Abstract
Proapoptotic B-cell lymphoma 2 (BCL-2) antagonist/killer (BAK) and BCL-2–associated X (BAX) form toxic mitochondrial pores in response to cellular stress. Whereas BAX resides predominantly in the cytosol, BAK is constitutively localized to the outer mitochondrial membrane. Select BCL-2 homology domain 3 (BH3) helices activate BAX directly by engaging an α1/α6 trigger site. The inability to express full-length BAK has hampered full dissection of its activation mechanism. Here, we report the production of full-length, monomeric BAK by mutagenesis-based solubilization of its C-terminal α-helical surface. Recombinant BAK autotranslocates to mitochondria but only releases cytochrome c upon BH3 triggering. A direct activation mechanism was explicitly demonstrated using a liposomal system that recapitulates BAK-mediated release upon addition of BH3 ligands. Photoreactive BH3 helices mapped both triggering and autointeractions to the canonical BH3-binding pocket of BAK, whereas the same ligands crosslinked to the α1/α6 site of BAX. Thus, activation of both BAK and BAX is initiated by direct BH3–interaction but at distinct trigger sites. These structural and biochemical insights provide opportunities for developing proapoptotic agents that activate the death pathway through direct but differential engagement of BAK and BAX.
Keywords: apoptosis, BCL-2 family, SAHB, stapled peptide, photocrosslinking
B-cell lymphoma 2 (BCL-2) antagonist/killer (BAK) is a proapoptotic BCL-2 family member that resides in the mitochondrial outer membrane as a quiescent monomer until stimulated by cellular stress to undergo conformational activation and oligomerization (1, 2). Poration of the mitochondria by BAK leads to the release of critical signaling factors, such as cytochrome c (3, 4) and second mitochondria-derived activator of caspases (Smac)/Diablo (5), which drive the apoptotic process. First discovered in 1995 (6–8), BAK was found to share functional homology with BCL-2–associated (BAX), each containing an essential BCL-2 homology domain 3 (BH3) required for oligomerization-based killing activity (9). The solution structure of the BAK BH3 domain complexed to a surface groove on antiapoptotic BCL-extra large (BCL-XL) defined a protein-interaction paradigm that contributes to BCL-2 family regulation of mitochondrial apoptosis (10). Indeed, antiapoptotic suppression of BAK and BAX through sequestration of BH3 helices is such an effective means of blocking apoptosis that cancer cells hijack and amplify this natural regulatory mechanism to enforce pathologic cell survival.
Although BAK and BAX are an essential gateway to apoptosis (2) and when deleted give rise to severe developmental defects (11), a host of questions persist regarding the physiologic mechanisms that govern their inhibition, activation, and self-association. In the inactive state, BAX is a soluble, cytosolic, and monomeric protein whose hydrophobic surfaces are buried within its core, including its C-terminal membrane insertion helix that is entrapped in the canonical surface groove (12). In contrast, monomeric BAK is a membrane-resident protein whose C terminus is presumably dislodged from the canonical binding groove and inserted into the outer mitochondrial membrane, potentially rendering BAK more vulnerable to activation in the absence of additional security mechanisms. Although a matter of ongoing debate, the homeostatic suppression of BAK may involve structural stabilization of the inactive monomer as a result of conformationally restricted access to the canonical binding groove (13, 14) and/or constitutive interactions with antiapoptotic proteins (15), voltage-dependent anion channel 2 (16), and/or perhaps other unknown proteins.
Because a wealth of structural and biochemical studies have elucidated the mechanistic basis for antiapoptotic suppression of the conformationally activated, BH3-exposed forms of BAX and BAK (10, 15, 17–19), this aspect of BAX/BAK regulation is widely accepted and has formed the basis for major efforts to disrupt such interactions pharmacologically to reactivate apoptosis in human cancer (20–22). However, the mechanisms by which BAK and BAX are activated in the first place have been the focus of much debate. The hypothesis that BH3-only proteins could activate multidomain proapoptotics such as BAX through direct and transient binding dates back to the very discovery of BH3-only protein BH3-interacting domain death agonist (BID), based on its interaction with BCL-2 and BAX (23). Subsequent studies documented the capacity of select BH3-only proteins and BH3 peptides to induce recombinant BAX-mediated pore formation in liposomal and mitochondrial systems (24, 25), with hydrocarbon-stapled peptide helices corresponding to the BID and BCL-2–interacting mediator of cell death (BIM) BH3 domains used to detect and quantitate direct binding interactions with BAX (26). Structural and biochemical analyses of the interaction between full-length recombinant BAX and a stapled BIM BH3 helix revealed a noncanonical BH3 interaction site at the confluence of BAX α-helices 1 and 6, a trigger site for BH3-mediated direct BAX activation (Fig. S1) (27). Upon engagement by a triggering BH3 ligand, BAX undergoes a major conformational change that includes allosteric release of its C-terminal helix for mitochondrial translocation and exposure of its BH3 domain, which both propagates BAX activation and induces functional oligomerization within the mitochondrial outer membrane (28).
In contrast to BAX, full-length BAK has been refractory to protein expression and purification (15), precluding the corresponding structural and biochemical studies to dissect its interactions and activation mechanisms using fully intact recombinant protein. Instead, truncated forms of BAK have been generated and used in a series of studies that documented (i) dimeric structures of BAK truncates 23–185 and 16–186 (13, 14), (ii) binding of select BH3 domains to the exposed canonical pocket of BAKΔC (29) and N- and C-terminal calpain-cleaved BAK (cBAK) (14), (iii) autoactive and ligand-stimulated pore-forming functionality of an artificially membrane-tethered form of BAKΔC (30), and (iv) BH3 ligand-induced activation of BAK truncates in liposomal- and mitochondrial-release assays (29, 31, 32). The application of select BH3 peptides to native BAK-containing mouse liver mitochondria (33) and of in vitro transcribed/translated and tagged BH3-only proteins to mitochondria isolated from wild-type and mutant BAK-reconstituted Bax−/−Bak−/− mouse embryonic fibroblasts also supports a direct activation mechanism for BAK-mediated mitochondrial apoptosis (34, 35). Applying such reconstitution and mutagenesis approaches, the BH1 and BH3 domains of BAK were deemed essential to the homo-oligomerization process (35). An insertional cysteine-mutagenesis and disulfide crosslinking approach further implicated the involvement of two binding interfaces, BAK BH3 domain in the canonical groove and α6/α6 interactions, in propelling BAK poration via an oligomerization of dimers mechanism (36, 37), a model also supported by electron paramagnetic analysis (30). In the absence of definitive structural support for this model, it remains unknown just how an oligomer of dimers would integrate into a membrane bilayer using its once-buried hydrophobic surfaces.
To advance the structural and biochemical dissection of the BAK activation pathway, we sought to address the longstanding challenge of generating full-length BAK by mutagenesis-based enhancement of protein solubility. Here, we report the successful production and application of full-length recombinant BAK to interrogate its pore-forming properties and the mechanism of BAK activation. We used an unbiased photoreactive BH3 helix crosslinking and mass spectrometry (MS) approach (38) to map and compare the BH3 binding modes that account for direct BAK and BAX activation.
Results
Production of Full-Length, Monomeric BAK.
