, the structural similarity of the strained conformations of NAD+, and the common acidic residue in the substrate protein needed to fix the N-ribose suggest that all ARTs make use of a common reaction mechanism. The high-resolution structure and simple reaction mechanism reported here will provide an understanding of the reaction. Finally, the unique complex structure also provides valuable information needed to design inhibitors of ART family proteins (38).
Materials and Methods
Sample preparation, crystallization, NAD+ soaking, data collection, structure determination, assay of ADP ribosylation, and assay of NADase activity using FPLC were carried out as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff at KouEnerugii ButsurigakuKenkyusho-Photon Factory (KEK-PF) for data collection and T. Yoshida for reading of the manuscript. This work was supported in part by a Strategic Research Foundation Grant-Aided Project for Private Universities; by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan; and by Grants-in-Aid for Scientific Research, MEXT of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.G.M. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4GY2 [apo-Ia-actin], 4H03 [NAD+-Ia-actin], and 4H0T [Ia-ADPR-actin]; and 4H0V, 4H0X, and 4H0Y [NAD+-Ia (E378S, E380A, E380S)-actin]).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217227110/-/DCSupplemental.
See Commentary on page 4163.
References
- 1.Ueda K, Hayaishi O. ADP-ribosylation. Annu Rev Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- 2.Gill DM, Pappenheimer AM, Jr, Brown R, Kurnick JT. Studies on the mode of action of diphtheria toxin. VII. Toxin-stimulated hydrolysis of nicotinamide adenine dinucleotide in mammalian cell extracts. J Exp Med. 1969;129(1):1–21. doi: 10.1084/jem.129.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moss J, Vaughan M. Mechanism of action of choleragen. Evidence for ADP-ribosyltransferase activity with arginine as an acceptor. J Biol Chem. 1977;252(7):2455–2457. [PubMed] [Google Scholar]
- 4.Gill DM, Meren R. ADP-ribosylation of membrane proteins catalyzed by cholera toxin: Basis of the activation of adenylate cyclase. Proc Natl Acad Sci USA. 1978;75(7):3050–3054. doi: 10.1073/pnas.75.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katada T, Ui M. Direct modification of the membrane adenylate cyclase system by islet-activating protein due to ADP-ribosylation of a membrane protein. Proc Natl Acad Sci USA. 1982;79(10):3129–3133. doi: 10.1073/pnas.79.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moss J, Garrison S, Oppenheimer NJ, Richardson SH. NAD-dependent ADP-ribosylation of arginine and proteins by Escherichia coli heat-labile enterotoxin. J Biol Chem. 1979;254(14):6270–6272. [PubMed] [Google Scholar]
- 7.Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem. 1980;255(22):10717–10720. [PubMed] [Google Scholar]
- 8.Aktories K, Weller U, Chhatwal GS. Clostridium botulinum type C produces a novel ADP-ribosyltransferase distinct from botulinum C2 toxin. FEBS Lett. 1987;212(1):109–113. doi: 10.1016/0014-5793(87)81566-1. [DOI] [PubMed] [Google Scholar]
- 9.Aktories K, et al. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322(6077):390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- 10.Vandekerckhove J, Schering B, Bärmann M, Aktories K. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 1987;225(1–2):48–52. doi: 10.1016/0014-5793(87)81129-8. [DOI] [PubMed] [Google Scholar]
- 11.Lang AE, et al. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science. 2010;327(5969):1139–1142. doi: 10.1126/science.1184557. [DOI] [PubMed] [Google Scholar]
- 12.Aktories K, Wegner A. Mechanisms of the cytopathic action of actin-ADP-ribosylating toxins. Mol Microbiol. 1992;6(20):2905–2908. doi: 10.1111/j.1365-2958.1992.tb01749.x. [DOI] [PubMed] [Google Scholar]
- 13.Aktories K, Lang AE, Schwan C, Mannherz HG. Actin as target for modification by bacterial protein toxins. FEBS J. 2011;278(23):4526–4543. doi: 10.1111/j.1742-4658.2011.08113.x. [DOI] [PubMed] [Google Scholar]
- 14.Han S, Craig JA, Putnam CD, Carozzi NB, Tainer JA. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat Struct Biol. 1999;6(10):932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- 15.Tsuge H, et al. Crystal structure and site-directed mutagenesis of enzymatic components from Clostridium perfringens iota-toxin. J Mol Biol. 2003;325(3):471–483. doi: 10.1016/s0022-2836(02)01247-0. [DOI] [PubMed] [Google Scholar]
