Significance
This article concerns the control of competence for bacterial genetic transformation. Competence is transient in the human pathogen Streptococcus pneumoniae, involving the specific expression of ∼100 genes that are turned ON suddenly and OFF almost as abruptly. Although the mechanism rendering all cells in a culture simultaneously competent is well understood, how competence stops has remained unknown. Here, we unravel the mechanism of shut-off, describing the discovery that a key recombination protein, DprA, exerts a negative control on competence through direct, physical interaction with the master regulator of competence, the response regulator ComE, to abolish transcription from ComE-activated promoters.
Abstract
Natural bacterial transformation is a genetically programmed process allowing genotype alterations that involves the internalization of DNA and its chromosomal integration catalyzed by the universal recombinase RecA, assisted by its transformation-dedicated loader, DNA processing protein A (DprA). In Streptococcus pneumoniae, the ability to internalize DNA, known as competence, is transient, developing suddenly and stopping as quickly. Competence is induced by the comC-encoded peptide, competence stimulating peptide (CSP), via a classic two-component regulatory system ComDE. Upon CSP binding, ComD phosphorylates the ComE response-regulator, which then activates transcription of comCDE and the competence-specific σX, leading to a sudden rise in CSP levels and rendering all cells in a culture competent. However, how competence stops has remained unknown. We report that DprA, under σX control, interacts with ComE∼P to block ComE-driven transcription, chiefly impacting σX production. Mutations of dprA specifically disrupting interaction with ComE were isolated and shown to map mainly to the N-terminal domain of DprA. Wild-type DprA but not ComE interaction mutants affected in vitro binding of ComE to its promoter targets. Once introduced at the dprA chromosomal locus, mutations disrupting DprA interaction with ComE altered competence shut-off. The absence of DprA was found to negatively impact growth following competence induction, highlighting the importance of DprA for pneumococcal physiology. DprA has thus two key roles: ensuring production of transformants via interaction with RecA and competence shut-off via interaction with ComE, avoiding physiologically detrimental consequences of prolonged competence. Finally, phylogenetic analyses revealed that the acquisition of a new function by DprA impacted its evolution in streptococci relying on ComE to regulate comX expression.
Originally discovered in the human pathogen Streptococcus pneumoniae (the pneumococcus) (1), bacterial transformation is a genetically programmed process allowing alteration of genotypes via homologous recombination. Bacterial transformation is regarded as a substitute for sexual reproduction (2), which is crucial for genetic diversity in eukaryotes but is lacking in prokaryotes. Transformation is phylogenetically widespread in bacteria (3), proceeding through the internalization of single-strand (ssDNA) derived from exogenous double-stranded DNA, requiring an evolutionarily conserved multiprotein DNA uptake apparatus (4). Integration of internalized DNA into the chromosome then relies on homology search (i.e., matching DNA in the chromosome) catalyzed by the universal bacterial recombinase RecA, assisted by the transformation-dedicated protein DprA. This cytosolic protein, which is also widely distributed in bacteria, ensures the loading of RecA onto transforming ssDNA (5).
In several species, including S. pneumoniae and Bacillus subtilis, the ability to internalize ssDNA, also called competence, is not permanent, as synthesis and assembly of the uptake apparatus is a transient, regulated process. Competence regulatory circuits are not universal but rather represent adaptations to every species’ lifestyle as exemplified by the S. pneumoniae and B. subtilis models (6). In pneumococcal cultures, competence or X-state, which is considered as an SOS substitute (7), develops abruptly during exponential growth phase in response to a competence-stimulating heptadecapeptide, CSP, encoded by comC (8). A dedicated ABC transporter, ComAB, matures and exports the comC-encoded pre-CSP (9). CSP acts through a receptor, the membrane histidine kinase (HK) ComD, to activate its cognate response regulator (RR), ComE (10). The ComDE pair belongs to the two-component signaling systems (TCS) (11) and a classic mode of activation, involving autophosphorylation of the CSP-sensor HK, ComD, and the subsequent transphosphorylation of its cognate RR transcription activator, ComE, recently received indirect support (12). ComE∼P activates both comAB and comCDE operons, establishing a positive feedback loop, which results in a sudden rise in extracellular CSP levels, rendering all cells in a culture simultaneously competent. ComE∼P activates another key gene, comX, which encodes the competence-specific σ factor, σX, the only alternative σ of S. pneumoniae (13). Genes under direct σX control, which are collectively known as late com genes (as opposed to the ComE-activated, early com genes), include those coding for the DNA uptake machinery and proteins dedicated to the processing of transforming DNA, such as DprA (14, 15).
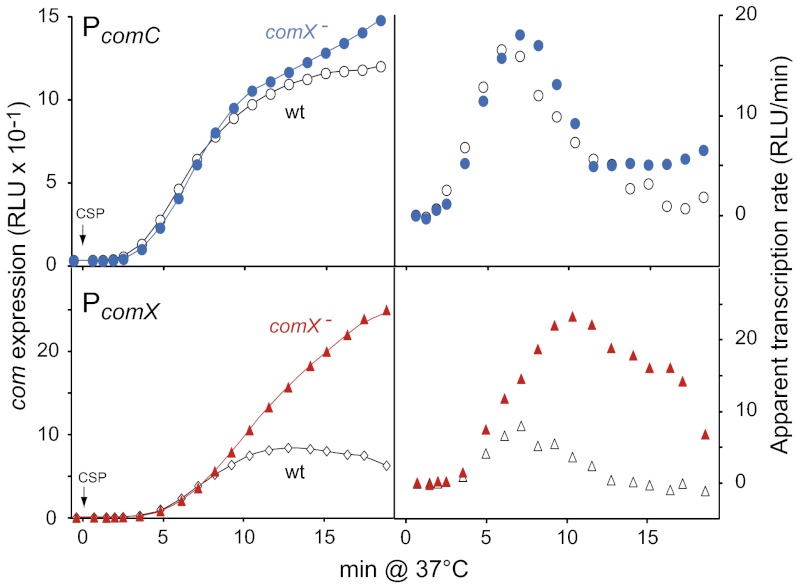
Although the development of pneumococcal competence is sudden, its cessation is almost as abrupt, hence the notion of “peak of competence” for pneumococcal transformation. Recent measurements of the apparent transcription rate of the comCDE operon indicated that comCDE expression was detectable 2.5 min after CSP addition, peaked after 8.5–9 min, then decreased ∼3.5-fold within the next 4.5–5 min (12) (Fig. 1). This shut-off phase is followed by a long period, about 60–80 min, during which cells are unresponsive to the presence of high concentrations of CSP in the medium (refractory period) (16–18). The mechanisms for the shut-off phase have remained unknown and simple signal termination occurring by dephosphorylation of the RR, as observed in many TCS (19), cannot account for the blind-to-CSP period unless it is combined with prevention of further HK activation. A first clue was obtained when it was observed that inactivation of comX resulted in higher expression of a comX::lacZ transcriptional fusion during the competence induction phase, at the time of competence shut-off and during the refractory period (13). These observations suggested that either ComX or the product of a late com gene (i.e., a gene depending on σX for expression) inhibits comX gene expression during the shut-off phase and CSP unresponsive period.
Fig. 1.
A Late com protein is required for comX transcription shutoff. Comparison of comC and comX expression in response to CSP in wild-type and comX− cells confirms influence of ComX on comX expression. (Left) Promoter activities; (Right) apparent rates of transcription [expressed as relative luminescence unit (RLU) increase per arbitrary time unit] calculated at a given time point, t1, by substracting RLU value at t0 from RLU value at t1, as previously described (12). CSP (100 ng/mL−1) was added after 10-min incubation in C+Y (SI Materials and Methods). Strains used: (Upper) comC::luc R2002 (comX−) and R1960; (Lower) comX::luc R2256 (comX−) and R2250. CSP was added at 0 min.