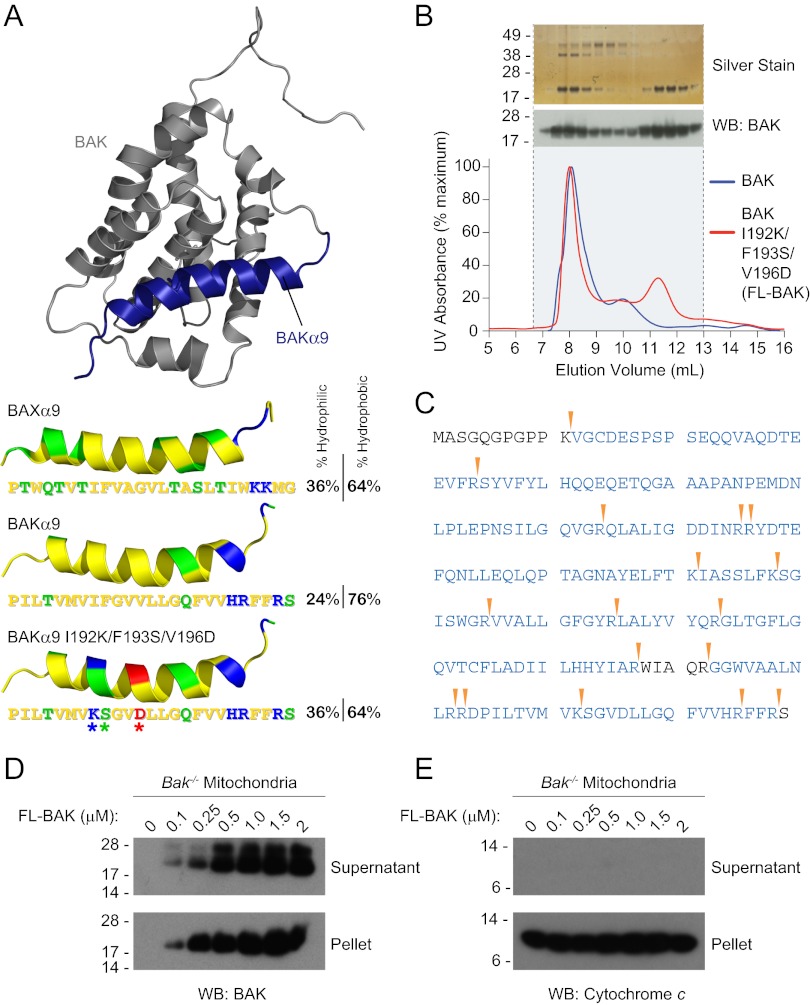
A prominent distinction between BAX and BAK is the respective cytosolic vs. mitochondrial outer membrane localization, which is driven, at least in part, by the disposition of the proteins’ C-terminal α-helices. The BAX α9 helix is bound tightly to its canonical binding groove (12), whereas the corresponding α-helix in BAK is constitutively released for mitochondrial outer membrane insertion. This structural and functional difference between the two α-helices is reflected by BAK α9 being comparatively more hydrophobic than BAX α9 (Fig. 1A). We reasoned that the increased hydrophobicity of BAK’s C terminus may account for its membrane tropism and propensity to aggregate upon bacterial protein expression, precluding the isolation of full-length monomer. To surmount this challenge, we modified the surface of BAK α9 to approximate the amphipathic character of BAX α9, converting the hydrophobic Phe193 residue of BAK into a hydrophilic Ser and replacing the hydrophobic Ile192 and Val196 pair with charged residues Lys and Asp, respectively, yielding a surface salt bridge (Fig. 1A). We used the pTYB4 vector to generate a chitin-binding domain fusion protein of murine BAK I192K/F193S/V196D [hereafter referred to as “full-length BAK” (FL-BAK)]. The fusion protein was expressed in Escherichia coli BL21 bacteria, isolated by chitin column chromatography, and subjected to DTT-based intein tag removal, as reported (12, 26). The elution profiles of wild-type vs. triply mutagenized FL-BAK were compared by size-exclusion chromatography (SEC). The wild-type construct yielded predominantly an oligomeric peak accompanied by a small dimeric peak, whereas the α9-mutagenized construct additionally produced the desired monomeric peak (Fig. 1B). The monomeric fraction (11- to 12-mL elution volume) migrated as a 23-kDa protein by SDS PAGE, and the identity of FL-BAK protein was confirmed by anti-BAK Western analysis (Fig. 1B) and MS/MS of the electrophoresed band (Fig. 1C). Importantly, we confirmed that this triple mutagenesis of BAK did not alter its functional activity, as evidenced by equivalent decreased viability of etoposide and staurosporine (STS)-treated Bax−/−Bak−/− immortalized baby mouse kidney epithelial (iBMK) (39) cells upon expression of wild-type or the triply mutant FL-BAK (Fig. S2).
Fig. 1.
Expression and purification of recombinant, full-length, and monomeric BAK. (A) The α9 helix of BAK was subjected to triple mutagenesis (I192K/F193S/V196D) to increase its hydrophilicity and thereby better match that of soluble, full-length BAX. The pictured model structure of BAK was calculated based on the solution structure of BAX using Modeller software (51), with BAK α9 colored blue. The hydrophobic, hydrophilic, positively charged, and negatively charged residues of the BAK and BAX α9 helices are colored yellow, green, blue, and red, respectively. (B) A comparison of the SEC elution profiles of BAK and FL-BAK demonstrates that mutagenesis enabled the isolation of a monomeric species (11- to 12-mL fractions). Silver staining and anti-BAK Western analysis of the electrophoresed FL-BAK fractions documented the isolation of monomeric BAK protein (∼23 kDa). (C) The identity of the isolated monomeric protein was confirmed to be FL-BAK by MS analysis. Tryptic sites are indicated by the arrowheads, and the FL-BAK sequences identified by LC-MS/MS are colored blue. (D) Upon exposure to isolated Bak−/− mouse liver mitochondria, FL-BAK dose-responsively translocated to the mitochondria-containing pellet fraction, as detected by anti-BAK Western analysis. (E) Autotranslocation of FL-BAK to the mitochondrial fraction did not induce cytochrome c release from the pellet to the supernatant, as measured by anti-cytochrome c Western analysis.
With recombinant, monomeric FL-BAK in hand, we first sought to evaluate its biochemical behavior in aqueous solution containing isolated Bak−/− mouse liver mitochondria (MLM). Recombinant, monomeric BAX requires ligand stimulation for mitochondrial translocation (28), but we observed that FL-BAK autotranslocated to MLM in a dose-responsive fashion (Fig. 1D). In contrast to ligand-stimulated BAX translocation (26, 28), the autotranslocation of FL-BAK was not accompanied by cytochrome c release (Fig. 1E). These data highlight the successful production of full-length monomeric BAK, which exhibits a natural preference for partitioning to mitochondria. Strikingly, this translocation is not accompanied by mitochondrial outer membrane permeabilization, suggesting that a further activation step is required.
Direct Activation of FL-BAK by Truncated BID and Its BH3 Helix.
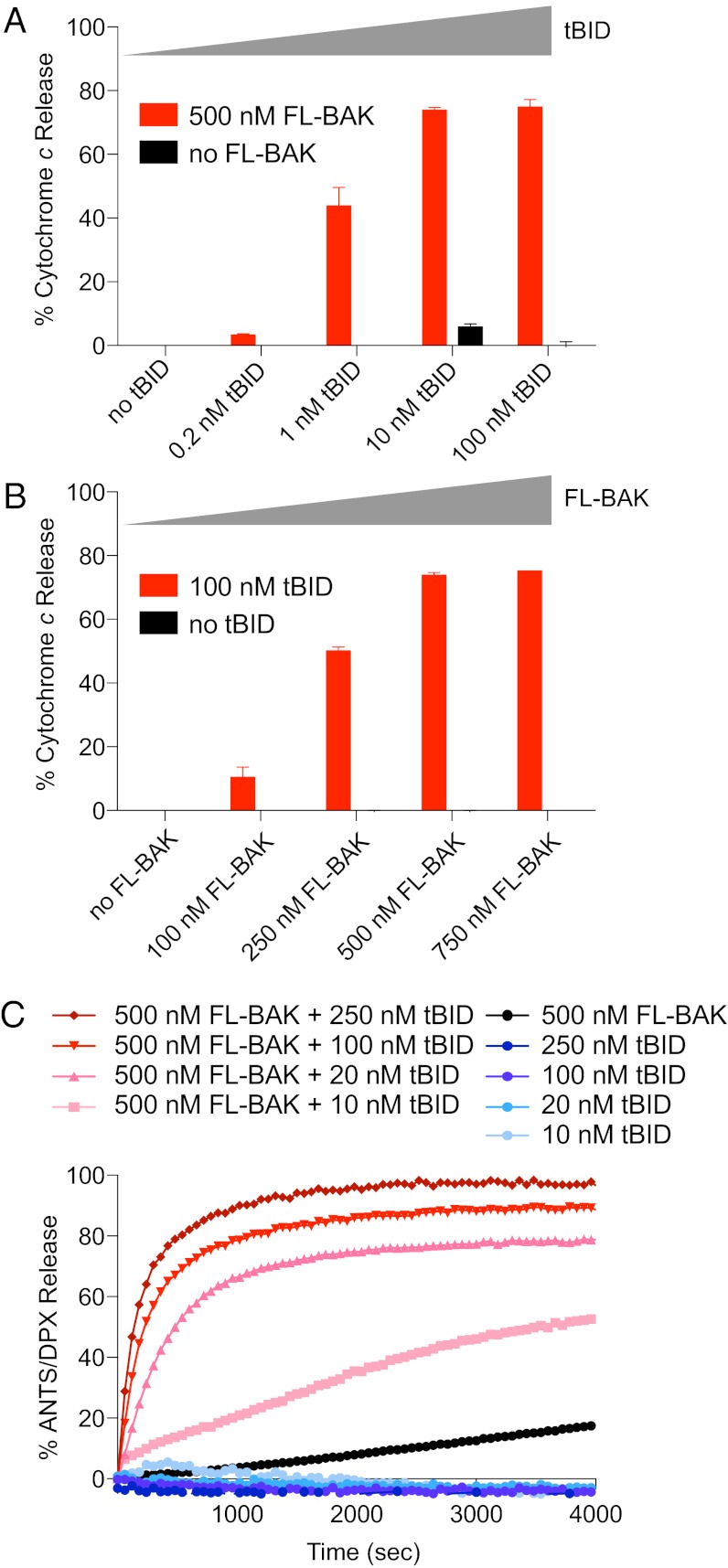
To determine if FL-BAK was functional, we subjected the mixture of FL-BAK and Bak−/− MLM to the activator BH3-only protein, truncated BID (tBID). We observed reciprocal dose-responsive activation of tBID-induced FL-BAK–mediated cytochrome c release, indicative of functionally intact BAK protein (Fig. 2 A and B). Because the MLM experimental framework does not distinguish between tBID-induced direct activation of FL-BAK and derepression of FL-BAK as a result of antiapoptotic inhibition by tBID, we also used a reductionist liposomal system to evaluate FL-BAK release activity in the absence of other mitochondrial factors. Incubating liposomes containing 8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt (ANTS)/p-xylene-bis-pyridinium bromide (DPX) (fluorophore/quencher) with tBID or FL-BAK alone yielded little to no release of entrapped fluorophore, but the combination of tBID and FL-BAK induced dose- and time-responsive liposomal release (Fig. 2C). SEC analysis of the liposomal reaction mixtures documented conversion of FL-BAK from a monomeric to an oligomeric form upon tBID exposure, with the majority of tBID remaining monomeric, consistent with the proposed “hit-and-run” mechanism for BH3-induced direct BAX/BAK activation (1) (Fig. S3). We further confirmed by chemical crosslinking analysis that, in each case, tBID-induced and FL-BAK–mediated liposomal and mitochondrial release was accompanied by oligomerization of FL-BAK (Fig. S4).
