Abstract
The number of nicotinic acetylcholine receptors (AChRs) present in the plasma membrane of muscle and neuronal cells is limited by the assembly of individual subunits into mature pentameric receptors. This process is usually inefficient, and a large number of the synthesized subunits are degraded by endoplasmic reticulum (ER)-associated degradation. To identify cellular factors required for the synthesis of AChRs, we performed a genetic screen in the nematode Caenorhabditis elegans for mutants with decreased sensitivity to the cholinergic agonist levamisole. We isolated a partial loss-of-function allele of ER membrane protein complex-6 (emc-6), a previously uncharacterized gene in C. elegans. emc-6 encodes an evolutionarily conserved 111-aa protein with two predicted transmembrane domains. EMC-6 is ubiquitously expressed and localizes to the ER. Partial inhibition of EMC-6 caused decreased expression of heteromeric levamisole-sensitive AChRs by destabilizing unassembled subunits in the ER. Inhibition of emc-6 also reduced the expression of homomeric nicotine-sensitive AChRs and GABAA receptors in C. elegans muscle cells. emc-6 is orthologous to the yeast and human EMC6 genes that code for a component of the recently identified ER membrane complex (EMC). Our data suggest this complex is required for protein folding and is connected to ER-associated degradation. We demonstrated that inactivation of additional EMC members in C. elegans also impaired AChR synthesis and induced the unfolded protein response. These results suggest that the EMC is a component of the ER folding machinery. AChRs might provide a valuable proxy to decipher the function of the EMC further.
Genes coding for nicotinic acetylcholine receptor (AChR) subunits form an evolutionarily conserved family, which includes a large repertoire of ionotropic receptors gated by acetylcholine (ACh). In mammals, there are at least 16 different genes encoding AChR subunits. Each subunit is made up of a large extracellular domain, four transmembrane segments (M1–M4), and a variably long intracellular loop between M3 and M4. Individual subunits assemble into homo- or heteropentameric receptors with distinct pharmacological and biophysical properties (reviewed in ref. 1). Interestingly, the assembly of AChRs seems rather slow and inefficient, which is a feature common to several multimeric transmembrane complexes, such as GABAA receptors (2) or cystic fibrosis transmembrane conductance regulator (CFTR) receptors (3). In vertebrate muscle cells, only 30% of the synthesized α subunits finally assemble into a mature pentameter (4). In neurons, a significant number of AChRs are also found in intracellular compartments (5). Although the physiological significance of AChR intracellular retention is not completely understood, it potentially has diverse pathological implications. For example, a rare mutation in the human β4 AChR subunit has been linked to amyotrophic lateral sclerosis (6), and this mutation was shown to impair the export of α4β4 AChRs from the endoplasmic reticulum (ER) (7). ER retention of AChRs was also proposed to influence ER stress and the unfolded protein response (UPR) in dopaminergic neurons (8). In the smoker’s brain, an enhancement of AChR expression likely contributes to tobacco addiction. This up-regulation involves a chaperone activity of nicotine, which enhances the assembly of specific receptor species and their export from the ER, together with a reduced degradation of AChR subunits by the proteasome system (9–11).
The overall efficiency of AChR biosynthesis is dependent on multiple processes in the ER and in early secretory compartments (reviewed in ref. 12). AChR subunits enter the ER cotranslationally in an unfolded state via the Sec61 translocon. Transient interactions with ER resident components of the biosynthetic machinery, such as BiP (13, 14), calnexin (15, 16), and ERp57 (17), then influence the folding process and protect immature subunits from degradation. Folding subunits are exposed to a variety of enzymes that catalyze posttranslational modifications (including glycosylation and palmitoylation) and regulate protein expression (18–20). Assembly of the subunits is sequential, and intermediates of certain receptors, such as muscle AChRs, have sufficient t1/2s to be isolated and characterized (reviewed in ref. 21). ER retention or retrieval signals prevent the exit of unassembled or partially assembled AChR subunits. For example, an ER retention signal present in the M1 transmembrane segment of the subunits is masked by receptor assembly to allow ER exit (22). Unassembled subunits are eventually degraded through the ER-associated degradation (ERAD) machinery (23). ER export signals have also been demonstrated to enhance the recruitment of receptors into ER exit sites (24). In addition, degradation signals present in the cytoplasmic M3–M4 subunit loop (25, 26) must be masked for ER exit licensing. The cytoplasmic M3–M4 loop is also involved in interactions with adaptor proteins, such as the small cytoplasmic chaperone 14-3-3 (27) or the UBXD4 protein (28).
Despite its physiological and pathological importance, relatively few cellular factors are known to control AChR biosynthesis at early steps. This might be due to the difficulty in identifying labile interactions with partially assembled receptors or because receptors interact with large cellular complexes. Genetic screens in model organisms provide a means to identify relevant components in biological networks involved in this specific process. Specifically, the nematode Caenorhabditis elegans is a powerful model organism with which to dissect AChR biosynthesis genetically. ACh is the main excitatory neurotransmitter in C. elegans, and at least 29 genes encode AChR subunits (29). Two types of ionotropic AChRs, heteromeric levamisole-sensitive AChRs (L-AChRs) and homomeric nicotine-sensitive AChRs (N-AChRs), are present at the neuromuscular junction (NMJ), the best-characterized cholinergic synapse in the worm (30). L-AChRs are heteropentameric receptors that can be activated by the nematode-specific cholinergic agonist levamisole. Levamisole causes hypercontraction of C. elegans body-wall muscles (BWMs) and death of WT worms at high concentrations (31, 32). Genetic screens for resistance to levamisole have identified the structural subunits of the receptor, including three α subunits (LEV-8, UNC-38, and UNC-63) and two non-α subunits (LEV-1 and UNC-29). The second type of receptor, N-AChR, is activated by nicotine and is insensitive to levamisole. It is most likely homomeric, composed of five ACR-16 subunits (33, 34).
Apart from receptor subunits, genetic screens identified three auxiliary proteins (RIC-3, UNC-50, and UNC-74) required for receptor biosynthesis. RIC-3 was identified in a suppressor screen of neuronal degeneration caused by a gain-of-function mutation in the C. elegans AChR composed of DEG-3 and DES-2 subunits (35). RIC-3 is a small integral membrane protein located in the ER that is required for the assembly of at least five distinct AChR receptors in C. elegans, including L-AChRs and N-AChRs (35–37). RIC-3 is conserved in flies and mammals, and it can either promote or inhibit the expression of AChRs and 5-HT3 receptors in heterologous systems (reviewed in ref. 12). Recent analysis suggests that RIC-3 activity depends on its expression level; at low levels, RIC-3 behaves as an ER chaperone promoting AChR assembly, whereas high expression levels cause ER retention through long-lived interactions with AChRs (38). UNC-50 was identified in a screen for resistance to levamisole. UNC-50 is the ortholog of GMH1, a transmembrane protein conserved from yeast to humans, which interacts with a guanine nucleotide exchange factor of the small G protein Arf and potentially regulates COPI-dependent trafficking events. UNC-50 localizes to the Golgi apparatus and promotes the targeting of L-AChRs to the plasma membrane by preventing routing and degradation in lysosomes (39). UNC-74 is a predicted thioredoxin necessary for L-AChR expression, but its role has not been fully characterized. Interestingly, expression of L-AChRs in Xenopus oocytes not only requires the five receptor subunits but the three auxiliary proteins RIC-3, UNC-50, and UNC-74 as well (36). Recently, the ER resident transmembrane proteins NRA-2/nicalin (nicastrin-like protein) and NRA-4/nodal modulator (NOMO), were proposed to regulate L-AChR subunit composition and stoichiometry (40).
To identify additional components required for AChR assembly and folding, we designed a genetic screen for mutants with only partially decreased sensitivity to the cholinergic agonist levamisole, because screens for complete resistance to levamisole are likely saturated. Such mutants completely paralyze on high levamisole concentration within a few hours but subsequently adapt within 12–16 h and recover motility in contrast to WT worms (41, 42). Here, we identify and characterize in vivo the ER membrane protein complex-6 (emc-6) gene. We demonstrate that EMC-6 is required for the proper assembly of AChRs through the function of an evolutionarily conserved complex involved in ER homeostasis.