Here we report that inactivation of the late com gene dprA strongly affects competence shut-off, revealing a recombination protein as the key agent of the rapid reversal of competence induction. We show that although lack of DprA up-regulates comX transcription, its engineered expression at an early stage of competence reduces expression of comX, in the absence of any other late com gene product. A combination of genetic, biochemical, and phylogenetic analyses allows us to conclude that that DprA plays two key roles in competence and genetic transformation of S. pneumoniae and closely related streptococci. The function of DprA as a transformation-dedicated RecA loader renders it essential for the production of transformants, and its specific interaction with ComE ensures the phyiologically important shut-off of competence. The latter action constitutes an original regulatory mechanism; to the best of our knowledge, this report of antagonization of an RR by a recombination protein is unique.
Results
Differential Impact of comX Inactivation on comCDE and comX Transcription.
Although inactivation of comX was reported to result in higher expression of a comX::lacZ fusion during the competence induction phase (13), it was recently found to have a very limited effect on expression of the comCDE operon (12). Taken together these observations suggested a differential behavior of the two ComE-dependent promoters (PE), PcomX and PcomC; however, they did not involve a direct comparison. This result prompted us to compare PcomX and PcomC expression in parallel in wild-type and comX− cells, using luc transcriptional fusions at each locus. Light emission, which results from activity of the luc-encoded Firefly luciferase, was recorded every minute after CSP addition (SI Materials and Methods). Similar subsidence in comC transcription was confirmed in both genetic backgrounds (Fig. 1). On the other hand, although comX transcription kinetics paralleled that of comC in wild-type cells, in comX− cells the apparent transcription rate of comX continued to increase for 2.5–3 min before decreasing slowly, resulting in an overall expression level of comX ∼fourfold higher 19 min after CSP addition (Fig. 1). These observations were thus fully consistent with the hypothesis of a role of ComX (σX) or a late com gene product in the deceleration of PcomX transcription (13), though still confirming that the deceleration of PcomC transcription occurred independently of ComX (12).
Inactivation of the Late com Gene dprA Affects comX Transcription Shut-Off.
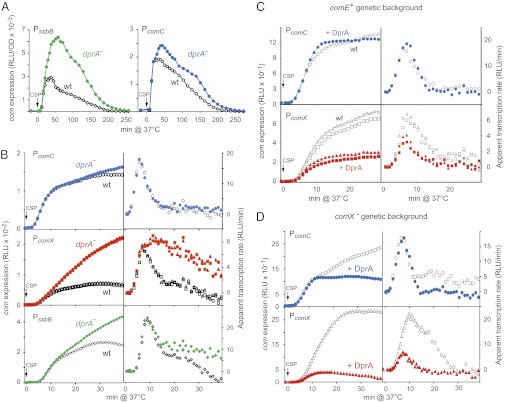
Serendipitously, while studying the role of the late com gene product DprA in transformation, dprA− cells were observed to display altered competence kinetics (20). Compared with wild-type, dprA− cells exhibited strongly enhanced expression of the late com gene ssbB, but only a small increase in PcomC expression (Fig. 2A). Direct comparison of comC, comX, and ssbB expression in wild-type and dprA− cells confirmed this hypothesis, dprA inactivation strongly increasing comX expression but having limited impact on comC expression (Fig. 2B). A specific increase in comX expression because of the absence of DprA was also observed in dprA− cells harboring single amino acid changes in comE, resulting in the up-regulation of competence, comED58E and comER120S (Fig. S1 A and B), the former a phosphorylmimetic mutant of ComE (12) and the latter a mutant displaying an extended competence period (21). Taken together, these data were consistent with an alteration of the shut-off of comX transcription in the dprA mutant.
Fig. 2.
Direct involvement of DprA in the control of comX expression. (A) Comparison of ssbB and comC expression in wild-type and dprA− cells reveals a strong influence of DprA on ssbB expression. Strains used: (Left) ssbB::luc R2018 (dprA−) and R1502; (Right) comC::luc R2017 (dprA−) and R1960. (B) Inactivation of dprA most strongly affects comX expression. PcomC, PcomX, and PssbB transcription was compared as described in Fig. 1. Strains used: (Top) comC::luc R2017 (dprA−) and R1960; (Middle) comX::luc R2240 and R2241 (both dprA−), and R2200 and R2218 (both dprA+); (Bottom) ssbB::luc R2018 (dprA−) and R1502. (C) Comparison of PcomX and PcomC transcription in wild-type cells with and without ectopic expression of dprA as an early com gene. Strains used: comC::luc R1998 (CEPE-dprA) and R1960 (Upper); comX::luc R2259 and R2260 (both CEPE-dprA+), and R2200 and R2218 (both without CEP) (Lower). See also Fig. S1C. (D) DprA inhibits PcomX transcription independently of any other late com gene product. Same experiment as in C but using comX− cells. Strains used: (Upper) comC::luc R2003 (CEPE-dprA) and R2002; (Lower) comX::luc R2265 (CEPE-dprA) and R2256.
Expression of DprA as an Early com Gene Product Accelerates the Shut-Off of comX.
To further document the impact of DprA on shut-off, we inserted a copy of the dprA gene at an ectopic chromosomal position, under the control of the PcomC promoter (SI Materials and Methods). Examination of PcomC and PcomX expression in otherwise wild-type cells harboring this construct revealed that expression of DprA as an early com gene product strongly reduced transcription from PcomX, yet having no detectable effect on PcomC (Fig. 2C). A similar result was observed in comER120S cells (Fig. S1C). These data suggested that DprA has a direct impact on PcomX transcription.
To establish whether DprA alone was sufficient to turn off comX transcription, the impact of dprA expression as an early com gene was evaluated in comX− cells. A very strong inhibition of PcomX expression was observed indicating that DprA inhibits comX transcription independently of any other late com gene product (Fig. 2D). A more limited inhibition of PcomC transcription was also observed (Fig. 2D), suggesting that DprA plays a role in the shut-off of all PE promoters.
DprA Targets ComE∼P.
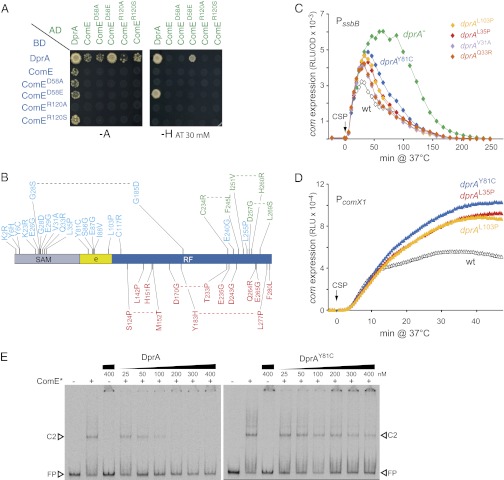
To determine whether PE promoter shut-off involved a direct interaction between ComE and DprA, yeast two-hybrid assays (Y2H) were performed as described previously (5). Interaction between DprA and the phosphorylmimetic mutant ComED58E strongly activated the interaction reporter genes in yeast under the most stringent conditions (i.e., on synthetic medium lacking histidine supplemented with 30 mM 3-aminotrioazole) (Fig. 3A). This interaction is bidirectional, which is quite infrequent in Y2H (i.e., 5% or less of Y2H interactions are reciprocal), strongly supporting a genuine physical interaction between DprA and ComED58E. Weaker interaction phenotypes were detected between DprA and the wild-type and mutant forms of ComE if plates were incubated in synthetic medium lacking adenine (Fig. 3A). These results suggest that a direct interaction of DprA with ComE could participate in the shut-off of pneumococcal competence. Importantly, in view of its strong interaction with the phosphorylmimetic ComED58E mutant, DprA could preferentially interact with ComE∼P, allowing it to interfere with the CSP signal-transduction cascade and block transcription of the early com genes, particularly comX.