Fig. 2.
The BH3-only protein tBID triggers FL-BAK–dependent mitochondrial and liposomal release. (A and B) tBID dose-responsively triggered FL-BAK–mediated cytochrome c release from isolated Bak−/− mitochondria, as measured by cytochrome c ELISA. Likewise, FL-BAK dose-responsively induced cytochrome c release upon exposure to fixed-dose tBID. No release was observed upon exposure of the mitochondria to tBID or FL-BAK alone. Experiments were performed in duplicate and were repeated at least three times using independent FL-BAK preparations with similar results. Data are mean ± SD. (C) tBID dose-responsively induced FL-BAK–mediated liposomal release of entrapped fluorophore, whereas tBID or FL-BAK alone had little to no effect. Data are representative of at least three independent experiments with similar results.
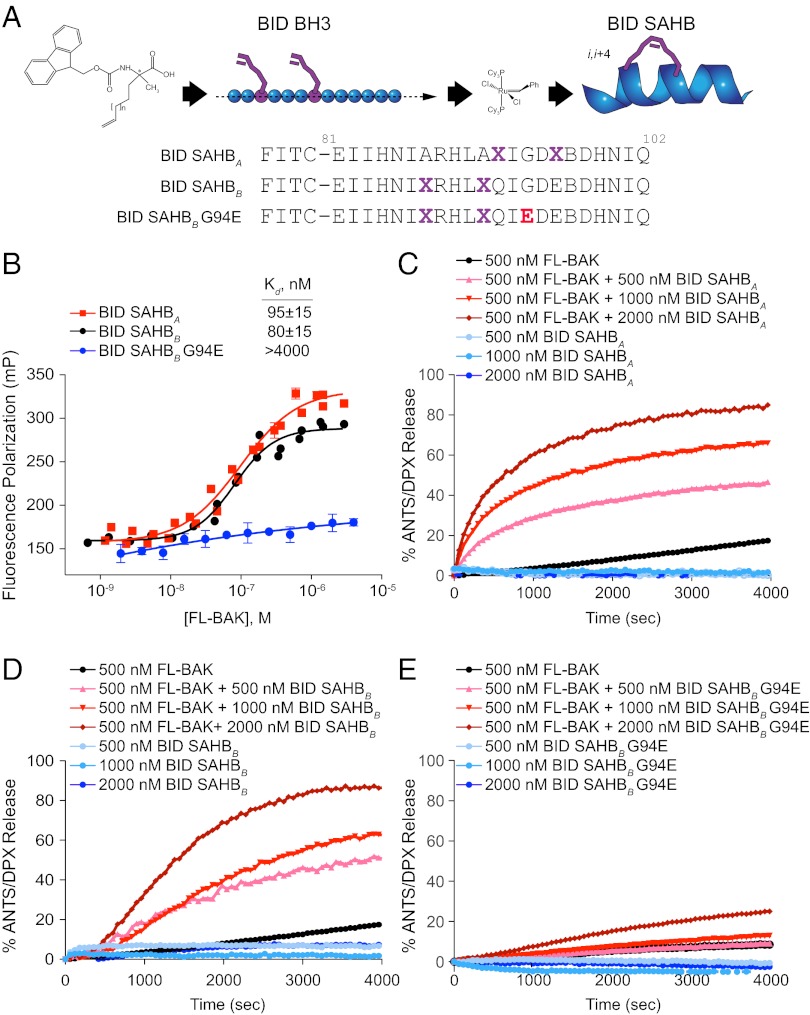
To link the FL-BAK–triggering activity of tBID to its BH3 domain, we generated stabilized α-helices of BCL-2 domains (SAHBs) modeled after the BID BH3 helix (20, 26) for functional analysis (Fig. 3A). We first performed fluorescence polarization (FP) binding analyses using FITC-BID SAHBs and FL-BAK, and we detected and quantified the direct interaction between BID BH3 and FL-BAK. Although two differentially stapled BID SAHBs engaged FL-BAK with Kd values of 80–95 nM, single G94E point mutagenesis of the α-helical interface abrogated binding activity (Fig. 3B). Correspondingly, BID SAHBs dose- and time-responsively activated FL-BAK–mediated liposomal release (Fig. 3 C and D), whereas little to no activity was observed for the point mutant SAHB (Fig. 3E). Taken together, these data document that recombinant and monomeric FL-BAK is functionally active and can be induced to porate membranes as a result of direct and measurable interactions with BID BH3.
Fig. 3.
BID SAHBs bind to FL-BAK in a sequence-dependent manner and directly trigger FL-BAK activation. (A) SAHBs corresponding to the BH3 motif of the BH3-only protein BID were generated by substituting nonnatural amino acids bearing olefin tethers at i, i+4 positions followed by ruthenium-catalyzed olefin metathesis. G94E point mutagenesis of the hydrophobic binding interface yielded a negative control SAHB for biochemical studies. X, stapling amino acid; B, norleucine (substituted for methionine to avoid thioether-based interference with ruthenium catalysis). (B) BID SAHBs A and B bound to FL-BAK with nanomolar affinity, whereas G94E point mutagenesis abrogated the interaction, as measured by FP assay. Experiments were performed at least in duplicate and were repeated three times with independent preparations of FL-BAK. Data are mean ± SEM. (C–E) BID SAHBs A and B dose-responsively induced FL-BAK–mediated liposomal release of entrapped fluorophore, whereas BID SAHBB G94E, or BID SAHBs or FL-BAK alone, had little to no effect. Data are representative of at least three independent experiments with similar results.
Photoreactive BID SAHBs Localize the BH3 Trigger Site on BAK.