Results
Disruption of the Evolutionarily Conserved Gene emc-6 Confers Partial Resistance to the Cholinergic Agonist Levamisole.
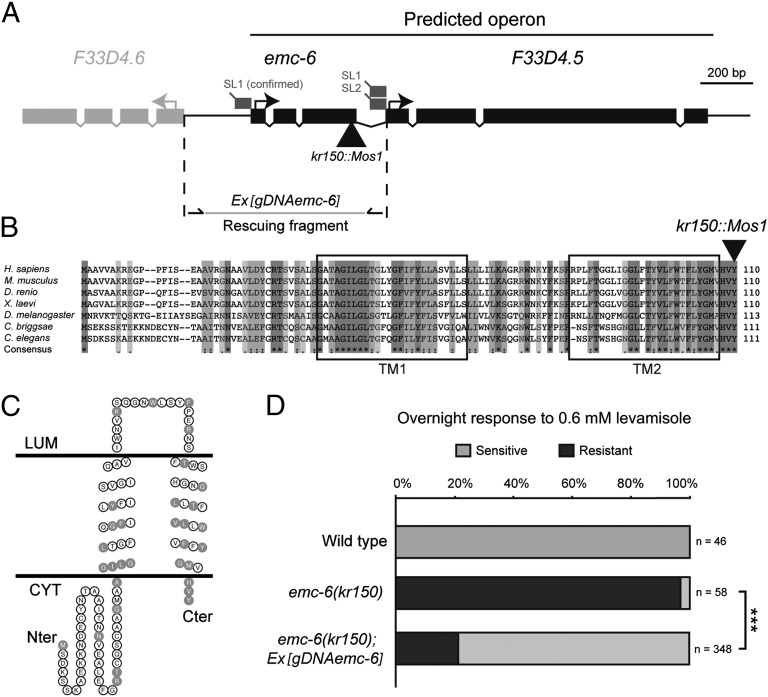
To identify new components involved in AChR metabolism, we used Mos1-mediated insertional mutagenesis to generate mutants resistant to the cholinergic agonist levamisole (42). A strain carrying the mutant allele kr150 was isolated based on its ability to adapt to overnight exposure to 0.6 mM levamisole. A Mos1 insertion was identified in kr150 by inverse PCR in the ORF F33D4.7 (Fig. 1A). This gene is conserved from yeast to humans (Fig. 1B and Table S1), and it was tentatively named emc-6 because of its homology with the yeast gene EMC6 (43). Based on cDNA sequencing, the C. elegans emc-6 messenger is transspliced to the splice leader SL1 and encodes a predicted 111-aa protein with two transmembrane domains (Fig. 1C). Topology prediction programs suggest that the N-terminal region of the protein is cytosolic. The decreased sensitivity to levamisole of kr150 mutants was rescued by providing a 900-bp genomic fragment containing the emc-6 coding region (Fig. 1 A and D), thus demonstrating that mutating emc-6 causes the partial resistance to levamisole in this mutant.
Fig. 1.
emc-6 encodes a conserved transmembrane protein. (A) Structure of the emc-6 genomic locus. emc-6 is predicted to be the upstream gene of an operon (www.wormbase.org). Black boxes represent the coding regions, and gray boxes represent the 5′ untranslated region. The black triangle illustrates the site of Mos1 insertion in kr150. SL1, SL1 transspliced leader; SL2, SL2 transspliced leader. (B) ClustalX alignment of C. elegans EMC-6 with orthologs from nematodes, fly, and vertebrates. Predicted transmembrane regions (TM1 and TM2) are boxed. Residues conserved in all species are highlighted in dark gray, residues with strongly similar properties (scoring >0.5) are highlighted in medium gray, and residues with weakly similar properties (scoring ≤0.5) are highlighted in light gray. The position of the kr150 mutation is indicated by a triangle. (C) EMC-6 topology model based on TMHMM and Topo2 predictions. Residues conserved in all species are labeled in gray. Cter, C-terminus; CYT, cytoplasm; LUM, lumen; Nter, N-terminus. (D) Expression of an emc-6 genomic fragment rescues levamisole resistance in kr150 mutants. Gray bars illustrate the percentage of dead animals after overnight exposure to 0.6 mM levamisole, and black bars illustrate the percentage of surviving animals. Three independent transgenic lines were tested. n, number of animals tested. ***P < 0.001, Fisher exact probability test.
Nonquantitative RT-PCR analysis demonstrated that emc-6 transcript remains in kr150 mutants. Insertion of the Mos1 transposon causes the replacement of the conserved ultimate amino acid by four unrelated residues followed by a STOP codon, a cryptic polyadenylation site being used in the Mos1 sequence (Fig. S1A). Quantitative RT-PCR analysis indicated, however, that the level of this emc-6 mRNA was severely reduced compared with the WT [21 ± 5% at L2 stage and 8 ± 1% at L4 stage (mean ± SEM); n = 4] (Fig. S1B). These data suggested that emc-6(kr150) is a strong loss-of-function mutant but may not be a null allele (details are provided in Fig. S2). Our attempts to generate a second allele of emc-6 by noncomplementation screening after EMS mutagenesis (30,155 F1 animals screened) was unsuccessful, suggesting that kr150 is a particular hypomorphic allele of emc-6 and that emc-6 might be an essential gene.
Apart from resistance to levamisole, the kr150 mutation is associated with general phenotypic defects, including embryonic lethality, growth retardation, and reduced brood size (Fig. S2). Unfortunately, the rescue of these phenotypes could not be quantitatively scored because the transgene expressing emc-6 from a genomic fragment was, by itself, causing partial embryonic lethality and significant developmental defects. Because adaptive resistance to levamisole is usually not associated with developmental phenotypes (e.g., lev-10, lev-9, oig-4, molo-1 mutant), these data suggest that emc-6 supports general functions apart from its role in cholinergic neurotransmission.
EMC-6 Is Required in Muscle for L-AChR Expression.
To characterize the emc-6 expression pattern, we built a transcriptional reporter expressing GFP under the control of emc-6 regulatory sequences. Surprisingly, we observed that transgenes driving GFP expression under the control of the 263-bp promoter sequence used for emc-6 rescue experiments induced lethality and did not transmit GFP expression after one generation. Despite significant efforts, we failed to establish stable expression of this transcriptional reporter and could only acquire pictures of mosaic F1 animals (Fig. S3A). In these mosaic animals, GFP was detected in almost all tissues, including neurons and muscle cells. The very unusual toxicity of a small promoter fragment suggested that it might trigger sequence-specific epigenetic mechanisms. To bypass this technical issue, we characterized the expression pattern of the Caenorhabditis briggsae ortholog of emc-6 (Cb-emc-6) in C. elegans. Cb-EMC-6 and C. elegans Ce-EMC-6 protein sequences are 98% identical. Expression of the genomic locus of Cb-emc-6 in emc-6(kr150) was sufficient to rescue the levamisole resistance of emc-6(kr150) mutants (Fig. S3B). Interestingly, transgenic expression of Cb-emc-6 was nontoxic in C. elegans. We therefore expressed a nuclear GFP under the control of the C. briggsae-promoting sequence of emc-6 in C. elegans (Fig. S3C). We were able to detect GFP expression in every tissue from the threefold stage to adulthood. Altogether, these results suggest that emc-6 is ubiquitously expressed.