Fig. 3.
DprA–ComE interaction is crucial for competence shut-off. (A) Y2H assays revealing physical interactions of DprA with wild-type and ComE mutant proteins. Cells expressing full-length DprA, wild-type or mutant ComE as GAL4 binding domain (BD) fusions (Left) were mated with cells expressing these proteins as GAL4 activation domain (AD) fusions (top line). Binary interactions were revealed by growth of diploid cells on selective medium lacking either adenine (−A) or histidine (−H) supplemented with 30 mM 3-aminotrioazole (AT). The previously documented self-interaction of DprA (5) served as positive control. (B) Position in dprA of mutations abolishing DprA–ComE interaction in Y2H. These mutations are shown in blue. Previously characterized mutations abolishing interaction with RecA and self-interaction (23) are shown in green and red, respectively. For each mutation, the wild-type residue is indicated first, followed by its position, then the mutant residue. Limits of DprA structural domains (SAM; Rossmann fold, RF; RF extension, e) are indicated. (C) Mutations abolishing DprA–ComE interaction affect the shutoff of ssbB expression. ssbB::luc strains used: R1502 (dprA+), R2018 (dprA−), R2178 (dprAL103P), R2179 (dprAL35P), R2180 (dprAY81C), R2181 (dprAV31A), and R2182 (dprAQ33R). (D) Mutations abolishing DprA-ComE interaction affect the shut-off of comX expression. comX1::luc strains used: R2218 (dprA+), R2454 (dprAL103P), R2455 (dprAL35P), and R2456 (dprAY81C). See also Fig. S2A. (E) DprA but not the DprA–ComE interaction mutant DprAY8IC inhibits the binding of ComED58E-R120S to PcomC promoter in vitro. ComED58E-R120S (ComE*) binding was assayed in the presence of the indicated concentrations of DprA or DprAY81C as described in Materials and Methods. C2, type 2 complex; FP, free probe. See also Fig. S2E.
Further evidence for interaction of DprA with ComED58E in pneumococcal cells was obtained through investigation of the impact of dprA inactivation on comX expression. Recently, comED58E cells were shown to exhibit wild-type transformation frequencies in the absence of both CSP and ComD, demonstrating that ComED58E triggers its own production and activates transcription of the early com promoters (12). We found that inactivation of dprA in otherwise ΔcomCD comED58E cells resulted in 40–70% increase in comX expression compared with dprA+ cells (Fig. S2A). The increase in comX expression resulting from ComD inactivation (Fig. S2A) was previously attributed to the sequestration of ComED58E by ComD because of the binary interaction between ComED58E and ComD in Y2H (12). Similarly, in the absence of ComD, we attribute the further increase in comX expression observed when dprA is inactivated to the relief of sequestration of ComED58E by DprA.
DprA–ComE Interaction Is Crucial for Competence Shut-Off.
A Y2H screen for DprA mutants unable to self-interact yet retaining full capacity to interact with RecA, and vice-versa, allowed the identification of DprA residues important for self-interaction and for interaction with RecA (22, 23). This screen also included a search for dprA mutations affecting DprA–ComE interaction, but also maintaining ability of DprA to self-interact and interact with RecA (22). Such mutations were obtained and located, revealing a striking clustering in the 5′ moiety of dprA, which contrasted with the 3′ location of mutations affecting DprA self-interaction and interaction with RecA (Fig. 3B). Notably, a majority of these mutations were located in the N-terminal sterile α-motif (SAM) domain, a region of DprA conserved at the structural but not the primary sequence level (5, 23).
Several of these mutations were then introduced at the dprA chromosomal locus to investigate their effect on the shut-off of competence. None of the mutations affected transformation frequency, suggesting full functionality of the mutant proteins for DNA processing. Although some mutations had no effect (dprAK23R, dprAE26G, dprAG28D, and dprAE87G), others resulted in ssbB shut-off kinetics intermediate between those in wild-type and dprA-null cells, the DprAY81C mutant protein exhibiting the strongest impact on competence shut-off (Fig. 3C). Shut-off alteration was confirmed through analysis of comX::luc kinetics (Fig. 3D and Fig. S2B). We concluded that a direct interaction between DprA and ComE is required to shut off expression of the early com genes, particularly comX.
DprA Inhibits the Binding of ComE to Early com Promoters in Vitro.
Next, before investigating the effect of DprA on in vitro binding of ComE to its early com promoter target, we wished to evaluate the stoichiometry of DprA and ComE, knowing there are ∼87,000 ComE monomers per fully induced cell (12). Calculations for DprA resulted in an estimate of ∼6,320 and ∼8,350 molecules per cell, respectively, 10 and 20 min after CSP addition (Fig. S2C). Thus, a DprA to ComE ratio of ∼0.1 is sufficient for DprA to start inhibiting PcomX transcription.
Previous measurements of apparent Kd through EMSA established that among various ComE proteins assayed in vitro, the ComED58E-R120S protein exhibited the lowest Kd for a PcomC promoter fragment, although binding to a PcomX promoter fragment was similar for all ComEs and much weaker than to PcomC (12). The various ComE proteins did not only display differences in Kds but also in the type of retarded complexes that were formed. For example, whatever the promoter fragment, the ComED58E-120S protein formed mainly C2 complexes migrating more slowly than the major species (C1) observed with other ComEs (12). We investigated the impact of DprA and DprAY81C on the binding of ComED58E-R120S to a PcomC promoter fragment (Fig. 3E). The presence of 200 nM DprA essentially abolished the binding observed with 50 nM ComED58E-R120S. In constrast, up to 400 nM DprAY81C had no detectable effect on ComED58E-R120S binding. As a control, DprA and DprAY81C were shown to be equally efficient in binding a dT100 oligonucleotide in vitro (Fig. S2D), indicating that purified DprAY81C was fully active.
The effect of DprA on the binding of wild-type ComE to PcomC and PcomX fragments was also investigated. In both cases, DprA significantly reduced or abolished ComE binding (Fig. S2E). Thus, in vitro data were fully consistent with DprA-dependent shut-off of early com gene transcription requiring direct interaction between DprA and ComE, and resulting from inhibition of ComE binding to its promoter targets.
C/C Dimerization of DprA Is Crucial for Competence Shut-Off.