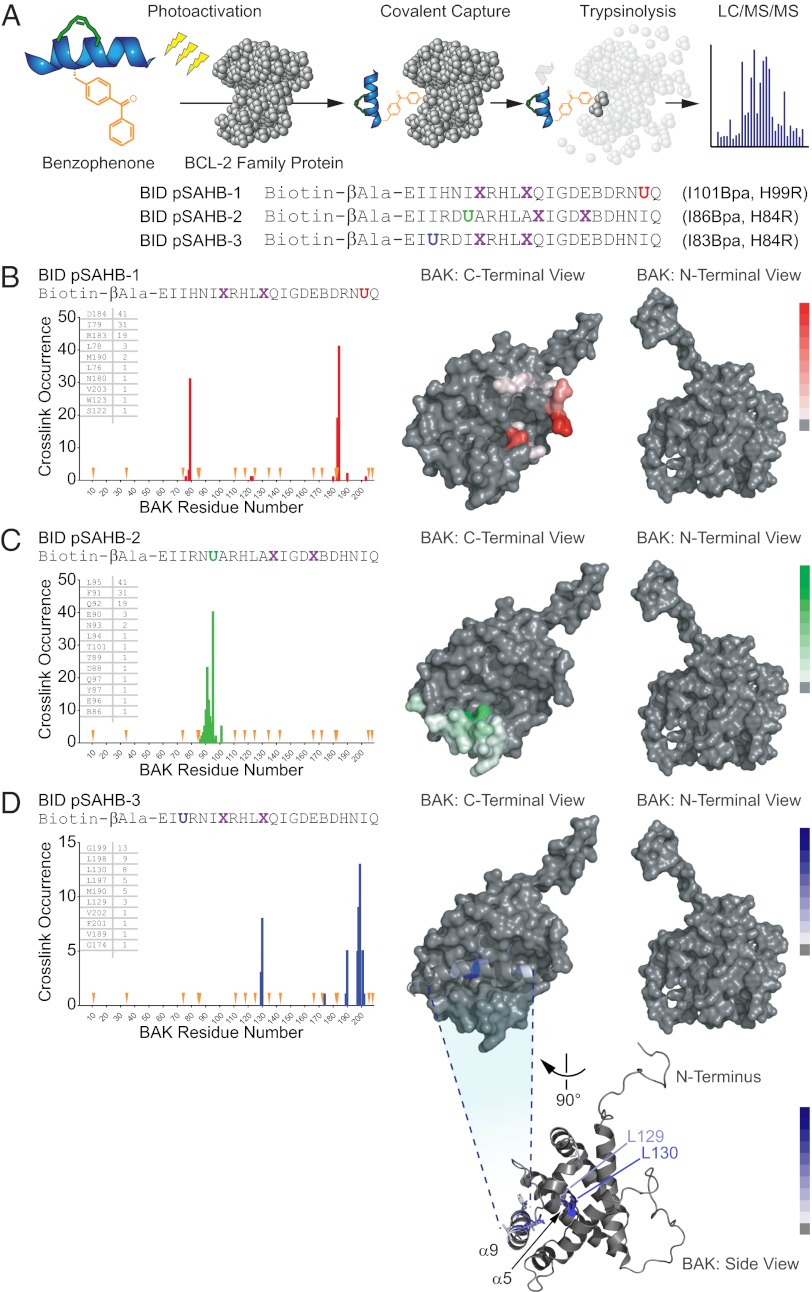
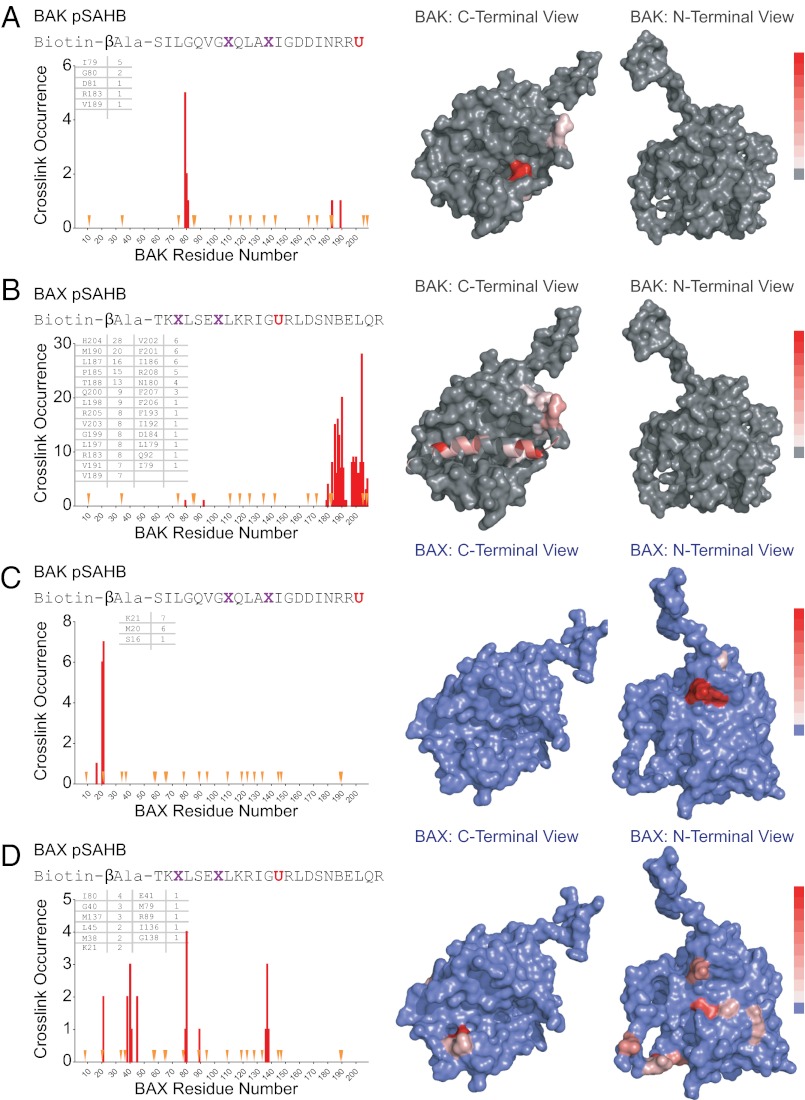
We previously developed photoreactive SAHBs (pSAHBs) containing differentially localized benzophenone moieties as chemical tools for covalently trapping protein interactors and explicitly localizing the sites of intercalation by MS (38). pSAHBs are ideally suited for rapid binding-site identification in the absence of protein solution or crystal structures. Here, we synthesized and deployed a panel of BID pSAHBs (Fig. 4A) to locate the BH3 interaction site on FL-BAK. To validate the binding specificity and utility of BID pSAHBs 1–3, we first conducted crosslinking analyses with the C-terminally truncated form of antiapoptotic BCL-XL (BCL-XLΔC), for which definitive structures are known. We found that the differentially placed benzophenone moieties in each BID pSAHB crosslinked to discrete subregions of the canonical BH3 binding pocket, corresponding precisely to the established structures of BH3 helix/BCL-XLΔC complexes [e.g., Protein Data Bank (PDB) IDs 2YJ1, 3FDL, 1BXL, and 3PL7] (Fig. S5 A–D).
Fig. 4.
Photoreactive BID pSAHBs localize the BH3 trigger site on BAK to its C-terminal BH3-binding pocket. (A) pSAHBs modeled after the BH3 domain of BID were generated for protein capture and binding-site identification by replacing select native residues with 4-benzoyl-L-phenylalanine (Bpa), followed by ring-closing metathesis of olefinic nonnatural amino acids installed at i, i+4 positions. To facilitate tryptic digestion of pSAHBs into shorter and more identifiable fragments by MS, single arginine substitutions were made as indicated, taking advantage of natural sites of homology between the human and mouse BID BH3 domains (i.e., H84R, H99R). X, stapling amino acid; B, norleucine; U, Bpa. (B–D) BID pSAHBs 1–3 were incubated individually with FL-BAK, and the mixtures were subjected to UV irradiation, electrophoresis, excision of the crosslinked protein, trypsin proteolysis, and LC-MS/MS analysis. The plots depict the frequency of crosslinked sites identified across the FL-BAK polypeptide sequence. Crosslinked residues are mapped onto a calculated model structure of FL-BAK (based on sequence homology to BAX) and colored according to the frequency of occurrence for each pSAHB (1, red; 2, green; 3, blue). For BID pSAHBs 1 and 2, which do not crosslink to residues of the C-terminal helix of FL-BAK, α9 has been removed from the structure to better visualize the crosslinked residues at the surface of the canonical BH3-binding pocket. For BID pSAHB-3, a side view of FL-BAK is also shown to demonstrate the localization of crosslinked residues at both the surface of the canonical BH3-binding pocket and the inner surface of α9. The latter dataset shows that, after α9 displacement, previously unexposed and inward-facing α9 residues become available for interaction with BID pSAHB-3.
Having validated the fidelity of BID pSAHBs 1–3, we next subjected FL-BAK to the crosslinking analysis. Walking the benzophenone moiety from the C to the N terminus of BID pSAHB led to sequential crosslinking of discrete residue clusters along the canonical BH3-binding pocket of FL-BAK (Fig. 4 B–D). Interestingly, BID pSAHB-3, which contained the most N-terminal benzophenone substitution, also crosslinked to select internal face residues of BAK α9, suggesting that the BID BH3 helix directly displaces the C-terminal helix, whose hydrophobic face becomes exposed for interaction (Fig. 4D). Importantly, we confirmed that BID pSAHB engagement of the canonical BH3-interaction site likewise occurred in the membrane context. We generated and purified FL-BAK–embedded and ANTS/DPX-loaded liposomes, confirming by proteinase K digestion and anti-BAK Western blot that FL-BAK was susceptible to proteolysis and thus surface exposed (Fig. S6A). The FL-BAK–containing liposomes preserved membrane integrity, as reflected by the release of fluorophore upon Triton X-100 lysis (Fig. S6B), and underwent dose-responsive, ligand-triggered release upon exposure to tBID (Fig. S6C). Having validated the functional reconstitution of FL-BAK into liposomes, we then incubated the liposomes with BID pSAHB-1 in the presence of UV light and subjected the mixture to our MS-based crosslinking analysis. Although the crosslinking and site-identification method was less efficient in the liposomal context, discrete residues of the FL-BAK canonical pocket were again detected by BID pSAHB-1 photoaffinity labeling (Fig. S6D). Thus, we find that, whether performed in solution or in the membrane environment, BID SAHB directly engages residues of the canonical BH3-binding groove of FL-BAK.