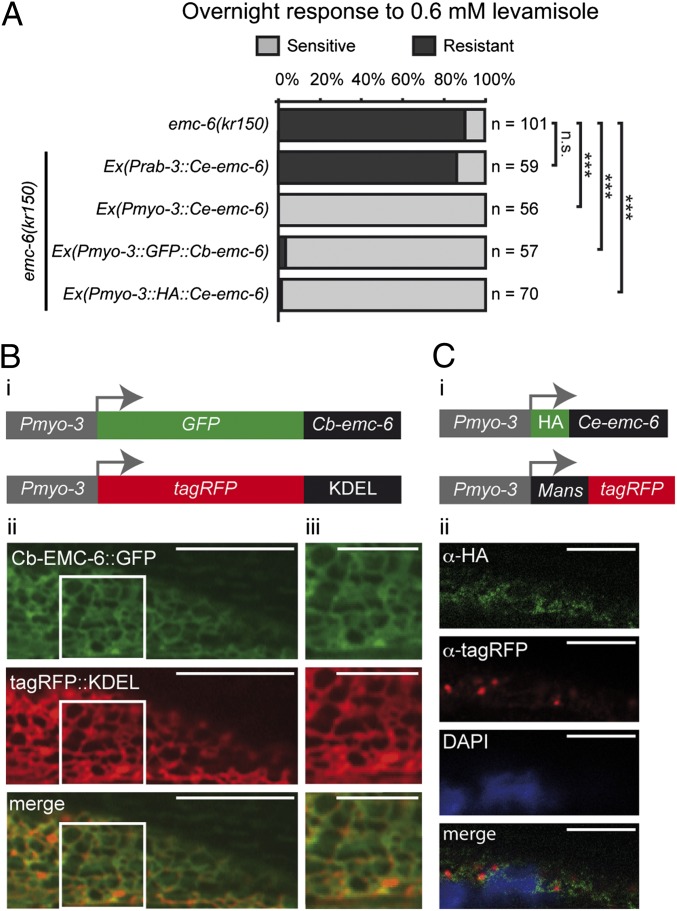
To test in which tissue EMC-6 was required for L-AChR expression, we used the muscle-specific Pmyo-3 promoter or the neuron-specific Prab-3 promoter to express the emc-6 cDNA in emc-6(kr150) mutants (Fig. 2A). Unlike the Prab-3::emc-6 construct, Pmyo-3::emc-6 fully rescued levamisole resistance of emc-6(kr150) mutants. This indicates that emc-6 functions in BWMs to promote L-AChR surface expression.
Fig. 2.
emc-6 is required in muscle cells for L-AChR function and localizes to the ER. (A) Expression of an emc-6 cDNA and GFP::Cb-EMC-6 or HA::Ce-EMC-6 translational fusion in muscle (Pmyo-3 promoter) but not in neurons (Prab-3 promoter) rescues levamisole resistance of emc-6(kr150) mutants. n, number of tested animals. n.s. (nonsignificant), P > 0.05; ***P < 0.001, Fisher exact probability test. (B) GFP::Cb-EMC-6 fusion proteins localize to the ER. (i) GFP::Cb-EMC-6 and tagRFP::KDEL translational fusions used for expression in BWM (Pmyo-3 promoter). (ii) Live images of GFP::Cb-EMC-6 and tagRFP::KDEL displaying a reticular staining pattern throughout the cytoplasm surrounding the nucleus. (Scale bars, 5 μm.) (iii) Magnified view of the reticulum network. (Scale bars, 2.5 μm.) (C) HA::Ce-EMC-6 fusion does not colocalize with a Golgi-resident tagRFP-tagged Mannosidase II protein (MANS::tagRFP). (i) HA::Ce-EMC-6 and MANS::tagRFP used for expression in BWMs (Pmyo-3 promoter). (ii) Immunostaining with anti-HA and anti-tagRFP antibodies. DAPI staining of cell nuclei. (Scale bars, 5 μm.)
EMC-6 Localizes to the ER.
To assay the subcellular localization of the EMC-6 protein, we expressed an N-terminally tagged EMC-6 construct in the BWMs. As mentioned above, transgenes driving Ce-emc-6 expression were extremely toxic, and we could only generate a single transgenic line, prompting us to express a tagged version of C. briggsae EMC-6 also. BWM expression of either HA::Ce-EMC-6 or GFP::Cb-EMC-6 rescued the emc-6(kr150) levamisole resistance phenotype (Fig. 2A). With both constructs, tagged EMC-6 localized to a perinuclear intracellular network reminiscent of ER. Consistently, GFP::Cb-EMC-6 colocalized with tagRFP fused to the ER retention signal KDEL (44) (Fig. 2B). Conversely, the Golgi marker α-MannosidaseII-tagRFP (45) did not colocalize with HA::Ce-EMC-6 (Fig. 2C). These results demonstrate that both C. briggsae and C. elegans EMC-6 primarily localize to the ER.
emc-6 Is Required for Surface Expression of Ligand-Gated Ion Channels at NMJs.
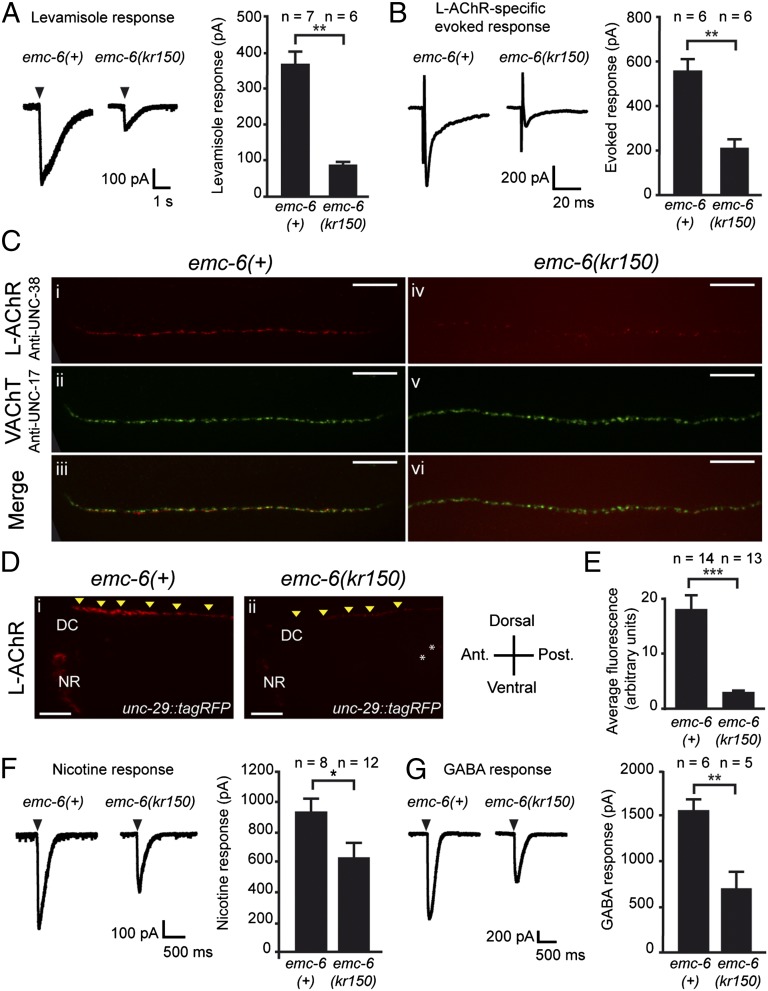
The resistance of emc-6(kr150) mutants to levamisole suggested that disrupting emc-6 impairs L-AChR function. To analyze this defect, we first recorded the electrophysiological response of BWM cells to pressure-applied levamisole and observed a 73% decrease in the response of emc-6(kr150) mutants compared with the WT (Fig. 3A). To evaluate the number of L-AChRs present at the synapse, we quantified the L-AChR–dependent evoked response after stimulation of cholinergic motor neurons and found a 66% reduction in emc-6(kr150). These data suggest that the amount of functional L-AChRs is severely reduced in emc-6(kr150) but that the remaining receptors are still clustered at NMJs (Fig. 3B).
Fig. 3.