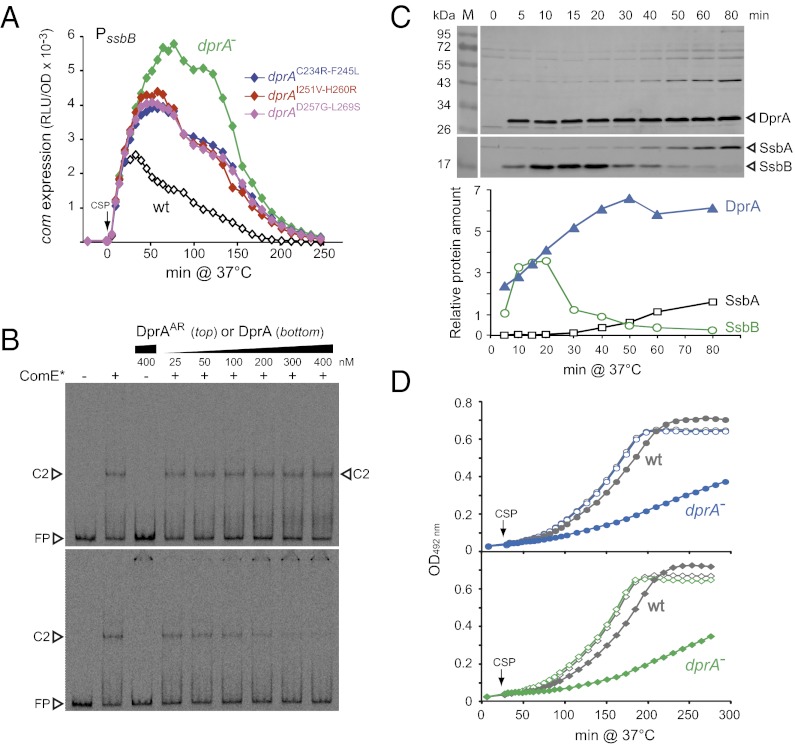
Tail-to-tail (C/C) dimerization of DprA was recently shown to be crucial for formation of nucleocomplexes in vitro and for genetic transformation. Presumably, dimerization is required to stabilize DprA–ssDNA complexes, facilitating the subsequent loading of RecA onto ssDNA (23). The DprA monomeric mutant, DprAH260A-L269R (DprAAR), thus exhibited a 2-log transformation defect despite displaying full ability to interact with RecA. To establish whether C/C dimerization of DprA was also important for competence shut-off, we compared expression of the late com gene ssbB after CSP addition in cells harboring mutations of dprA previously shown to abolish DprA self-interaction in Y2H, dprAC234R-F245L, dprAI251V-H260R, and dprAD257G-L269S (23). Compared with wild-type and dprA-null cells, DprA self-interaction mutants exhibited an intermediate shutoff phenotype (Fig. 4A). Moreover, similar to DprAY81C and in contrast to DprA, the DprAAR protein did not affect in vitro binding of ComED58E-R120S to PcomC (Fig. 4B). We concluded that, in addition to physical interaction with ComE, C/C dimerization of DprA is required to shut-off transcription of the early com genes.
Fig. 4.
DprA dimerization requirement, intrinsic stability and impact on postcompetence growth. (A) Mutations abolishing C/C dimerization of DprA affect the shutoff of ssbB expression. ssbB::luc strains used: R1502 (dprA+), R2018 (dprA−), R2209 (dprAC234R-F245L), R2210 (dprAI251V-H260R), and R2211 (dprAD257G-L269S). (B) DprA, but not the C/C dimerization mutant DprAAR, inhibits the binding of ComED58E-R120S to early com promoters in vitro. Same experimental set-up as in Fig. 3E. (C) DprA stability readily accounts for the competence shut-off phase and the subsequent CSP refractory period. DprA levels (Lower curve) were analyzed with a BioImager (LAS-4000; Fujifilm) after Western blotting (Upper) using extracts from R1502 cells induced with CSP (25 ng/mL−1) and harvested at the indicated time after CSP addition. Levels of the unstable competence-induced (SsbB) and stable house-keeping (SsbA) ssDNA-binding proteins were monitored in parallel, as controls. (D) Growth of dprA-null cells is severely impeded by prolonged competence resulting from lack of DprA-mediated shut-off. OD492 nm was monitored in parallel with light emission in the experiment shown in Fig. 2A, in cultures treated with CSP (filled symbols); open symbols correspond to control cultures without CSP. Strains used: (Upper) R2017 (dprA−) and R1960 (wt); (Lower) R2018 (dprA−) and R1502 (wt).
DprA Stability May Underlie the CSP Unresponsive Period.
Because ComE and ComD are stable proteins (12, 24), DprA itself should be rather stable to fulfill a direct role in competence shut-off via its interaction with ComE. This theory prompted us to document DprA stability via Western blotting, revealing that DprA is as stable as ComE. In contrast to the competence-induced ssDNA-binding protein SsbB, which appeared unstable, the amount of DprA was maximal 40 min after CSP addition and remained at the same level at least until 80 min after CSP (Fig. 4C). This feature is fully consistent with DprA being responsible for the blind-to-CSP period that typically follows the peak of competence in pneumococcal cultures.
DprA-Driven Competence Shut-Off Is Crucial for Pneumococcal Physiology.
Evidence was obtained that the DprA-dependent shut-off of competence is physiologically important. Thus, it was repeatedly observed that although growth rate of dprA− cells was identical to wild-type, induction of competence resulted in a severe growth defect of mutant cells (Fig. 4D). To further document the importance of the competence regulatory role of DprA, we assayed the transfer by transformation of a dprA-null mutation (dprA::spc21C, conferring resistance to spectinomycin, SpcR) into a series of cup (for competence up) mutant strains (e.g., comER120S or comDD299N) (Table 1). These comDE mutants, previously selected on the basis of up-regulated comCDE expression, exhibited enhanced spontaneous competence development, resulting in some cases in altered cell viability (21). Although dprA::spc21C was readily introduced into four of the cup mutants, it could not be integrated in six other mutants, as deduced from the ∼3-log reduction in frequency of the SpcR transformants compared with a reference marker. We concluded that the latter cup mutations were synthetic-lethal with the dprA knock-out. This conclusion was reinforced by the finding that the low-frequency SpcR clones obtained in the cup strain trt (comDD299N) corresponded to double transformants that simultaneously integrated dprA::spc21C and ejected the trt mutation (Fig. S3A). It is of note that DprA was not essential in the comED58E cells, despite the proposed sequestering of ComED58E by DprA (Fig. S2A), implying that the phosphorylmimetic mutation leads to competence levels lower than those attained with other cup mutations.
Table 1.
Synthetic lethality of dprA and cup mutations
| Nature of the cup mutation* | Mutation permissive for dprA inactivation† | Mutation non permissive for dprA inactivation‡ |
| Point mutation | cup4 (comDE214K), cup6 (comDT290A), comDD58E | trt1 (comDD299N), cup3 (comDT233I), cup5 (comDY312CK), cup11 (comDQ226L), cup10 (comER120S), |
Potential dprA::spc (SpcR) transformants appeared only after prolonged incubation (41 h) as microhemolysis zones (e.g., Fig. S3A), which did not grow further and could not be recovered for growth in liquid medium.
*These mutations have been previously described (21).
†Frequency of dprA::spc transformants with normal colony size of ∼0.2 relative to the rpsL41 (SmR) reference marker (i.e., close to the expected value).
‡Approximately 3 log reduction in the frequency of dprA::spctransformants with normal colony size (e.g., Fig. S3A) compared with the permissive situation.
To further investigate the basis for the observed synthetic lethality, we launched a screen for mariner minitransposon insertions that would suppress lethality (i.e., insertions allowing survival of otherwise lethal dprA-trt double mutant cells). Fifteen independent suppressive insertions were characterized (SI Materials and Methods) and found to be located in the comAB operon (Fig. S3B), resulting in defective CSP export and therefore abolition of spontaneous competence development. This result unambiguously established that the up-regulation of spontaneous competence, a consequence of the trt mutation, cannot be tolerated in the absence of DprA. We concluded that uncontrolled competence is detrimental to pneumococcal cells and that DprA plays a crucial role in competence regulation, hence in maintenance of cell physiology, by ensuring its shut-off.
Involvement in Competence Shut-Off Impacts Streptococcal DprA Evolution.