Activator pSAHBs Detect Distinct Trigger Sites for BAK and BAX.
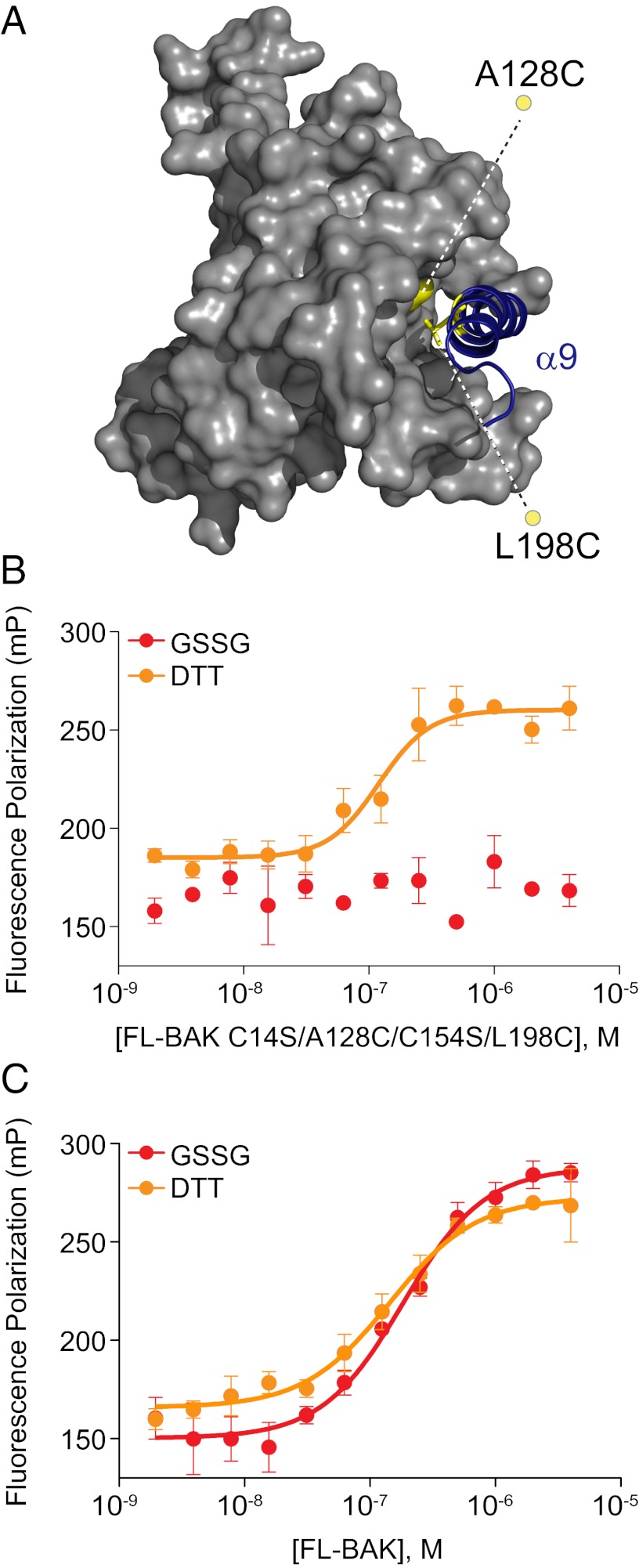
Because photoactivated benzophenone crosslinking may favor stable over transient BH3 interactions, we sought to interrogate further the BID pSAHB crosslinking results to rule out the possibility of an alternative BH3 interaction site on BAK, as discovered for BAX by NMR analysis (27). First, we mutated Leu198 of α9 and Ala128 of α5 to cysteine residues and converted native cysteines C14 and C154 to serines to generate a FL-BAK construct in which α9 can be reversibly locked into its canonical binding pocket based on redox conditions (Fig. 5A). FITC-BID SAHBB bound to FL-BAK C14S/A128C/C154S/L198C under reducing conditions (10 mM DTT), but no interaction was observed under oxidizing conditions [2 mM glutathione disulfide (GSSG)] (Fig. 5B). Of note, the reducing and oxidizing conditions had no effect on the interaction between FITC-BID SAHBB and the parental FL-BAK construct (Fig. 5C). Thus, the data suggest that when the canonical BH3-binding pocket is blocked by locking the C terminus in place, no further binding can occur between BID SAHB and FL-BAK. This result stands in striking contrast to the biochemical- and NMR-based analyses of the interaction between BIM SAHB and BAX, in which covalent tethering of the BAX C terminus to its canonical pocket had no disruptive effect on BIM SAHB binding, which was maintained at the α1/α6 trigger site (28).
Fig. 5.
The binding interaction between BID SAHB and FL-BAK requires access to the canonical BH3-binding pocket. (A) To determine if the BID SAHB/FL-BAK interaction is explicitly dependent on displacement of α9 (blue) and exposure of the canonical pocket, FL-BAK residues A128 of α5 and L198 of α9 were mutated to cysteines (yellow), and native cysteines C14 and C154 were mutated to serines, yielding an α9-tethered FL-BAK construct in which the C-terminal helix is reversibly locked into its binding pocket by redox conditions. (B) Oxidized FL-BAK (2 mM GSSG) showed no interaction with FITC-BID SAHBB, whereas binding activity was completely restored upon addition of reducing agent (10 mM DTT), as assessed by FP assay. Experiments were performed at least in duplicate and were repeated at least two times with independent preparations of FL-BAK. Data are mean ± SEM. (C) In contrast, redox conditions have no effect on the binding interaction between FITC-BID SAHBB and FL-BAK. Experiments were performed in at least duplicate and repeated at least twice with independent preparations of FL-BAK. Data are mean ± SEM.
Next, we subjected recombinant full-length BAX to BID pSAHB crosslinking analysis to determine if pSAHBs were capable of detecting the α1/α6 trigger site. Indeed, we find that BID pSAHBs crosslinked to a series of residues located within the previously defined BH3 trigger site at the N-terminal face of BAX (Fig. S7 A and B). Interestingly, BID pSAHB-2 also crosslinked to discrete residues within the canonical BH3-binding pocket of BAX (Fig. S7B). These data suggest that, once BAX is triggered at the α1/α6 site, which was defined previously by biochemical and NMR studies as the initiating interaction for BAX activation (28), compatible BH3 binding at the canonical groove upon allosteric release of BAX α9 can also occur.