EMC-6 is required for surface expression of ionotropic receptors. (A) Response to pressure ejection of levamisole in voltage-clamped ventral BWMs is reduced in emc-6(kr150) [mean ± SEM (WT: 367 ± 37 pA, n = 7; emc-6(kr150): 87 ± 8 pA, n = 6); P = 0.0012]. Arrowheads mark application onset. (B) L-AChR–dependent evoked currents recorded from BWMs after ventral nerve cord stimulation are decreased in emc-6(kr150). Electrically evoked responses were obtained in a unc-49(e407); acr-16(ok789) genetic background to eliminate GABAA receptor and N-AChR contributions (WT: 553 ± 50 pA, n = 6; emc-6(kr150): 210 ± 31 pA, n = 6; P = 0.0022). (C) L-AChR expression is decreased at NMJs of emc-6(kr150) mutants (i, iii, iv, and vi), whereas presynaptic differentiation is unaffected (ii, iii, v, and vi). L-AChRs are labeled using anti–UNC-38. Cholinergic boutons are labeled using an anti-vesicular acetylcholine transporter UNC-17 (VAChT) antibody. (Scale bars, 10 μm.) (D) Visualization of L-AChRs in living worms using a knock-in UNC-29–tagRFP L-AChR subunit engineered by MosTIC homologous recombination. DC, dorsal cord; NR, nerve ring. Arrowheads indicate L-AChR clusters at the dorsal nerve cord. Intestinal autofluorescence is indicated with an asterisk. (Scale bars, 10 μm.) (E) UNC-29–tagRFP fluorescence is reduced in emc-6(kr150) mutants (WT: 18.1 ± 2.5 arbitrary units, n = 14; emc-6(kr150): 2.9 ± 0.3 arbitrary units, n = 13; ***P < 0.001, Mann–Whitney test). (F) EMC-6 is required for N-AChR function. Response to pressure ejection of nicotine (WT: 931 ± 78 pA, n = 8; emc-6(kr150): 633 ± 94 pA, n = 12; P = 0.023). Arrowheads mark application onset. (G) EMC-6 is required for GABAA receptor function. Response to pressure ejection of GABA (WT: 1552 ± 118 pA, n = 6; emc-6(kr150): 710 ± 172 pA, n = 5; P = 0.0043). Arrowheads mark application onset.
To test if these electrophysiological phenotypes might reflect a synaptogenesis defect, we stained cholinergic boutons by immunofluorescence. The staining pattern of the vesicular acetylcholine transporter UNC-17 was indistinguishable between emc-6(kr150) and WT animals. By contrast, the staining of L-AChRs was dramatically decreased in emc-6(kr150) mutants (Fig. 3C). To quantify the synaptic decrease of the L-AChRs in emc-6(kr150), we generated a knock-in strain carrying the unc-29 L-AChR subunit endogenously tagged with tagRFP using the MosTIC technique (46). Consistent with immunostaining of L-AChRs, we observed an 82% decrease of the amount of L-AChR at the NMJ (Fig. 3 D and E). The remaining L-AChRs still formed synaptic clusters in emc-6(kr150) mutants. Altogether, these electrophysiological and histological results indicate that emc-6 is required for the surface expression of L-AChRs.
In WT animals, BWMs sense excitatory cholinergic inputs through heteromeric L-AChRs and homomeric ACR-16–containing N-AChRs, as well as through inhibitory GABAergic input through the UNC-49 GABAA receptors. To decipher the specificity of the emc-6(kr150) phenotype at NMJs, we recorded the electrophysiological responses of BWM to pressure-ejected nicotine, which stimulates only ACR-16–containing N-AChRs, (Fig. 3F) or to GABA (Fig. 3G). We found a 36% decreased response to nicotine and a 57% decreased response to GABA in emc-6(kr150) mutants compared with WT animals. These results demonstrate that emc-6 is required for the surface expression of at least three different types of multimeric ion channels.
emc-6 Stabilizes L-AChR Subunits Before or During Receptor Assembly.
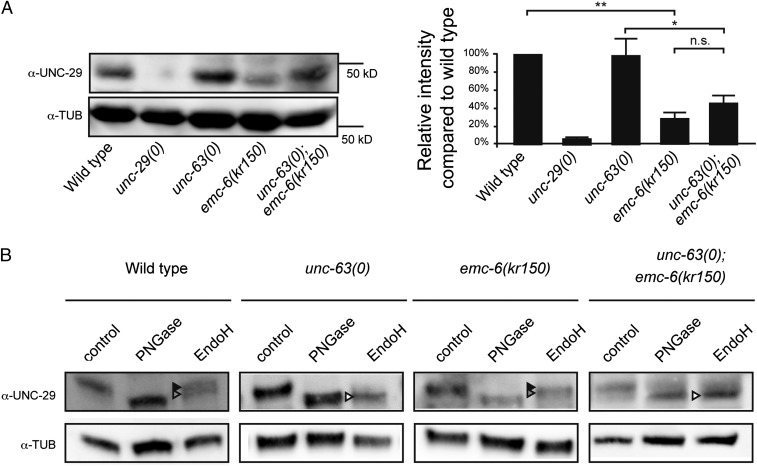
To test if the reduction of L-AChR at the NMJ of emc-6(kr150) mutants reflected a decrease in total receptor expression, we quantified the amount of the UNC-29 L-AChR subunit by Western blot analysis (Fig. 4A). UNC-29 expression was decreased by 72% in emc-6(kr150) compared with the WT. Decreased L-AChR expression could be caused at transcriptional or translational steps. To test if emc-6 was required for gene expression, we measured the mRNA levels of unc-29, unc-38, and unc-63, the three essential L-AChR subunits, and the mRNA levels of acr-16 and unc-49, the constitutive subunits of homomeric N-AChRs and GABAA receptors, respectively (Fig. S4 A and B). Compared with the WT, we did observe a slight but not significant decrease of these receptors’ mRNA levels in emc-6(kr150) mutants.
Fig. 4.
emc-6 is required before or during L-AChR assembly. (A) L-AChR expression is reduced in emc-6(kr150) mutants. Levels of unassembled UNC-29 L-AChR subunits detected in unc-63(kr13) are further decreased in unc-63(0); emc-6(kr150) double mutants. UNC-29 levels are quantified by Western blot using anti–UNC-29 antibodies and normalized to tubulin levels. The migration profile of UNC-29 subunit appears to be slightly different in emc-6(kr150) compared with other genotypes, which might reflect slight changes of UNC-29 glycosylation. Seven independent experiments were quantified (mean ± SEM). n.s. (not significant), P > 0.05; *P < 0.05; **P < 0.01 after Holm correction, Mann–Whitney–Wilcoxon test. (B) Remaining L-AChRs exit the ER in emc-6(kr150). Treatments with EndoH or N-Glycosidase F (PNGase) were performed on protein extracts of mixed-stage animals before SDS/PAGE analysis. Black arrowheads indicate glycosylated forms resistant to EndoH, gray arrowheads indicate partially glycosylated forms partially resistant to EndoH, and white arrowheads indicate unglycosylated forms sensitive to EndoH.
To test if EMC-6 was required for L-AChR biosynthesis within the ER, we analyzed the effect of emc-6 inactivation on unassembled L-AChR subunits retained in the ER. If EMC-6 is required for the folding or stability of unassembled L-AChR subunits, impairing emc-6 function should reduce the level of remaining unassembled subunits. Conversely, if EMC-6 is required after the assembly of the receptor, we predict that ER-retained unassembled subunits would not be affected by emc-6(kr150) mutation. We have previously demonstrated that L-AChR subunits do not traffic to the plasma membrane when the essential UNC-63 subunit is absent (38). In unc-63(0) mutants, unassembled UNC-29 subunits accumulate in the ER and are readily detected by Western blot (Fig. 4A). In unc-63(0); emc-6(kr150) double mutants, the amount of UNC-29 was decreased [compare with unc-63(0)] to a similar extent as in emc-6(kr150) single mutants (compared with WT) (Fig. 4A), suggesting that emc-6 is indeed required for the stability of ER-retained unassembled subunits.
Recently, Rer1, a Golgi-ER retrieval receptor, was demonstrated to control the expression of vertebrate muscle AChRs by ensuring the retention of unassembled subunits in the ER (47). To test the possibility that EMC-6 might also be involved in the control of the ER exit of receptor subunits, we analyzed the UNC-29 glycosylation profile in unc-63(0) and emc-6(kr150) (Fig. 4B). Mature N-glycosylations on membrane proteins are resistant to Endoglycosidase H (EndoH) cleavage after trafficking through the secretory pathway, whereas the nascent sugar side chains synthesized in the ER are cleaved by EndoH. By contrast, peptide-N-glycosidase F (PNGase) eliminates all carbohydrates from N-linked glycosylation. To distinguish between mature and immature L-AChRs, we analyzed the digestion profiles of the UNC-29 subunit after treatments by EndoH or PNGase. In the WT, UNC-29 is resistant to EndoH, whereas it is fully sensitive in unc-63(0) in agreement with ER retention of the remaining unassembled L-AChR subunits (the doublet that was observed in the WT likely corresponds to partially glycosylated subunits generated during L-AChR maturation processes, in addition to fully resistant mature species). In emc-6(kr150), the remaining UNC-29 subunits were also resistant to EndoH, similar to the WT, indicating that these subunits were properly matured in the absence of EMC-6. In contrast, UNC-29 was fully sensitive to EndoH in the double mutant unc-63(0); emc-6(kr150), indicating that even in the absence of EMC-6, unassembled subunits are still retained in the ER.