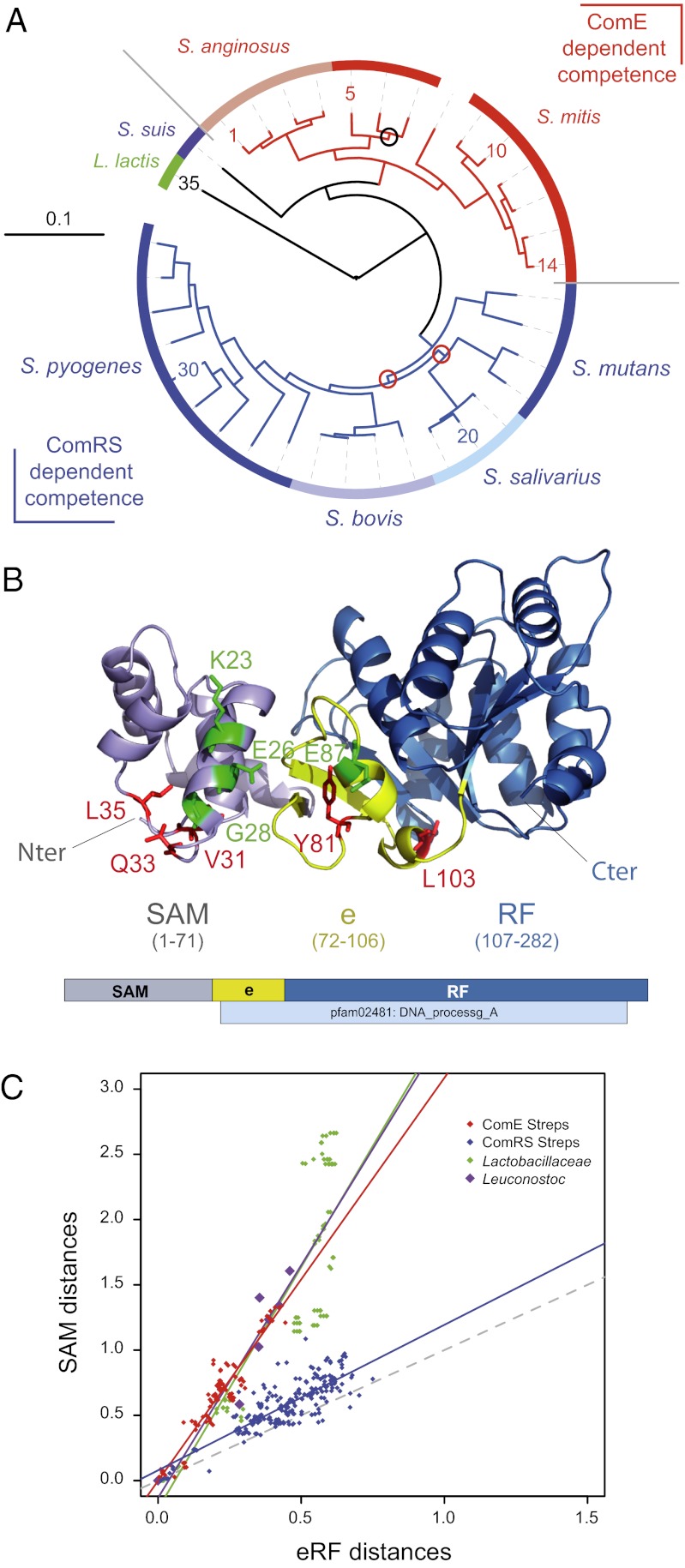
Phylogenetic analyses of the ComD-ComE TCS led to the conclusion that many streptococci lack ComDE orthologs, and therefore use widely different regulatory cascades to control competence (25). This conclusion was fully supported by the recent identification of a competence regulatory circuit involving an Rgg transcriptional activator, ComR, and a peptide pheromone encoded by comS (26–29). Thus, although streptococci belonging to the Streptococcus anginosus and Streptococcus mitis groups, which includes S. pneumoniae, presumably rely on ComDE orthologs to regulate comX expression, those belonging to the Streptococcus pyogenes, Streptococcus bovis, Streptococcus salivarius, and Streptococcus mutans groups use instead ComRS systems (Fig. 5A) (30). On the other hand, the DprA family was previously defined at the primary sequence level by the Pfam02481 (5), but was recently proposed to consist, at the structural level, of a larger entity comprising an extended Rossmann fold (eRF), which entirely overlaps Pfam02481, preceded by a SAM fold (23) (Fig. 5B). The structural conservation of a SAM, together with the lack of primary sequence conservation of the N terminus of DprA proteins, were consistent with a differential rate of evolution of the SAM and eRF DprA ancestral domains.
Fig. 5.
Involvement in competence shut-off impacts DprA structure. (A) A phylogenetic tree of streptococci was generated as described in the SI Materials and Methods. Tree color code: red, species with ComE-regulated competence; blue, species with ComRS-regulated competence; green, identifies Lactococcus lastis. All internal branches received the maximum support with both parametric methods and all had 100% boostrap support, except three branches (98%, black circle; 88%, red circles). Digits identifying species follow the relative order of leaves in the different subtrees, starting from 1 (Streptococcus intermedius) and increasing in a clockwise orientation, with 10, 14, 20, 30, and 35 corresponding, respectively, to Strepotococcus peroris, S. pneumoniae, Streptococcus thermophilus, Streptococcus porcinus, and Lactococcus lactis (used as an outgroup). For other digits, the correspondence with species names is given in Table S1. (B) Ribbon schematic and linear representation of S. pneumoniae DprA structure. DprA 3D image was generated with PyMOL Molecular Graphics System The conserved primary sequence defining the DNA processing A family (pfam02481) is shown below the linear map. Domain color code: violet, SAM; blue, RF; yellow, e (RF extension). Note that ComE interaction residues are located on the same face, with residue changes affecting competence shut-off (shown in red) preferentially found in flexible loops and those not impacting shutoff (shown in green) in α-helices. (C) Comparative evolution of SAM and eRF domains among streptococci with and without ComE-regulated competence. The evolutionary distances between species were computed from SAM and eRF sequence alignments as described in the SI Results and Discussion. Each dot corresponds to distances computed for a given species pair. Same color code as in A for streptococci; green, Lactobacillaceae; purple, Leuconostoc species. Colored solid lines represent regression curves for each group of species. Slopes = 1.12 (r = 0.82) for ComRS streptococci and 3.09 (r = 0.95), 3.57 (r = 0.92), and 3.71 (r = 0.88) for ComE streptococci, Leuconostoc sp., and Lactobacillaceae, respectively. Gray dotted line indicates the line of slope 1, expected if domains evolved at identical rates.
The present finding that DprA plays a key role in the shut-off of pneumococcal competence, requiring a direct interaction with ComE, together with the observation that a majority of mutations abolishing DprA–ComE interaction were located in the SAM (Fig. 3B), led us to investigate whether acquisition of this regulatory function had an evolutionary impact on DprA structure. DprA sequences from streptococci, as well as Lactobacillaceae and Leuconostoc species used as controls, were retrieved. SAM and eRF sequences were aligned, trees computed, and evolutionary distances of DprA domains between species calculated from the trees as described in SI Materials and Methods. The plot of the distance computed on the SAM domain against the distance computed on the eRF domain for each species pair revealed a striking bimodal distribution among streptococci (Fig. 5C). The ComE-dependent group thus behaved similarly to Lactobacillaceae and Leuconostoc species in showing an accelerated evolution (3.1- to 3.7-fold) of the SAM domain compared with the eRF domain. In contrast, the ComRS-dependent group exhibited a rate of evolution of the SAM domain almost identical to that of the eRF domain. Although the latter observation was puzzling (Fig. S4), the bimodal distribution of streptococci with respect to SAM domain evolution matching the presence/absence of ComE regulated competence was consistent with the hypothesis that the acquisition of a new function by DprA had an impact on its evolution.