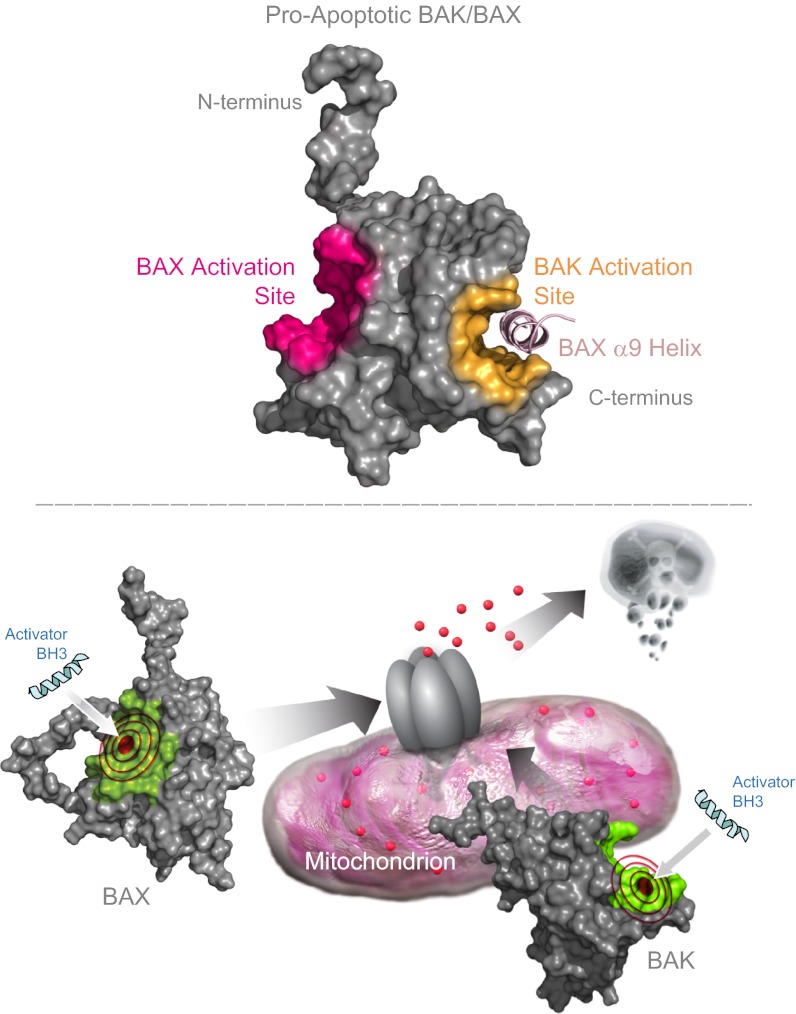
To document that this paradigm holds for another activator BH3 helix, we generated and applied a BIM pSAHB in crosslinking analyses of both FL-BAK and BAX. Again, we found that BIM pSAHB crosslinked exclusively to residues of the canonical BH3-binding pocket on BAK (Fig. S7C), whereas a series of α1, α1–α2 loop, and α6 residues also crosslinked only in BAX (Fig. S7D). Finally, to explore whether the distinct trigger sites for initiating direct activation of BAK/BAX by BID/BIM BH3 helices are invoked during the self-propagation and/or oligomeric self-association steps of BAK/BAX activation (28, 40, 41), we generated BAK and BAX pSAHBs corresponding to the BAK and BAX BH3 helices for crosslinking analyses. Consistent with the results observed for BID and BIM pSAHBs, BAK and BAX pSAHBs crosslinked exclusively to residues of the canonical BH3-binding pocket of FL-BAK (Fig. 6 A and B), whereas the same pSAHBs crosslinked either additionally or exclusively to residues of the α1/α6 trigger site on BAX (Fig. 6 C and D). Whether the identified BAK BH3 in groove interaction reflects a propagation mechanism defined by nucleation of dimers that then oligomerize through a second stable binding interface, as previously proposed (36, 37), or a linear autoactivation mechanism that ultimately produces a membrane-embedded oligomer of conformationally altered monomers remains to be structurally determined. In the former case, the identified crosslinks between BAK pSAHB and the canonical pocket of FL-BAK likewise could reflect a stable component of the homo-dimer or homo-oligomer of BAK. Taken together, these data suggest that the initiating step for direct BAK and BAX activation, whether triggered by activator BH3-only domains or propelled by BAK/BAX BH3 domains, employs distinct BH3-binding interfaces on the two proapoptotic multidomain proteins—the canonical groove for FL-BAK and the noncanonical α1/α6 site for full-length BAX.
Fig. 6.
The BAK/BAX BH3 helices likewise engage distinct trigger surfaces on FL-BAK and BAX. (A and B) BAK and BAX pSAHBs were incubated individually with FL-BAK, and the mixtures were subjected to UV irradiation, electrophoresis, excision of the crosslinked protein, trypsin proteolysis, and LC-MS/MS analysis. The plots depict the frequency of crosslinked sites identified across the FL-BAK polypeptide sequence. Crosslinked residues are mapped onto the calculated model structure of FL-BAK (gray) and colored according to the frequency of occurrence as in Fig. 4. Because BAK pSAHB crosslinks to FL-BAK α9 were not identified, the C-terminal helix was removed from the calculated FL-BAK structure to better visualize the crosslinked residues at the surface of the canonical BH3-binding pocket. (C and D) When exposed to full-length BAX, the identical BAK and BAX pSAHBs crosslinked to a series of residues at the α1/α6 trigger site, with no canonical site crosslinks observed for BAK pSAHB and a select few identified for BAX pSAHB. The plots depict the frequency of crosslinked sites identified across the BAX polypeptide sequence. Crosslinked residues are mapped onto the structure of BAX (blue) and colored according to the frequency of occurrence as in Fig. 4. Because pSAHB crosslinks to BAX α9 were not evident, the C-terminal helix was removed from the BAX structure to better visualize the BAX pSAHB-crosslinked residues at the surface of the canonical BH3-binding pocket. X, stapling amino acid; B, norleucine; U, Bpa.
Discussion
The oligomerization of BAK and BAX in the mitochondrial outer membrane is one of the ultimate control points for mitochondrial apoptosis (2). As such, the mechanisms that underlie BAK/BAX regulation and channel formation remain a high-priority focus in cell death research. The production of recombinant BCL-2 family proteins has been an essential step in both dissecting their biochemical functions and translating the insights into potential therapies to modulate cell death in human disease. For example, bacterial expression and purification of antiapoptotic BCL-XL, with the C terminus removed and an unstructured loop trimmed, led to a fundamental understanding of how BCL-2 survival proteins trap the BH3 helices of proapoptotic proteins to block cell death (10, 42) and provided a blueprint for the development of ABT-263, the first selective BCL-2/BCL-XL inhibitor to advance to clinical testing in cancer (21, 43, 44). Because of the membrane-based activity of many BCL-2 family members, which is facilitated by a hydrophobic infrastructure including a C-terminal membrane insertion helix, the production of sufficient soluble, full-length, and monomeric recombinant protein for biochemical and structural analyses has been a longstanding challenge. The C-termini of BCL-2 family proteins appear to play a key role in structural integrity, membrane trafficking, and functional insertion (12, 28, 45, 46), and therefore, the analysis of full-length constructs is ideal. For example, our understanding of the functional dynamics of BCL-W and BAX has been advanced greatly by the structural and biochemical analyses of the corresponding full-length proteins (12, 28, 47–49). Although the cytosolic disposition of select multidomain BCL-2 family proteins has favored bacterial expression and purification of full-length monomeric protein, membrane-localized members have been especially refractory.
To overcome the longstanding challenge of producing full-length, monomeric BAK, we undertook a surface mutagenesis approach to increase the hydrophilicity of its hydrophobic C terminus. In doing so, we developed an FL-BAK construct amenable to bacterial expression, affinity chromatography, and FPLC-based purification in monomeric form. What has been learned from interrogating recombinant FL-BAK? First, in contrast to full-length recombinant BAX, soluble FL-BAK autotranslocates to isolated mitochondria, demonstrating a differential membrane tropism that is consistent with the relative subcellular distributions of BAX and BAK in vivo. Potential differences in affinity of the BAX and BAK α9 helices for their respective binding pockets, leading to distinct propensities to release α9 for membrane insertion, could account for this phenomenon. Second, translocation of FL-BAK, in and of itself, does not permeabilize the mitochondria, indicating that a definitive activation step is required either in the form of a direct trigger or inhibition of suppressive heterotypic protein interactions. Third, the combined use of FL-BAK and BID SAHB enabled the quantitation of a direct and sequence-dependent interaction between BAK and an activator BH3 helix. The BH3-only protein tBID or its essential BH3 helix in the form of BID SAHB activates and functionally oligomerizes FL-BAK in the absence of any other factors, definitively validating a direct activation mechanism for BAK. Fourth, biochemical occlusion of the canonical BH3-binding pocket by disulfide tethering of the BAK C-terminal helix abrogates the binding interaction between BID SAHB and FL-BAK, suggesting that, in contrast to BAX, the primary site for initiating direct BAK activation lies at the C-terminal face of the protein. Finally, with FL-BAK and BAX in hand, combined with the application of photoreactive SAHBs, we were able to accomplish comparative structural mapping of BH3 binding sites on these essential executioner proteins, revealing distinct trigger sites for initiating and propelling their activation.
Installation of photoreactive benzophenone moieties along the sequence of BH3 peptide helices enables an unbiased analysis of covalent intercalation sites on a target protein using proteomic methods (38). The capacity to map binding interfaces rapidly and accurately with this combined chemical biology and MS/MS strategy provides an alternative and complementary approach to bridge the often multiyear gap until such interfaces are characterized by definitive structural methods such as NMR or X-ray crystallography. In this case, the application of BH3-only direct activator helices (26–28, 40), such as BID, BIM, BAK, and BAX pSAHBs, all pointed to an exclusive site of direct FL-BAK interaction at its canonical binding groove. In striking contrast, the very same pSAHBs consistently crosslinked to an entirely distinct BH3-binding site localized to the opposite side of the full-length BAX protein, previously characterized by a battery of NMR and biochemical analyses as the trigger site for direct BAX activation (27, 28). Thus, to initiate BAK and BAX activation, membrane-localized BAK and cytosolic BAX use distinct triggering surfaces. Interestingly, a subset of activator pSAHBs showed dual crosslinking to both the α1/α6 and canonical BH3-binding sites on BAX, suggesting that once triggered at the N terminus, release of the BAX C terminus exposes a second BH3-compatible binding interface at the canonical groove. Indeed, this concept of sequential BH3-only binding interactions to drive BAX activation is supported by a prior stepwise analysis of tBID-induced BAX oligomerization (35). Taken together, our data suggest that activator BH3-engagement at the canonical pocket in the context of the mitochondrial outer membrane may represent a common mechanism for promoting BAK and BAX activation, with the α1/α6 triggering mechanism of BAX representing a unique afferent step required to regulate the activation and mitochondrial translocation of cytosolic BAX. The ultimate goal of these direct mechanisms for BAK and BAX activation is membrane integration in the form of oligomeric pores, whose structures remain to be defined. Indeed, the development of full-length forms of BCL-2 family members, such as BAK, is an essential step in advancing our structural and biochemical understanding of the protein interaction dynamics that dictate their proapoptotic function in the membrane environment. Further, our characterization of the differential trigger sites for BAK and BAX provides a blueprint for the development of selective pharmacologic agents, such as recently described for BAX (50), to directly activate the death program in diseases of pathologic cell survival.