Altogether, these results suggest that EMC-6 is not involved in the regulation of the ER exit of AChRs and point to a stabilizing role of EMC-6 during receptor subunit maturation and assembly within the ER.
emc-6 Is a Component of the EMC, a Conserved Complex Required for L-AChR Maturation in C. elegans.
In yeast, the EMC-6 ortholog (EMC6p) forms a physical complex with five additional proteins (43). This complex localizes to the ER and was accordingly named the “ER membrane protein complex.” Inactivation of this complex triggers ER stress and activation of the UPR pathway. It was also recently linked to the ERAD machinery in human cell lines (48). The EMC members EMC1 to EMC6 are conserved from yeast to humans (Table S1). We tentatively named the C. elegans orthologs emc-1 through emc-6 (corresponding ORFs are shown in Table S1).
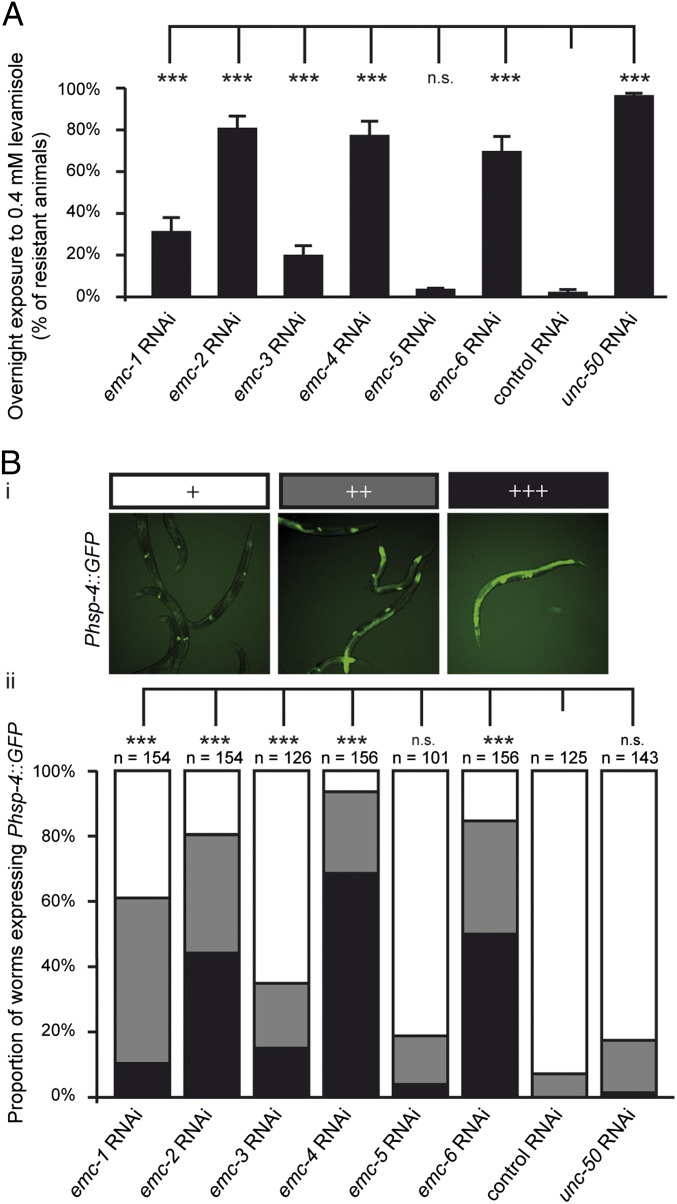
The L-AChR biosynthesis defects in emc-6(kr150) might be caused by a global dysfunction of the EMC. Alternatively, the very selective kr150 mutation might have unmasked a specific contribution of the EMC-6 protein for multimeric transmembrane receptor biosynthesis. To test these hypotheses, we inactivated the emc-1 through emc-6 genes by RNAi because no mutations exist in any of the C. elegans EMC member orthologs except for emc-6(kr150). Inactivation of EMC members led to various phenotypic defects, including embryonic lethality and developmental arrest (Table S2). Strikingly, inactivation of emc-1, emc-2, emc-3, emc-4, and emc-6 caused significant resistance to levamisole compared with worms fed with an empty RNAi vector (Fig. 5A). The lack of effect of emc-5 RNAi might reflect a specific function of EMC-5 in the EMC or heterogeneity of EMC composition according to its substrate.
Fig. 5.
EMC is required for L-AChR expression and ER homeostasis. (A) EMC inactivation by RNAi leads to resistance to levamisole. Bars represent the percentage of resistant animals after overnight exposure to 0.4 mM levamisole following preembryonic RNAi in eri-1(mg366). Four to 13 independent experiments were performed (20–45 animals per condition per experiment) (mean ± SEM). n.s. (not significant), P > 0.05; ***P < 0.001 after Holm correction, Mann–Whitney–Wilcoxon test. (B) EMC inactivation triggers the UPR. (i) Representative pictures of young adults expressing GFP under the control of the hsp-4 promoter reporting the extent of the UPR (intensity: +, white; ++, gray; +++ black). (ii) Bar graph represents UPR in worms exposed to control (empty vector), emc, or unc-50 RNAi. RNAi was performed preembryonically by feeding the parent WT worms with bacteria expressing dsRNA. n, number of animals tested. Four to eight independent experiments were pooled. n.s. (not significant), P > 0.05; ***P < 0.001 after Bonferroni correction, Fisher exact test.
These data provide indirect support for the existence of an EMC in C. elegans and suggest that the entire complex is required to ensure proper maturation of L-AChRs.
C. elegans EMC Is Involved in ER Homeostasis.
The EMC was identified as a complex involved in ER homeostasis in yeast and in ERAD in human cells. To test if EMC inactivation leads to ER stress in C. elegans, we used the UPR reporter Phsp-4::gfp, whose expression is strongly induced in the gut and hypodermis by UPR activation (49). Inactivation of emc-1, emc-2, emc-3, emc-4, and emc-6 by RNAi was associated with significant ER stress (Fig. 5B). Inactivation of emc-5 seemed to have no significant effect on UPR activation. To test whether UPR activation was a side effect of decreased L-AChR function, we inactivated unc-50 by RNAi and saw no ER stress induction despite strong resistance to levamisole (Fig. 5 A and B).
Because EMC inactivation caused ER stress, we asked whether the L-AChR biosynthesis defects of kr150 mutants could be an indirect consequence of ER stress. We therefore tested whether down-regulation or overactivation of the UPR pathway would decrease L-AChR expression. Mutants of the two major components of the UPR pathway in C. elegans, ire-1 and xbp-1 (49), were fully sensitive to levamisole and had normal levels of L-AChRs at the NMJ based on immunostaining (Fig. S5 A and B). Similarly, the upr-1(zc6) mutation, which leads to constitutive activation of the UPR pathway (49), did not cause resistance to levamisole. These results demonstrate that C. elegans EMC inactivation triggers ER stress and activation of the UPR independent of its role in L-AChR biosynthesis.