Discussion
Dual Role for Pneumococcal DprA in Competence and Transformation.
The present work adds a unique facet to S. pneumoniae DprA activities in showing that this DNA processing protein fulfils another biologically important function, playing a direct role in the shut-off of competence. S. pneumoniae DprA was first described as the prototype for a new subfamily of so-called recombination mediator proteins, dedicated to bacterial transformation (5). DprA ensures the loading of RecA onto ssDNA to form presynaptic filaments (Fig. 6A). This loading activity was recently shown to require both direct interaction with RecA and C/C dimerization of DprA, the latter being crucial for formation of stable nucleocomplexes in vitro (23). C/C dimerization and RecA interaction residues and surfaces were found to overlap, localizing in the C-terminal RF domain of DprA. We now show that DprA is responsible for the shut-off of pneumcoccal competence. This role requires both its direct interaction with the master competence regulator, ComE, and the ability to form C/C dimers. Interestingly, ComE interaction residues were predominantly localized in the N-terminal SAM domain of DprA (Fig. 3B).
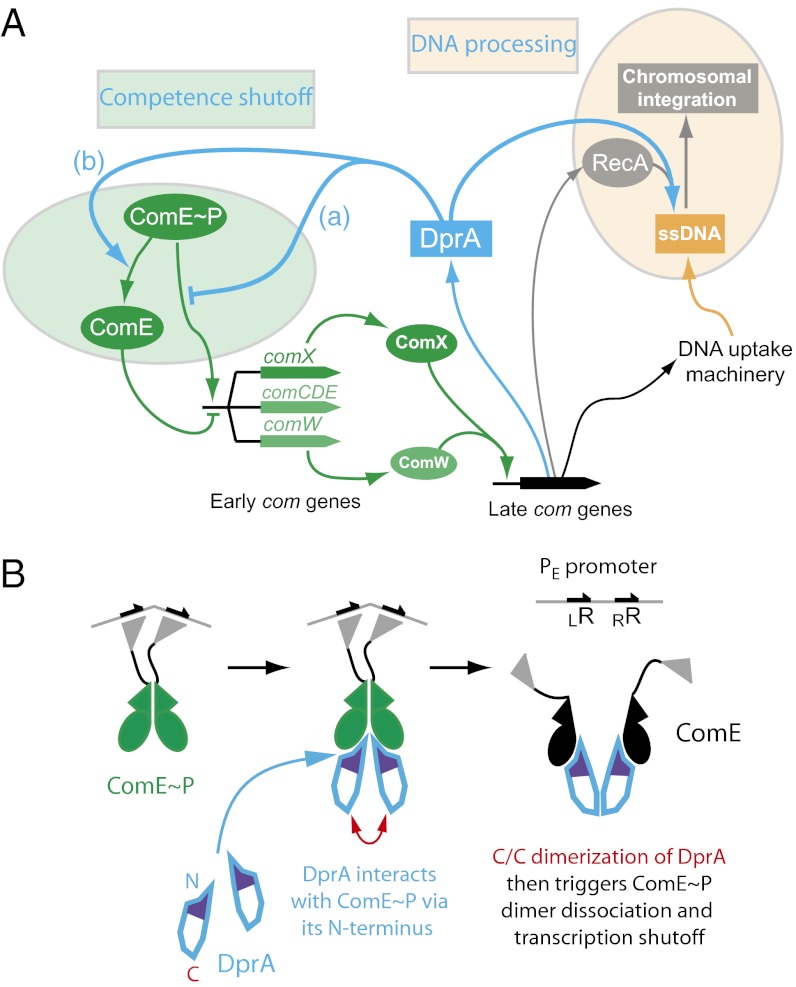
Fig. 6.
A dual role for DprA: RecA loading and competence shut-off. (A) DprA was first recognized as playing a key role in the processing of internalized ssDNA as a transformation-dedicated RecA loader. The present study documents its direct involvement in the shut-off of competence. We propose that DprA similarly affects PcomX and PcomW, controlling, respectively, expression of the competence-specific σX factor and ComW, which is crucial for σX activation and stabilization. DprA could act as an antiactivator (a) or affect ComE phophorylation state (b) (Discussion). (B) Diagrammatic representation of the involvement of DprA C/C dimerization in the shutoff of PE promoters. DprA could first interact with ComE∼P as a monomer, through its N terminus. C/C dimerization of two monomers could then trigger ComE∼P dimer dissociation. As RR phosphorylation mediates dimerization, which then promotes transcriptional activation (12), ComE∼P dimer dissociation is likely to result in transcription shut-off. Possible alternatives are evoked in the Discussion.
DprA and ComE Activities Converge to Shut Off Early com Promoters.
Recent results shed light on the mechanism of shut-off of pneumococcal competence and the subsequent blind-to-CSP period that had remained unknown for several decades. Thus, the shut-off of PcomC was recently proposed to be intrinsic to ComDE, subsidence of comCDE transcription readily occurring in the absence of any late com gene product (12). Based on the finding that nonphosphorylated ComE efficiently bound PcomC in vitro and acted as a repressor of spontaneous competence development, it was concluded that ComE accumulating in response to CSP efficiently outcompeted ComE∼P for binding to PcomC, thus preventing further transcription. Evidence was also obtained for a differential transcriptional activation and repression of PcomC and PcomX, the ComE/ComE∼P ratio, and the relative affinities of ComE and ComE∼P for PE promoters dictating the individual patterns of expression (12). This differential behavior was confirmed in this study through the demonstration that the shut-off of PcomX was more affected than that of PcomC by a late com gene product (Fig. 1), identified as DprA (Fig. 2, and Figs. S1 and S2).
We propose that in the absence of DprA, ComE∼P concentration or availability increases (see below), resulting in prolonged PcomX transcription but having little effect on PcomC. This differential effect correlates with the recently documented hierarchy of PE promoters (12). Because of lower affinity of ComE for PcomX than for PcomC, shut-off of the latter occurs first. Then, once ComE concentration has reached the critical threshold for PcomC repression, an increase in ComE∼P availability resulting from dprA inactivation can only have a more limited effect, as observed. The prominent effect of DprA is therefore to accelerate the shut-off of PcomX. The PcomW promoter presents kinetics and characteristics very similar to those of PcomX (12). Notably, among ComE-binding boxes in PE promoters, which consist of a 9-bp direct repeat separated by a stretch of 12 bp, PcomW contains the most divergent left repeat, and the least conserved right repeat is that of PcomX. Therefore, although the effect of dprA inactivation on PcomW expression was not documented, we propose that DprA accelerates its shut-off as well (Fig. 6A).
To summarize, we propose that DprA shifts the ComE/ComE∼P ratio in favor of repression of PE promoters, with a hierarchy depending on the relative strength of each promoter. Because both ComX and ComW are fairly unstable (31), DprA-dependent shut-off of PcomX and PcomW promoters in fine ensures the rapid cessation of σX-driven expression of the late com genes, including dprA itself (Fig. 6A).
Mechanism of DprA-Dependent Inhibition of ComE∼P Driven Transcription.
The preferential acceleration of PcomX shutoff by DprA is most consistent with DprA targeting ComE∼P. Results of Y2H showing that DprA interacts more strongly with the phosphorylmimetic ComED58E protein than with other ComEs (Fig. 3A) and evidence for sequestration of ComED58E by DprA in comD-null cells (Fig. S2A) strongly support this view. In light of these observations, we do not favor the hypothesis that DprA acts as an inhibitor of the ComD HK, as one would expect the protein to preferentially bind ComE rather than ComE∼P. We envision instead two types of mechanisms for the DprA-dependent shut-off of PE promoters. DprA could either act as an antiactivator, antagonizing interaction of ComE∼P with its promoter targets (Fig. 6 A, a), or affect ComE phosphorylation state (Fig. 6 A, b). Either mechanism would account for our observations. Although according to our estimates there is an excess of ComE over DprA, the effective ratio could well be in favor of DprA if its target is ComE∼P rather than ComE, the former presumably representing a minor fraction of total RR molecules in the cell.