Methods
Expression and Purification of FL-BAK.
Production of monomeric FL-BAK was achieved by introducing the point mutations I192K/F193S/V196D into the native mouse BAK sequence (National Center for Biotechnology Information: NP_031549.2) (Fig. 1A), which was subcloned into the pTYB4 vector (New England BioLabs) using NcoI and SmaI restriction sites. PCR-based site-directed mutagenesis was confirmed by DNA sequencing. The chitin-binding domain fusion construct of FL-BAK was expressed in BL21(DE3) E. coli and induced with 1 mM isopropylthio-β-galactoside at 18 °C overnight. Collected bacterial pellets were resuspended in lysis buffer (20 mM Tris, 250 mM NaCl, pH 7.6), lysed by microfluidization (M-110L; Microfluidics), and centrifuged at 45,000 rpm for 1 h at 4 °C (Beckman L-90K, rotor type 70 Ti). The cleared cellular lysates were subjected to chitin affinity resin (New England BioLabs) chromatography followed by overnight on-bead intein-tag cleavage using 50 mM DTT. FL-BAK was eluted with lysis buffer, concentrated, and then subjected to SEC (GE Healthcare Life Sciences) at 4 °C using 20 mM Hepes (pH 7.8), 150 mM KCl buffer conditions. FL-BAK and its cysteine-mutant derivative were used upon isolation without further storage. FL-BAK C14S/A128C/C154S/L198C was maintained in the oxidized state by exposure to 2 mM GSSG; reduction of the disulfide tether was accomplished by 1 h incubation in 10 mM DTT. Recombinant full-length BAX and BCL-XLΔC were produced as described (12, 26), and tBID was purchased from R&D Systems. Protein concentrations were determined by Bradford assay (Bio-Rad).
Structure Modeling.
A model structure of BAK was generated based on the solution structure of BAX (PDB ID 1F16) (12) using Modeller software (51) and was evaluated and displayed using PROCHECK_NMR (52) and PYMOL (53).
SAHB Synthesis and Characterization.
Peptide synthesis, hydrocarbon stapling by olefin metathesis, and derivatization with FITC and biotin were performed according to our established methods (54, 55). All peptides were purified by LC/MS to >95% purity and quantified by amino acid analysis. pSAHBs were generated as described (38) by incorporating Fmoc-4-benzoyl-L-phenylalanine (EMD Biosciences) at the indicated locations (U) in the peptide sequence.
Mitochondrial Translocation Assay.
Bak−/− mitochondria were isolated from the livers of Bak−/− mice as previously reported (26), resuspended in experimental buffer [125 mM KCl, 10 mM Tris⋅3-(N-morpholino)propanesulfonic acid (Mops) (pH 7.4), 5 mM glutamate, 2.5 mM malate, 1 mM KPO4, 10 μM EGTA-Tris (pH 7.4)], and then treated with the indicated concentrations of freshly purified FL-BAK. After 90 min incubation at room temperature followed by tabletop centrifugation at 5,500 × g, the supernatant and pellet fractions were separated and subjected to SDS/PAGE and immunoblotting using BAK NT (Millipore) and cytochrome c (BD Pharmingen) antibodies.
Mitochondrial Cytochrome c Release Assay.
Cytochrome c release assays were performed as previously reported (26). Briefly, Bak−/− mitochondria were incubated with the indicated concentrations of FL-BAK and tBID for 45 min at room temperature in experimental buffer (see above). The pellet and supernatant fractions were isolated by centrifugation at 3,200 rpm, and cytochrome c was quantitated using a colorimetric ELISA (R&D Systems). Percent cytochrome c released into the supernatant (%cytocsup) from releasable mitochondrial pools was calculated according to the following equation: %cytoc = [(cytocsup − cytocbackgr)/(cytoctotal − cytocbackgr)]*100, where background release represents cytochrome c detected in the supernatant of vehicle-treated samples and total release represents cytochrome c measured in 1% (vol/vol) Triton X-100–treated samples.
Liposomal Release Assay.
The liposomal release assay was adapted from a previously described method (48). Large unilamellar vesicles (LUVs) were generated from a lipid mixture containing 48 mol% phosphatidylcholine, 28 mol% phosphatidylethanolamine, 10 mol% phosphatidylinositol, 10 mol% dioleoyl phosphatidylserine, and 4 mol% tetraoleoyl cardiolipin (Avanti Polar Lipids). The lipid mixture (1-mg aliquots) was prepared from chloroform stocks and dried as a thin film in glass test tubes under nitrogen gas and then under vacuum for 15 h. The fluorescent dye ANTS (6.3 mg) and the quencher DPX (19.1 mg) were added to 1 mg of dry lipid film, and the mixture was resuspended in assay buffer (200 mM KCl, 1 mM MgCl2, 10 mM Hepes, pH 7.0). After five freeze-thaw cycles (liquid nitrogen/water bath), the lipid mixtures were extruded through a 100-nm nucleopore membrane (Whatman) using an Avanti mini extruder, followed by gravity flow SEC using a crosslinked Sepharose CL-2B column (Sigma Aldrich). LUVs (10 μL) were treated with the indicated concentrations of FL-BAK, tBID, and BID SAHBs, singly and in combination, in 96-well format (Corning) in a total reaction volume of 100 μL. Fluorescence was measured from t = 0 (F0) to t = 2 h, and then Triton X-100 was added to a final concentration of 0.2% (vol/vol) followed by measurement of fluorescence for an additional 10 min to determine maximal release (F100). Percent ANTS/DPX release was calculated as [(F − F0)/(F100 − F0)] × 100.
FP Binding Assay.
FP assays were performed as previously described (56). Briefly, FITC-BID SAHBs (15 nM) were incubated with the indicated serial dilution of FL-BAK protein in binding buffer (50 mM Tris, 100 mM NaCl, pH 8.0) until equilibrium was reached. FP was measured using a SpectraMax M5 microplate reader (Molecular Devices). Kds were calculated by nonlinear regression analysis of dose–response curves using Prism software (GraphPad).
Photoaffinity Labeling.
pSAHBs were mixed with FL-BAK or BAX at a 1:1 ratio (10 μM) in Buffer A (20 mM Tris, 250 mM NaCl, pH 7.6), incubated for 10 min, and then irradiated at 365 nm for 2 h on ice, in accordance with our previously reported method (38). Unreacted pSAHBs were removed by overnight dialysis at 4 °C in Buffer B (200 mM NaCl, 50 mM Tris, pH 7.4) using D-tube dialyzers with a molecular weight cutoff of 6–8 kDa (EMD Biosciences). Biotin capture was accomplished by incubating the reaction mixture with high-capacity streptavidin agarose (Thermo) in Buffer B. Streptavidin beads were washed three times with 1% (wt/vol) SDS in PBS (3×), 1 M NaCl in PBS and then three times with 10% (vol/vol) ethanol in PBS. Biotinylated proteins were eluted by boiling in 10% (wt/vol) SDS solution (Promega) containing 10 mg/mL D-biotin, subjected to SDS/PAGE, and then visualized by Coomassie staining (Simply Blue Safestain; Invitrogen). Bands corresponding to crosslinked species were excised, subjected to in-gel digestion with trypsin, and then analyzed by LC-MS/MS (see below).
MS Analysis.