Discussion
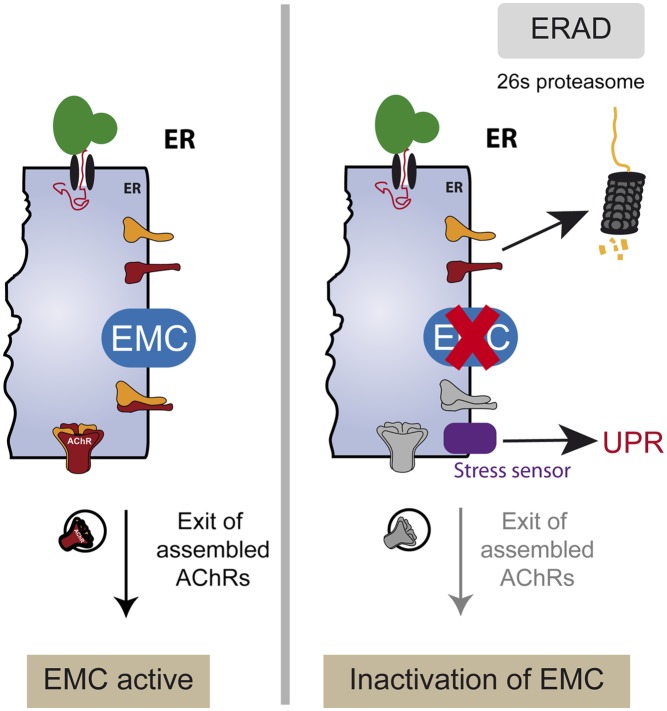
A genetic screen for mutants partially resistant to the cholinergic agonist levamisole identified emc-6, a previously uncharacterized gene coding for a small transmembrane protein of the ER. Our data show that EMC-6 is cell-autonomously required in muscle cells to ensure the expression of L-AChRs as well as two other multimeric ligand-gated ion channels. It likely functions within the recently identified EMC to stabilize unassembled L-AChR subunits and promote their assembly into mature receptors. More generally, the EMC seems to be essential for the viability of the organism and is required for maintaining ER homeostasis in vivo.
EMC Is an Evolutionarily Conserved Protein Complex.
emc-6 is the ortholog of the yeast gene EMC6, which was identified in 2009 by Jonikas et al. (43) over the course of a genome-wide identification of genes contributing to protein folding in the ER. Comprehensive analysis of the interactions between these genes identified a functional cluster of six poorly characterized genes subsequently named EMC1 to EMC6. Immunoprecipitation experiments demonstrated that these six proteins form an apparently stoichiometric complex. They are all predicted to be ER resident transmembrane proteins, except for EMC2. This complex was further identified independently by Kopito and colleagues (48) in 2012 in a high-throughput approach combining proteomics and functional genomics to define ERAD networks in human cells. Physical interactors of 25 baits involved in ERAD were identified after protein complex purification and liquid chromatography-tandem MS. Specifically, reciprocal interactions were detected among 10 proteins, including orthologs of the six yeast EMC members. The four additional proteins are not present in yeast but seem to be conserved in C. elegans (Table S1). Remarkably, almost nothing is known about the function of these genes, either in yeast or in mammals, and none of these genes have previously been studied in C. elegans.
Here, we have characterized EMC-6 and demonstrated that it is an ER protein likely expressed in all tissues. The shared phenotypes observed after inactivation of other emc genes suggests that EMC-6 functions together with other EMC members in C. elegans. For example RNAi inactivation of emc-1, emc-2, emc-4, or emc-6 causes embryonic lethality or developmental arrest, and inactivation of any emc gene except emc-5 decreased the expression of L-AChRs (Fig. 5A). The phenotypic differences that we observed after inactivating individual emc genes might arise for several nonexclusive reasons. First, they might reflect variation of RNAi efficiency for individual emc genes and the tissues in which they are expressed (50) because our phenotypic readout is at the organismal level. Second, the EMC might contain core components whose inactivation destabilizes the entire complex and has more drastic effects than when targeting peripheral components. Third, the composition of the complex might vary according to its substrates. Fourth, it is very likely that individual components contribute specific functions to the whole complex. Testing these different hypotheses will require the detailed characterization of each EMC subunit and the biochemical reconstitution of the complex.
EMC Requirement for ER Homeostasis Can Be Genetically Dissociated from Specific Functions for Cys-Loop Receptor Biosynthesis.
We showed that disruption of the EMC in C. elegans causes induction of the UPR (Fig. 5B). Several lines of evidence suggest that UPR induction in emc mutants is a direct consequence of protein maturation defects in the ER. First, EMC inactivation in yeast causes a clear UPR induction related to an accumulation of unfolded protein in the ER (51). Interestingly, Jonikas et al. (43) noticed that in yeast, “the pattern of genetic interactions of strains deleted for EMC members most closely resembled that seen in a strain overexpressing the misfolded transmembrane protein Sec61-2p (a mutated form of the Sec61 translocon) (52) and is similar to the pattern of a strain overexpressing the misfolded transmembrane protein KWS (53).” Second, induction of ER stress and UPR by tunicamycin treatment causes the transcriptional induction of 9 of the 10 EMC members in human cell lines, a typical feature of genes involved in protein folding and degradation (reviewed in ref. 54). Third, emc inactivation in C. elegans causes developmental defects that are usually not related to constitutive UPR activation (49), indicating that theses defects, as well as UPR activation, are likely to be downstream consequences of impairment of ER folding capacity in emc mutants.
Whether the EMC is similarly required for all transmembrane proteins or is more specifically involved in the biosynthesis of a subclass of proteins, such as ACh and GABA ionotropic receptors, is difficult to answer. However, we observed that the emc-6 mutation isolated in our genetic screen does not impair the synthesis of all transmembrane proteins. First, we monitored the expression of the AMPA-like glutamate receptor subunit GLR-1, which assembles into a tetrameric ligand-gated ion channel (55), and we saw no decrease in the emc-6(kr150) background compared with the WT (Fig. S6A). Second, we analyzed vulval development of animals carrying the emc-6(kr150) mutation, which provides indirect access to the functionality of the EGF receptor LET-23, a cell surface glycoprotein with a single transmembrane domain. Decreased activity of let-23 causes vulval induction defects (56). The lack of vulval defects in emc-6(kr150) mutants (Fig. S6B) suggests that the EGF receptor LET-23 is properly expressed. These data do not exclude the likely possibility that EMC-6 is required for the synthesis of other unidentified proteins, yet the emc-6(kr150) mutation unmasked some in vivo requirements of EMC-6 that are specific for the biosynthesis of GABA and ACh receptors and that all belong to the same superfamily of proteins called the Cys-loop receptors (reviewed in ref. 57).
EMC Stabilizes Unassembled Subunits of L-AChR.
We have shown in this work that loss of EMC function decreases L-AChR expression, likely by impairing the stability of unassembled subunits or assembly intermediates in the ER (Fig. 4). Interestingly, our results functionally connect the EMC to ERAD because AChR subunits are established integral-membrane ERAD substrates (23). We propose that the EMC regulates the amount of subunits available to assemble into mature L-AChRs either by behaving as a chaperone for unassembled or partially assembled subunits or by modulating ERAD of AChR subunits.
The best-characterized factors that are known to influence AChR subunit stability in the ER are ER chaperones. In Cos cells and in myotubes, binding of AChR subunits with BIP was proposed to act as a protective pathway to prevent misfolded subunits from altering receptor assembly (58). This hypothesis has been indirectly confirmed in C. elegans, where the interaction between BIP and unassembled L-AChR subunits is much stronger than with assembled L-AChRs (59). The calnexin and ERp57 ER-resident component of the biosynthetic machinery also form transient complexes with newly synthesized AChRs before full assembly occurs (17). The association of AChR subunits with ER proteins would decrease the rate of ERAD-dependent subunit degradation (17). By analogy, the EMC might provide a functional chaperone activity to stabilize or enhance the folding of partially assembled AChR subunits.
L-AChRs are the first proteins, to our knowledge, whose synthesis has been demonstrated to depend on EMC activity. The isolation of the emc-6(kr150) mutant may have been serendipitous. Alternatively, screening for L-AChR synthesis defects might be a very sensitive assay for EMC activity because of the very inefficient folding of these oligomeric transmembrane proteins. Specifically, AChR subunits contain an amphipathic transmembrane helix, M2, with a series of hydrophilic residues. In mature receptors, the M2 helices from each of the five subunits are organized around a pseudosymmetry axis and form a transmembrane cationic pore. It is not known how the hydrophilic face of the M2 helix of a partially assembled subunit is stabilized in the ER membrane before being buried in mature receptors. This topological issue is shared by all Cys-loop receptors, including the N-AChRs and UNC-49/GABAA receptor, which we have demonstrated also depend on the EMC for their synthesis in C. elegans. To gain further insight into the role of this complex in the biosynthesis of AChRs, it will be of great interest to identify other “substrates” of the EMC and test if the EMC indeed behaves as a chaperone for additional transmembrane proteins.
Materials and Methods
General Methods and Strains.