C/C dimerization of DprA has proven crucial for both the production of transformants (23) and the shut-off of competence (Fig. 4 A and B). In the first case, dimerization is presumably required to stabilize complexes between DprA and internalized ssDNA. In the light of this observation, the need for DprA dimerization in competence shut-off could indicate that DprA–ssDNA complexes are the active species. Therefore, to check whether DNA availability affects competence shut-off we tested the impact of the presence of exogenous DNA in liquid culture on competence kinetics but did not detect any effect. As an alternative, DprA dimerization could trigger disruption of ComE∼P dimers (Fig. 6B), which are presumably required for transcription activation (12). DprA could also interact with ComE∼P before its binding to DNA, its dimerization being required for stabilization of DprA–ComE∼P complexes, resulting in sequestration of ComE∼P or its locking in a conformation no longer compatible with DNA binding.
DprA-Driven Competence Shut-Off Is Physiologically Important.
The finding that the absence of DprA negatively impacts growth upon competence induction (Fig. 4D) highlights the importance of DprA’s control of competence for pneumococcal physiology. This conclusion is reinforced by the finding that dprA inactivation is not tolerated in several mutants displaying up-regulated competence (Table 1 and Fig. S3), suggesting that permanent activation of competence is toxic. Various reasons could account for the apparent need to tightly control competence in S. pneumoniae. First, unlike B. subtilis, pneumococcal competence develops during early exponential growth phase (i.e., in actively replicating and dividing cells). Differentiation to competence could require the slowing down of both processes, as in B. subtilis (32), but prolonging this pause may pose a potential challenge to survival. Second, the development of competence in only 10% of B. subtilis cells, a consequence of bistability, was proposed to serve to minimize risks (33). In contrast, there is no sign of bistability in pneumococcal competence, presumably because of the presence of several promoters contributing to maintenance of comCDE basal expression (34), and nearly all cells become competent. The whole population is thus at risk, which could account for the evolution of an efficient competence shut-off mechanism in this species (6).
The observation that DprA is required for postcompetence resumption of growth in pneumococcal cells might explain the previous finding that inactivation of DprA resulted in >104-fold reduction in transformation in S. pneumoniae but only 50- to 100-fold in B. subtilis (35). We envision the possibility that chromosomal transformation frequencies in cells lacking DprA are similar in both species but that potential transformants formed in S. pneumoniae are then lost because of the failure of dprA− cells to resume normal growth.
Investigation of DprA stability revealed the entire protein pool synthesized at competence is transmitted to daughter cells (Fig. 4C). DprA thus contrasts with other competence-induced proteins that are important for the processing of transforming DNA such as SsbB, which is required for the stabilization of internalized ssDNA (36), or CoiA. Both SsbB and CoiA are rapidly degraded once competence disappears (Fig. 4C) (37). We conclude that DprA stability being of no use for DNA processing is the consequence of its recruitment for the shut-off of competence, which has thus shaped its life span.
Finally, phylogenetic analyses of DprA conducted among streptococcal species to investigate whether the acquisition of this new function by DprA had an impact on the evolution of this domain led us to conclude that this unique role of DprA is not specific to S. pneumoniae but is likely to be shared by all streptococci relying on ComE as a master regulator of competence (Fig. 5).
Concluding Remarks.
The antagonization of an RR, ComE, by a recombination mediator protein, DprA, constitutes an original regulatory mechanism. Nevertheless, the involvement of a DNA processing protein in regulation of a global cellular response is not unprecedented. RecA is thus well known to be directly involved in SOS induction in Escherichia coli, with RecA-ssDNA nucleofilaments constituting the ultimate signal for DNA damaging conditions, and RecA interaction with the SOS repressor LexA triggering its autoproteolysis, thereby turning ON SOS. It is amazing that conversely, in S. pneumoniae and related streptococci, the RecA loader DprA was recruited to turn OFF competence, regarded as an SOS substitute.
Materials and Methods
Strains, Culture, and Transformation Conditions.
All of the strains and plasmids constructed in this work are listed, together with primers, in Table S2. Standard procedures for chromosomal transformation and growth media were used. These procedures are briefly described with appropriate references together with specific constructions in SI Materials and Methods.
Cloning, Purification, and Epression of Soluble DprA and ComE Proteins.
Recombinant plasmid pR502 allowing expression of a dprAY81C-intein fusion was constructed as follows. A dprAY81C fragment was amplified with primers dprA4-dprA2 on strain R2180 genomic DNA. The PCR product has a 5′ NdeI site (present in dprA4) and a NdeI site internal to dprA. The amplified product was digested with NdeI and inserted into NdeI-digested pR430, carrying dprA-intein. The orientation and sequence of the substituted dprAY81C fragment were verified to retain plasmid pR502. Expression and purification of DprA and DprAY81C were performed as previously described (5) using the IMPACT system (New England Biolabs), which allows isfopropyl-β-D-thiogalactopyranoside–inducible expression from a multicopy plasmid of the protein of interest fused through its C terminus to the dual tag intein-calmodulin binding domain.
E. coli BL21-Gold(DE3) strain (Stratagen) containing the relevant pKHS-comE or pKHS-comED58E-R120S plasmid, and plasmid pGKJE3, which allows expression of chaperones to prevent ComE aggregation, was used to purify soluble His-tagged ComE proteins through retention on a Ni-NTA column (Qiagen) and elution with imidazole (200 mM), as previously described (12). After centrifugal concentration, DprA and ComE proteins were further purified by gel filtration onto a Superdex Hiload 16/60 column.
Western-Blot Analysis of DprA Cellular Content.
For preparation of cell extracts, 20 mL R1501 culture in C+Y medium at OD550 0.07 (∼1.06 108 cfu/mL−1) was induced with CSP (25 ng/mL−1) and was incubated at 37 °C. After 10 and 20 min, 5 mL aliquots were taken. Cells were then collected by centrifugation; pellets were resuspended in 100 µL [Tris (10 mM, pH 8.0), EDTA (1 mM)] buffer and lysed for 10 min at 37 °C after addition of 4 µL [DOC (0.25%), SDS (0.5%)]. Next, 100 μL loading buffer was added and the extract was incubated for 5 min at 85 °C before loading onto a 12% (wt/vol) acrylamide-SDS gel. Proteins were transferred to Westran CS Membrane (Whatman) by semidry blotting using a GE Healthcare Blotting transfer unit (TE 77 PWR) for 90 min at 0.8 mA/cm−2 in Tris (48 mM)-Glycin (39 mM), SDS (0.037%), and MeOH [20% (vol/vol)].
Western-blots were performed as previously described (38), with 2-h incubation at room temperature with a 1/10,000th dilution of rabbit polyclonal antibodies raised against purified DprA peptides using the Eurogentec protocol (AS-DOUB-LX + AS-PCAP-RABBIT). The ECL Prime Western Blotting Detection System (GE Healthcare) and a BioImager were used for signal detection (30-s exposure). As a standard for determination of DprA cellular amounts in strain R1501, we used various concentrations of purified DprA protein mixed with extract of the DprA-null (R2017) mutant containing total protein amounts similar to those used for R1501 (Fig. S2C).
Promoter Binding Assay.