MS analysis of covalent pSAHB interaction sites was performed as previously described (38). Excised gel bands were destained, washed by dehydration, reduced with 20 mM DTT in 100 mM ammonium bicarbonate, washed again, and then alkylated by incubation with 10 mM iodoacetamide in the dark. After washing and dehydration with acetonitrile, gel slices were rehydrated overnight in trypsin digestion buffer [12.5 ng/mL sequencing grade modified trypsin (Promega) in 50 mM ammonium bicarbonate]. After digestion, samples were extracted from the gel with 50% (vol/vol) acetonitrile/5% (vol/vol) formic acid in water, the solvent was evaporated by SpeedVac (Thermo Fisher), and the samples were resuspended in 0.5% (vol/vol) trifluoroacetic acid in 1 M urea followed by desalting using C18 Stop and Go Extraction (STAGE) tips, constructed in house using Empore C18 solid phase extraction material (57). Samples were subjected to nano LC-MS/MS using a 20-cm column composed of a 100-μm i.d. fused silica capillary that was flame pulled in-house to produce an ∼5-μm tip, and packed with Maccel C18 Resin (3 μm particle size, 200Å pore size, The Nest Group, Inc.). Data were collected on an LTQ Orbitrap Discovery hybrid mass spectrometer (ThermoFisher) operated in data-dependent mode (38). The MS/MS spectra were assigned by searching with the SEQUEST algorithm (58) against a sequence-reversed database containing BAK, the corresponding mutated BAK sequences, BAX, BCLXLΔC, trypsin, and common keratin contaminants. Peptide false-detection rates (FDR) were limited to <5% by filtering peptide spectral matches according to their XCorr, mass accuracy, tryptic state, charge state, and peptide length. When filtering for crosslinked species, spectral matches corresponding to multiply crosslinked peptides or crosslinked keratins were considered contaminants and included in the FDR 5% limit. The resultant high-confidence sites of covalent modification were plotted by spectral count frequency across the polypeptide sequence.
Supplementary Material
Acknowledgments
We thank Eric D. Smith for graphical support, Marina Godes for technical assistance with mitochondrial isolation and purification, David Whitehead for protein production support, Eileen White for providing wild-type and Bax−/−Bak−/− iBMK cells, David Andrews for guidance on the liposomal assay system, and Steven Gygi and Wilhelm Haas for their advice and input on the MS analyses. This work was supported by National Institutes of Health Grants 5R01CA050239 and 5R01GM090299 and a Stand Up to Cancer Innovative Research Grant (to L.D.W.) and a Lander Fellowship in Cancer Chemical Biology and an American Association of University Women International Fellowship (to E.S.L.).
Footnotes
Conflict of interest statement: L.D.W. is a scientific advisory board member and consultant for Aileron Therapeutics.
This article is a PNAS Direct Submission.
See Author Summary on page 4168 (volume 110, number 11).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214313110/-/DCSupplemental.
References
- 1.Wei MC, et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14(16):2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 2.Wei MC, et al. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science. 2001;292(5517):727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jürgensmeier JM, et al. Bax directly induces release of cytochrome c from isolated mitochondria. Proc Natl Acad Sci USA. 1998;95(9):4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J, et al. Prevention of apoptosis by Bcl-2: Release of cytochrome c from mitochondria blocked. Science. 1997;275(5303):1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 5.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102(1):33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 6.Chittenden T, et al. Induction of apoptosis by the Bcl-2 homologue Bak. Nature. 1995;374(6524):733–736. doi: 10.1038/374733a0. [DOI] [PubMed] [Google Scholar]
- 7.Farrow SN, et al. Cloning of a bcl-2 homologue by interaction with adenovirus E1B 19K. Nature. 1995;374(6524):731–733. doi: 10.1038/374731a0. [DOI] [PubMed] [Google Scholar]
- 8.Kiefer MC, et al. Modulation of apoptosis by the widely distributed Bcl-2 homologue Bak. Nature. 1995;374(6524):736–739. doi: 10.1038/374736a0. [DOI] [PubMed] [Google Scholar]
- 9.Chittenden T, et al. A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein binding functions. EMBO J. 1995;14(22):5589–5596. doi: 10.1002/j.1460-2075.1995.tb00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: Recognition between regulators of apoptosis. Science. 1997;275(5302):983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 11.Lindsten T, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6(6):1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: Coregulation of dimer formation and intracellular localization. Cell. 2000;103(4):645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, et al. Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator. J Struct Biol. 2009;166(1):32–37. doi: 10.1016/j.jsb.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Moldoveanu T, et al. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell. 2006;24(5):677–688. doi: 10.1016/j.molcel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Willis SN, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315(5813):856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 16.Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science. 2003;301(5632):513–517. doi: 10.1126/science.1083995. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17(3):393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 18.Cheng EH, et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8(3):705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 19.Day CL, et al. Solution structure of prosurvival Mcl-1 and characterization of its binding by proapoptotic BH3-only ligands. J Biol Chem. 2005;280(6):4738–4744. doi: 10.1074/jbc.M411434200. [DOI] [PubMed] [Google Scholar]
- 20.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305(5689):1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435(7042):677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen M, et al. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci USA. 2007;104(49):19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. BID: A novel BH3 domain-only death agonist. Genes Dev. 1996;10(22):2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 24.Kuwana T, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17(4):525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Kuwana T, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111(3):331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 26.Walensky LD, et al. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24(2):199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Gavathiotis E, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455(7216):1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell. 2010;40(3):481–492. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai H, et al. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J Cell Biol. 2011;194(1):39–48. doi: 10.1083/jcb.201102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh KJ, et al. Conformational changes in BAK, a pore-forming proapoptotic Bcl-2 family member, upon membrane insertion and direct evidence for the existence of BH3-BH3 contact interface in BAK homo-oligomers. J Biol Chem. 2010;285(37):28924–28937. doi: 10.1074/jbc.M110.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landeta O, et al. Reconstitution of proapoptotic BAK function in liposomes reveals a dual role for mitochondrial lipids in the BAK-driven membrane permeabilization process. J Biol Chem. 2011;286(10):8213–8230. doi: 10.1074/jbc.M110.165852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du H, et al. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J Biol Chem. 2011;286(1):491–501. doi: 10.1074/jbc.M110.167148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letai A, et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2(3):183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 34.Kim H, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006;8(12):1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 35.Kim H, et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36(3):487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dewson G, et al. Bak activation for apoptosis involves oligomerization of dimers via their alpha6 helices. Mol Cell. 2009;36(4):696–703. doi: 10.1016/j.molcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 37.Dewson G, et al. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol Cell. 2008;30(3):369–380. doi: 10.1016/j.molcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Braun CR, et al. Photoreactive stapled BH3 peptides to dissect the BCL-2 family interactome. Chem Biol. 2010;17(12):1325–1333. doi: 10.1016/j.chembiol.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathew R, Degenhardt K, Haramaty L, Karp CM, White E. Immortalized mouse epithelial cell models to study the role of apoptosis in cancer. Methods Enzymol. 2008;446:77–106. doi: 10.1016/S0076-6879(08)01605-4. [DOI] [PubMed] [Google Scholar]
- 40.Llambi F, et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44(4):517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan C, et al. Auto-activation of the apoptosis protein Bax increases mitochondrial membrane permeability and is inhibited by Bcl-2. J Biol Chem. 2006;281(21):14764–14775. doi: 10.1074/jbc.M602374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muchmore SW, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381(6580):335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 43.Park CM, et al. Discovery of an orally bioavailable small molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J Med Chem. 2008;51(21):6902–6915. doi: 10.1021/jm800669s. [DOI] [PubMed] [Google Scholar]
- 44.Wilson WH, et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: A phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2010;11(12):1149–1159. doi: 10.1016/S1470-2045(10)70261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferrer PE, Frederick P, Gulbis JM, Dewson G, Kluck RM. Translocation of a Bak C-terminus mutant from cytosol to mitochondria to mediate cytochrome C release: Implications for Bak and Bax apoptotic function. PLoS ONE. 2012;7(3):e31510. doi: 10.1371/journal.pone.0031510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nechushtan A, Smith CL, Hsu YT, Youle RJ. Conformation of the Bax C-terminus regulates subcellular location and cell death. EMBO J. 1999;18(9):2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinds MG, et al. The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity. EMBO J. 2003;22(7):1497–1507. doi: 10.1093/emboj/cdg144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lovell JF, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135(6):1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 49.Wilson-Annan J, et al. Proapoptotic BH3-only proteins trigger membrane integration of prosurvival Bcl-w and neutralize its activity. J Cell Biol. 2003;162(5):877–887. doi: 10.1083/jcb.200302144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gavathiotis E, Reyna DE, Bellairs JA, Leshchiner ES, Walensky LD. Direct and selective small-molecule activation of proapoptotic BAX. Nat Chem Biol. 2012;8(7):639–645. doi: 10.1038/nchembio.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fiser A, Sali A. Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 53.DeLano WL. 2002. The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA) www.pymol.org. Accessed February 1, 2013.
- 54.Bird GH, Crannell CW, Walensky LD. Chemical synthesis of hydrocarbon-stapled peptides for protein interaction research and therapeutic targeting. Curr Protoc Chem Biol. 2011;3:99–117. doi: 10.1002/9780470559277.ch110042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bird GH, Bernal F, Pitter K, Walensky LD. Synthesis and biophysical characterization of stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pitter K, Bernal F, Labelle J, Walensky LD. Dissection of the BCL-2 family signaling network with stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:387–408. doi: 10.1016/S0076-6879(08)01623-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 58.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]