Unless indicated otherwise, strains were maintained at 20 °C on nematode growth medium (NGM) agar plates under standard conditions. OP50 Escherichia coli was generally used for feeding, except for strains exposed to RNAi, which were maintained on 1-mM isopropyl-β-D-thiogalactopyranoside (IPTG) plates with HT115 for feeding. The WT reference strain was N2 Bristol. Strains, molecular constructs, transgenes, and primers are described in SI Materials and Methods.
Levamisole Assay.
Assays for levamisole sensitivity after overnight exposure were performed as previously described (60). Further details are provided in SI Materials and Methods.
Electrophysiology.
Electrophysiological methods were as previously described by Richmond and Jorgensen (30) and Gendrel et al. (60). L-AChR evoked recording analysis was performed in a unc-49(e407); acr-16(ok789) background to eliminate currents arising from activation of GABAA receptors and N-AChRs.
Immunocytochemical Staining.
Worms were prepared by the freeze-crack method described previously (60). Further details are provided in SI Materials and Methods.
Western Blotting.
Mixed-stage populations were collected from OP50-seeded NGM plates. They were rinsed three times with water and allowed to sediment on ice to wash out bacteria. Pellets were solubilized in Laemmli buffer with 2% (vol/vol) β-mercaptoethanol, boiled at 90 °C for 10 min, sonicated for 1 min, and centrifuged for 5 min at 15,700 × g. For glycosylation profiles, samples were treated with denaturation buffer for 10 min at 90 °C, incubated with endoglycosidase H (EndoH; New England Biolabs) or PNGaseF (New England Biolabs) for 1 h at 37 °C, and subsequently treated with Laemmli buffer as described above. Membranes were imaged with a LAS4000 (GE Healthcare Life Sciences) luminescence detector and band intensity-quantified with LAS4000 software. Further details are provided in SI Materials and Methods.
Isolation of RNA and Quantitative Real-Time PCR Assay.
Synchronized first-stage larval (L1) cultures were obtained by treating young gravid adults with a bleaching solution for 5 min. Surviving eggs were allowed to reach the L1 stage during an overnight immersion in M9 solution. Total RNA was isolated from this synchronized population using the RNeasy Kit (Qiagen) according to the manufacturer’s instructions. All samples were treated with DNase (Fermentas). First-strand cDNA was synthesized from 200 ng of total RNA using an oligo(dT) primer and a Moloney murine leukemia virus reverse transcriptase (Fermentas) at 42 °C for 1 h. Quantitative PCR was performed using LightCycler 480 SYBR Green I Master (Roche). Primer sequences are listed in SI Materials and Methods. A relative quantification model, including kinetic PCR efficiency correction, was used to evaluate the relative expression ratio of the target genes (61). RNA levels were normalized to the three housekeeping genes cdc-42, pmp-3, and Y45F10D.4 according to the method of Hoogewijs et al. (62). Further details are provided in SI Materials and Methods.
RNAi.
RNAi was performed using the bacterial feeding protocol (63) with the following modifications. NGM agar plates containing 1 mM IPTG and 100 μg/mL ampicillin were seeded with bacteria expressing dsRNA. L4-staged hermaphrodites were placed onto these plates and grown at 20 °C. After 4 d, the F1 progeny of these worms were scored for phenotypes of interest. If not available in the library of Kamath and Ahringer (63), PCR was used to amplify fragments flanked by the T7 promoter at the 5′ and 3′ ends. Further details are provided in SI Materials and Methods.
Light Microscopy and Fluorescence Quantification.
For surface AChR quantification, animals were anesthetized with M9 buffer containing 100 mM sodium azide mounted on 2% (wt/vol) agarose in M9 pads and examined by epifluorescence using a Nikon macrozoom microscope equipped with a chilled CCD camera (stress induction) or a Leica DM5000B microscope with a spinning disk. Images were acquired using METAMORPH 7.1 software (Molecular Devices) and analyzed with IMAGEJ software (National Institutes of Health).
Supplementary Material
Acknowledgments
We thank L. Pintard, M. Labouesse, and E. Chevet for critical reading of the manuscript. We thank L. Briseño-Roa and M. Barkoulas for technical advice. We thank the Caenorhabditis Genetic Center and the International C. elegans Gene Knock-Out Consortium for strains. We thank T. Hoppe, M. Calfon, L. Pintard, and M.-A. Felix for critical reagents. M.R. was supported by the Neuropole francilien and by the Association pour la Recherche contre le Cancer. This work was funded by the Institut National de la Santé et de la Recherche Médicale (V.J.P.R. and J.-L.B.), the European Molecular Biology Organization (T.B.), the Centre National de la Recherche Scientifique (T.B.), Grant MH073156 from the National Institutes of Health (to J.E.R.), and the Association Française contre les Myopathies and Fondation pour la Recherche Médicale (J.-L.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.M.H. is a guest editor invited by the Editorial Board.
See Author Summary on page 4178 (volume 110, number 11).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216154110/-/DCSupplemental.
References
- 1.Millar NS, Gotti C. Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology. 2009;56(1):237–246. doi: 10.1016/j.neuropharm.2008.07.041. [DOI] [PubMed] [Google Scholar]
- 2.Gorrie GH, et al. Assembly of GABAA receptors composed of alpha1 and beta2 subunits in both cultured neurons and fibroblasts. J Neurosci. 1997;17(17):6587–6596. doi: 10.1523/JNEUROSCI.17-17-06587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen TJ, et al. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83(1):129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 4.Merlie JP, Lindstrom J. Assembly in vivo of mouse muscle acetylcholine receptor: Identification of an alpha subunit species that may be an assembly intermediate. Cell. 1983;34(3):747–757. doi: 10.1016/0092-8674(83)90531-7. [DOI] [PubMed] [Google Scholar]
- 5.Arroyo-Jim nez MM, et al. Ultrastructural localization of the alpha4-subunit of the neuronal acetylcholine nicotinic receptor in the rat substantia nigra. J Neurosci. 1999;19(15):6475–6487. doi: 10.1523/JNEUROSCI.19-15-06475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabatelli M, et al. Rare missense variants of neuronal nicotinic acetylcholine receptor altering receptor function are associated with sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18(20):3997–4006. doi: 10.1093/hmg/ddp339. [DOI] [PubMed] [Google Scholar]
- 7.Richards CI, et al. Trafficking of alpha4* nicotinic receptors revealed by superecliptic phluorin: Effects of a beta4 amyotrophic lateral sclerosis-associated mutation and chronic exposure to nicotine. J Biol Chem. 2011;286(36):31241–31249. doi: 10.1074/jbc.M111.256024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srinivasan R, et al. Pharmacological chaperoning of nicotinic acetylcholine receptors reduces the endoplasmic reticulum stress response. Mol Pharmacol. 2012;81(6):759–769. doi: 10.1124/mol.112.077792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sallette J, et al. Nicotine upregulates its own receptors through enhanced intracellular maturation. Neuron. 2005;46(4):595–607. doi: 10.1016/j.neuron.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 10.Govind AP, Walsh H, Green WN. Nicotine-induced upregulation of native neuronal nicotinic receptors is caused by multiple mechanisms. J Neurosci. 2012;32(6):2227–2238. doi: 10.1523/JNEUROSCI.5438-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rezvani K, Teng Y, Shim D, De Biasi M. Nicotine regulates multiple synaptic proteins by inhibiting proteasomal activity. J Neurosci. 2007;27(39):10508–10519. doi: 10.1523/JNEUROSCI.3353-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millar NS, Harkness PC. Assembly and trafficking of nicotinic acetylcholine receptors (Review) Mol Membr Biol. 2008;25(4):279–292. doi: 10.1080/09687680802035675. [DOI] [PubMed] [Google Scholar]
- 13.Blount P, Merlie JP. BIP associates with newly synthesized subunits of the mouse muscle nicotinic receptor. J Cell Biol. 1991;113(5):1125–1132. doi: 10.1083/jcb.113.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulson HL, Ross AF, Green WN, Claudio T. Analysis of early events in acetylcholine receptor assembly. J Cell Biol. 1991;113(6):1371–1384. doi: 10.1083/jcb.113.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelman MS, Chang W, Thomas DY, Bergeron JJ, Prives JM. Role of the endoplasmic reticulum chaperone calnexin in subunit folding and assembly of nicotinic acetylcholine receptors. J Biol Chem. 1995;270(25):15085–15092. doi: 10.1074/jbc.270.25.15085. [DOI] [PubMed] [Google Scholar]