The upstream regulatory region of comCDE (255 bp) was amplified with primer pair Oligo-LE/Oligo-RE. The PCR fragment was end-labeled with [γ-33P]-ATP (Perkin-Elmer) using T4 polynucleotide kinase (New England Biolabs) according to standard procedures (39) and purified on MicroSpin G-50 columns (GE Healthcare). Promoter binding assay was carried out in a total volume of 20 μL containing 50 mM NaCl, 50 mM Tris●HCl pH 7.5, 5% (vol/vol) glycerol, ∼5 nM end-labeled PcomC promoter fragment, 1 mM MgCl2, 0.15 μg Poly(dI-dC) (as nonspecific competitor), and a mixture of ComED58E-R120SHis protein (50 nM) and wild-type or mutant DprA proteins (at the concentrations indicated in Figs. 3E and 4B). Reaction assays were incubated at 37 °C for 30 min and reaction products were analyzed by electrophoresis in native TBE polyacrylamide gels [5% (wt/vol)]. After 110-min electrophoresis at 4 °C at constant voltage (20 V/cm−1) in 0.5× TBE buffer, gels were dried at 80 °C for 30 min and exposed to a PhosphorImager screen (Life Science Systems-Fujifilm Global). Quantification of free DNA and protein-bound DNA was performed using Multi Gauge 3.0 Analysis Software (Life Science Systems-Fujifilm Global).
Supplementary Material
Acknowledgments
We thank Maud Hertzog for generating DprA 3D images; Marc Prudhomme for discussions; and Nathalie Campo and Calum Johnston for editing this manuscript. This work was supported in part by grants from the Ministère Délégué à la Recherche et aux Nouvelles Technologies Programme Microbiologie 2003–2004; RB/CD/2003/09/001 (to J.-P.C. and P.N.); and the Agence Nationale de la Recherche Projet 2010 Blanc SVSE 3: PneumocoX (to J.-P.C. and P.P.). N.M. was the recipient of a PhD thesis fellowship accompanying the Programme Microbiologie 2003–2004 grant (09/2003-09/2006) and the Association pour la Recherche sur le Cancer (2007).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219868110/-/DCSupplemental.
References
- 1.Griffith F. The significance of pneumococcal types. J Hyg (Lond) 1928;27(2):113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JM, Dowson CG, Spratt BG. Localized sex in bacteria. Nature. 1991;349(6304):29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 3.Johnsborg O, Eldholm V, Håvarstein LS. Natural genetic transformation: Prevalence, mechanisms and function. Res Microbiol. 2007;158(10):767–778. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2(3):241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 5.Mortier-Barrière I, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130(5):824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 6.Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 7.Charpentier X, Polard P, Claverys JP. Induction of competence for genetic transformation by antibiotics: Convergent evolution of stress responses in distant bacterial species lacking SOS? Curr Opin Microbiol. 2012;15(5):570–576. doi: 10.1016/j.mib.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Håvarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92(24):11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hui FM, Zhou L, Morrison DA. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene. 1995;153(1):25–31. doi: 10.1016/0378-1119(94)00841-f. [DOI] [PubMed] [Google Scholar]
- 10.Pestova EV, Håvarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21(4):853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 11.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin B, et al. ComE/ComE∼P interplay dictates activation or extinction status of pneumococcal X-state (competence) Mol Microbiol. 2013;87(2):394–411. doi: 10.1111/mmi.12104. [DOI] [PubMed] [Google Scholar]
- 13.Lee MS, Morrison DA. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol. 1999;181(16):5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dagkessamanskaia A, et al. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: Competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol. 2004;51(4):1071–1086. doi: 10.1111/j.1365-2958.2003.03892.x. [DOI] [PubMed] [Google Scholar]
- 15.Peterson SN, et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol. 2004;51(4):1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 16.Hotchkiss RD. Cyclical behavior in pneumococcal growth and transformability occasioned by environmental changes. Proc Natl Acad Sci USA. 1954;40(2):49–55. doi: 10.1073/pnas.40.2.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomasz A, Hotchkiss RD. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc Natl Acad Sci USA. 1964;51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JD, Morrison DA. Modulation of competence for genetic transformation in Streptococcus pneumoniae. J Gen Microbiol. 1987;133(7):1959–1967. doi: 10.1099/00221287-133-7-1959. [DOI] [PubMed] [Google Scholar]
- 19.Huynh TN, Stewart V. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol Microbiol. 2011;82(2):275–286. doi: 10.1111/j.1365-2958.2011.07829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergé M. 2002. Compétence pour la transformation génétique chez la bactérie à Gram positif Streptococcus pneumoniae: Etude du gène tardif dprA. PhD dissertation (Université Paul Sabatier, Toulouse, France)
- 21.Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol. 2000;38(4):867–878. doi: 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]
- 22.Mirouze N. 2007. Identification du produit d'un gène tardif impliqué dans la régulation de la compétence et le processing de l’ADN lors de la transformation naturelle chez S. pneumoniae. PhD dissertation (Université Paul Sabatier, Toulouse, France)
- 23.Quevillon-Cheruel S, et al. Structure-function analysis of pneumococcal DprA protein reveals that dimerization is crucial for loading RecA recombinase onto DNA during transformation. Proc Natl Acad Sci USA. 2012;109(37):E2466–E2475. doi: 10.1073/pnas.1205638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ween O, Gaustad P, Håvarstein LS. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol Microbiol. 1999;33(4):817–827. doi: 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]
- 25.Martin B, Quentin Y, Fichant G, Claverys JP. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 2006;14(8):339–345. doi: 10.1016/j.tim.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Gardan R, Besset C, Guillot A, Gitton C, Monnet V. The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J Bacteriol. 2009;191(14):4647–4655. doi: 10.1128/JB.00257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fontaine L, et al. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol. 2010;192(5):1444–1454. doi: 10.1128/JB.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78(3):589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mashburn-Warren L, Morrison DA, Federle MJ. The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone. J Bacteriol. 2012;194(17):4589–4600. doi: 10.1128/JB.00830-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Håvarstein LS. Increasing competence in the genus Streptococcus. Mol Microbiol. 2010;78(3):541–544. doi: 10.1111/j.1365-2958.2010.07380.x. [DOI] [PubMed] [Google Scholar]
- 31.Sung CK, Morrison DA. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J Bacteriol. 2005;187(9):3052–3061. doi: 10.1128/JB.187.9.3052-3061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haijema BJ, Hahn J, Haynes J, Dubnau D. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol Microbiol. 2001;40(1):52–64. doi: 10.1046/j.1365-2958.2001.02363.x. [DOI] [PubMed] [Google Scholar]
- 33.Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56(3):615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin B, et al. Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol Microbiol. 2010;75(6):1513–1528. doi: 10.1111/j.1365-2958.2010.07071.x. [DOI] [PubMed] [Google Scholar]
- 35.Claverys JP, Martin B, Polard P. The genetic transformation machinery: Composition, localization, and mechanism. FEMS Microbiol Rev. 2009;33(3):643–656. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 36.Attaiech L, et al. Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: Maintenance of a reservoir for genetic plasticity. PLoS Genet. 2011;7(6):e1002156. doi: 10.1371/journal.pgen.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desai BV, Morrison DA. An unstable competence-induced protein, CoiA, promotes processing of donor DNA after uptake during genetic transformation in Streptococcus pneumoniae. J Bacteriol. 2006;188(14):5177–5186. doi: 10.1128/JB.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison DA, Mortier-Barrière I, Attaiech L, Claverys JP. Identification of the major protein component of the pneumococcal eclipse complex. J Bacteriol. 2007;189(17):6497–6500. doi: 10.1128/JB.00687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY), 2nd Ed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.