- 16.Schleberger C, et al. Structure and action of the binary C2 toxin from Clostridium botulinum. J Mol Biol. 2006;364(4):705–715. doi: 10.1016/j.jmb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Sundriyal A, Roberts AK, Shone CC, Acharya KR. Structural basis for substrate recognition in the enzymatic component of ADP-ribosyltransferase toxin CDTa from Clostridium difficile. J Biol Chem. 2009;284(42):28713–28719. doi: 10.1074/jbc.M109.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margarit SM, Davidson W, Frego L, Stebbins CE. A steric antagonism of actin polymerization by a Salmonella virulence protein. Structure. 2006;14(8):1219–1229. doi: 10.1016/j.str.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Han S, Arvai AS, Clancy SB, Tainer JA. Crystal structure and novel recognition motif of rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: Structural insights for recognition specificity and catalysis. J Mol Biol. 2001;305(1):95–107. doi: 10.1006/jmbi.2000.4292. [DOI] [PubMed] [Google Scholar]
- 20.Ménétrey J, et al. NAD binding induces conformational changes in Rho ADP-ribosylating clostridium botulinum C3 exoenzyme. J Biol Chem. 2002;277(34):30950–30957. doi: 10.1074/jbc.M201844200. [DOI] [PubMed] [Google Scholar]
- 21.Evans HR, et al. The crystal structure of C3stau2 from Staphylococcus aureus and its complex with NAD. J Biol Chem. 2003;278(46):45924–45930. doi: 10.1074/jbc.M307719200. [DOI] [PubMed] [Google Scholar]
- 22.Vogelsgesang M, Stieglitz B, Herrmann C, Pautsch A, Aktories K. Crystal structure of the Clostridium limosum C3 exoenzyme. FEBS Lett. 2008;582(7):1032–1036. doi: 10.1016/j.febslet.2008.02.051. [DOI] [PubMed] [Google Scholar]
- 23.Mueller-Dieckmann C, Ritter H, Haag F, Koch-Nolte F, Schulz GE. Structure of the ecto-ADP-ribosyl transferase ART2.2 from rat. J Mol Biol. 2002;322(4):687–696. doi: 10.1016/s0022-2836(02)00818-5. [DOI] [PubMed] [Google Scholar]
- 24.O’Neal CJ, Jobling MG, Holmes RK, Hol WG. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309(5737):1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- 25.Tsuge H, et al. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin. Proc Natl Acad Sci USA. 2008;105(21):7399–7404. doi: 10.1073/pnas.0801215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrey D, Honig B. GRASP2: Visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- 27.van Damme J, et al. Analysis of the catalytic site of the actin ADP-ribosylating Clostridium perfringens iota toxin. FEBS Lett. 1996;380(3):291–295. doi: 10.1016/0014-5793(96)00052-x. [DOI] [PubMed] [Google Scholar]
- 28.Ménétrey J, Flatau G, Boquet P, Ménez A, Stura EA. Structural basis for the NAD-hydrolysis mechanism and the ARTT-loop plasticity of C3 exoenzymes. Protein Sci. 2008;17(5):878–886. doi: 10.1110/ps.073398508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96(17):9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347(6288):44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 31.Kuppuraj G, Sargsyan K, Hua YH, Merrill AR, Lim C. Linking distinct conformations of nicotinamide adenine dinucleotide with protein fold/function. J Phys Chem B. 2011;115(24):7932–7939. doi: 10.1021/jp1118663. [DOI] [PubMed] [Google Scholar]
- 32.Jørgensen R, et al. Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature. 2005;436(7053):979–984. doi: 10.1038/nature03871. [DOI] [PubMed] [Google Scholar]
- 33.Jørgensen R, Wang Y, Visschedyk D, Merrill AR. The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep. 2008;9(8):802–809. doi: 10.1038/embor.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35(4):208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Laing S, Unger M, Koch-Nolte F, Haag F. ADP-ribosylation of arginine. Amino Acids. 2011;41(2):257–269. doi: 10.1007/s00726-010-0676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corda D, Di Girolamo M. Mono-ADP-ribosylation: A tool for modulating immune response and cell signaling. Sci STKE. 2002;2002(163):pe53. doi: 10.1126/stke.2002.163.pe53. [DOI] [PubMed] [Google Scholar]
- 37.Schreiber V, et al. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277(25):23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 38.Shniffer A, et al. Characterization of an actin-targeting ADP-ribosyltransferase from Aeromonas hydrophila. J Biol Chem. 2012;287(44):37030–37041. doi: 10.1074/jbc.M112.397612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.