- 16.Wanamaker CP, Green WN. N-linked glycosylation is required for nicotinic receptor assembly but not for subunit associations with calnexin. J Biol Chem. 2005;280(40):33800–33810. doi: 10.1074/jbc.M501813200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wanamaker CP, Green WN. Endoplasmic reticulum chaperones stabilize nicotinic receptor subunits and regulate receptor assembly. J Biol Chem. 2007;282(43):31113–31123. doi: 10.1074/jbc.M705369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green WN, Wanamaker CP. The role of the cystine loop in acetylcholine receptor assembly. J Biol Chem. 1997;272(33):20945–20953. doi: 10.1074/jbc.272.33.20945. [DOI] [PubMed] [Google Scholar]
- 19.Gehle VM, Walcott EC, Nishizaki T, Sumikawa K. N-glycosylation at the conserved sites ensures the expression of properly folded functional ACh receptors. Brain Res Mol Brain Res. 1997;45(2):219–229. doi: 10.1016/s0169-328x(96)00256-2. [DOI] [PubMed] [Google Scholar]
- 20.Drisdel RC, Manzana E, Green WN. The role of palmitoylation in functional expression of nicotinic alpha7 receptors. J Neurosci. 2004;24(46):10502–10510. doi: 10.1523/JNEUROSCI.3315-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wanamaker CP, Christianson JC, Green WN. Regulation of nicotinic acetylcholine receptor assembly. Ann N Y Acad Sci. 2003;998:66–80. doi: 10.1196/annals.1254.009. [DOI] [PubMed] [Google Scholar]
- 22.Wang J-M, et al. A transmembrane motif governs the surface trafficking of nicotinic acetylcholine receptors. Nat Neurosci. 2002;5(10):963–970. doi: 10.1038/nn918. [DOI] [PubMed] [Google Scholar]
- 23.Christianson JC, Green WN. Regulation of nicotinic receptor expression by the ubiquitin-proteasome system. EMBO J. 2004;23(21):4156–4165. doi: 10.1038/sj.emboj.7600436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan R, et al. Nicotine up-regulates alpha4beta2 nicotinic receptors and ER exit sites via stoichiometry-dependent chaperoning. J Gen Physiol. 2011;137(1):59–79. doi: 10.1085/jgp.201010532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller SH, Lindstrom J, Ellisman M, Taylor P. Adjacent basic amino acid residues recognized by the COP I complex and ubiquitination govern endoplasmic reticulum to cell surface trafficking of the nicotinic acetylcholine receptor alpha-Subunit. J Biol Chem. 2001;276(21):18384–18391. doi: 10.1074/jbc.M100691200. [DOI] [PubMed] [Google Scholar]
- 26.Ren X-Q, et al. Structural determinants of alpha4beta2 nicotinic acetylcholine receptor trafficking. J Neurosci. 2005;25(28):6676–6686. doi: 10.1523/JNEUROSCI.1079-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeanclos EM, et al. The chaperone protein 14-3-3eta interacts with the nicotinic acetylcholine receptor alpha 4 subunit. Evidence for a dynamic role in subunit stabilization. J Biol Chem. 2001;276(30):28281–28290. doi: 10.1074/jbc.M011549200. [DOI] [PubMed] [Google Scholar]
- 28.Rezvani K, et al. UBXD4, a UBX-containing protein, regulates the cell surface number and stability of alpha3-containing nicotinic acetylcholine receptors. J Neurosci. 2009;29(21):6883–6896. doi: 10.1523/JNEUROSCI.4723-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones AK, Sattelle DB. Functional genomics of the nicotinic acetylcholine receptor gene family of the nematode, Caenorhabditis elegans. Bioessays. 2004;26(1):39–49. doi: 10.1002/bies.10377. [DOI] [PubMed] [Google Scholar]
- 30.Richmond JE, Jorgensen EM. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat Neurosci. 1999;2(9):791–797. doi: 10.1038/12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis JA, Wu CH, Berg H, Levine JH. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95(4):905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleming JT, et al. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J Neurosci. 1997;17(15):5843–5857. doi: 10.1523/JNEUROSCI.17-15-05843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Touroutine D, et al. acr-16 encodes an essential subunit of the levamisole-resistant nicotinic receptor at the Caenorhabditis elegans neuromuscular junction. J Biol Chem. 2005;280(29):27013–27021. doi: 10.1074/jbc.M502818200. [DOI] [PubMed] [Google Scholar]
- 34.Francis MM, et al. The Ror receptor tyrosine kinase CAM-1 is required for ACR-16-mediated synaptic transmission at the C. elegans neuromuscular junction. Neuron. 2005;46(4):581–594. doi: 10.1016/j.neuron.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Halevi S, et al. The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. EMBO J. 2002;21(5):1012–1020. doi: 10.1093/emboj/21.5.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boulin T, et al. Eight genes are required for functional reconstitution of the Caenorhabditis elegans levamisole-sensitive acetylcholine receptor. Proc Natl Acad Sci USA. 2008;105(47):18590–18595. doi: 10.1073/pnas.0806933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jospin M, et al. A neuronal acetylcholine receptor regulates the balance of muscle excitation and inhibition in Caenorhabditis elegans. PLoS Biol. 2009;7(12):e1000265. doi: 10.1371/journal.pbio.1000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander JK, et al. Ric-3 promotes alpha7 nicotinic receptor assembly and trafficking through the ER subcompartment of dendrites. J Neurosci. 2010;30(30):10112–10126. doi: 10.1523/JNEUROSCI.6344-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eimer S, et al. Regulation of nicotinic receptor trafficking by the transmembrane Golgi protein UNC-50. EMBO J. 2007;26(20):4313–4323. doi: 10.1038/sj.emboj.7601858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almedom RB, et al. An ER-resident membrane protein complex regulates nicotinic acetylcholine receptor subunit composition at the synapse. EMBO J. 2009;28(17):2636–2649. doi: 10.1038/emboj.2009.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rapti G, Richmond J, Bessereau J-L. A single immunoglobulin-domain protein required for clustering acetylcholine receptors in C. elegans. EMBO J. 2011;30(4):706–718. doi: 10.1038/emboj.2010.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulin T, et al. Positive modulation of a Cys-loop acetylcholine receptor by an auxiliary transmembrane subunit. Nat Neurosci. 2012;15(10):1374–1381. doi: 10.1038/nn.3197. [DOI] [PubMed] [Google Scholar]
- 43.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4(5):815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 45.Rolls MM, Hall DH, Victor M, Stelzer EHK, Rapoport TA. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol Biol Cell. 2002;13(5):1778–1791. doi: 10.1091/mbc.01-10-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robert V, Bessereau J-L. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 2007;26(1):170–183. doi: 10.1038/sj.emboj.7601463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valkova C, et al. Sorting receptor Rer1 controls surface expression of muscle acetylcholine receptors by ER retention of unassembled alpha-subunits. Proc Natl Acad Sci USA. 2011;108(2):621–625. doi: 10.1073/pnas.1001624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christianson JC, et al. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14(1):93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 50.Simmer F, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1(1):E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9(6):861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365(6442):176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 53.Vashist S, Ng DTW. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165(1):41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 55.Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378(6552):78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 56.Ferguson EL, Sternberg PW, Horvitz HR. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature. 1987;326(6110):259–267. doi: 10.1038/326259a0. [DOI] [PubMed] [Google Scholar]
- 57.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440(7083):448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 58.Forsayeth JR, Gu Y, Hall ZW. BiP forms stable complexes with unassembled subunits of the acetylcholine receptor in transfected COS cells and in C2 muscle cells. J Cell Biol. 1992;117(4):841–847. doi: 10.1083/jcb.117.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gottschalk A, et al. Identification and characterization of novel nicotinic receptor-associated proteins in Caenorhabditis elegans. EMBO J. 2005;24(14):2566–2578. doi: 10.1038/sj.emboj.7600741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gendrel M, Rapti G, Richmond JE, Bessereau J-L. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature. 2009;461(7266):992–996. doi: 10.1038/nature08430. [DOI] [PubMed] [Google Scholar]
- 61.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]