Summary
The balance between self-renewal and differentiation must be tightly regulated in somatic stem cells to ensure proper tissue generation and to prevent tumor-like overgrowth. A Drosophila larval brain lobe consists of the central brain and the optic lobe, and possesses three well-defined neural stem cell lineages that generate differentiated cells in a highly reproducible pattern. Unambiguous identification of various cell types in these stem cell lineages is pivotal for studying the regulation of neural stem cells and progenitor cells at a single-cell resolution. This chapter will describe the methodology for collection and processing of larval brains for examination by fluorescence confocal microscopy.
Keywords: Neural stem cell, cell fate markers, Drosophila, larval central brain, larval optic lobe, fluorescence immunolocalization
1. Introduction
The central brain occupies the medial half of a fly larval brain lobe, and contains neural stem cells (called neuroblasts) that undergo repetitive asymmetric divisions to self-renew and to generate a neural progenitor cell with limited developmental potential (Sousa-Nunes et al., 2010). Two distinct larval brain neuroblast lineages (types I and II) can be unambiguously identified based on the progenitor progeny generated and the combination of cell fate markers expressed (Weng and Lee, 2011). A type I neuroblast divides asymmetrically to self-renew and to generate a neural progenitor cell called a ganglion mother cell (GMC), which divides once to produce two post-mitotic neurons (Doe, 2008; Egger et al., 2008; Knoblich, 2008). In contrast, a type II neuroblast divides to self-renew and to produce an immature intermediate neural progenitor cell (INP), which acquires restricted developmental potential during maturation and undergoes limited rounds of asymmetric divisions to regenerate and to produce GMCs (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008). While all neuroblasts express the molecular marker Deadpan (Dpn), a type I neuroblast also expresses Asense (Ase) whereas a type II neuroblast expresses PointedP1 (PntP1) (Bowman et al., 2008) (Komori and Lee, unpublished). An immature INP expresses a high level of PntP1, but following maturation, an INP expresses Dpn and Ase. Finally, a GMC shows nuclear localization of Ase and Prospero (Pros) whereas an immature neuron expresses Pros only.
The optic lobe occupies the lateral half of a fly larval brain lobe, and contains two single-cell layers of neuroepithelial stem cells that form the inner and the outer proliferation center (Sousa-Nunes et al., 2010; Weng and Lee, 2011). Neuroepithelial cells in the outer proliferation center are located on the surface of the optic lobe, and initially divide symmetrically to expand their population (Egger et al., 2007). In third larval instar, these neuroepithelial cells progressively differentiate into lamina precursor at the lateral edge and into medulla neuroblasts at the medial edge (Egger et al., 2010; Ngo et al., 2010; Reddy et al., 2010; Wang et al., 2011; Yasugi et al., 2010; Yasugi et al., 2008). The apical complex protein PatJ specifically labels all neuroepithelial cells while the expression of the Notch reporter E(spl)mγ-GFP labels differentiating neuroepithelial cells at the medial edge and medulla neuroblasts. Medullaneuroblasts express molecular markers including Dpn and Ase.
2. Materials
2.1. Reagents
Fix solution: 4% formaldehyde, 0.1M PIPES (pH=6.9), 0.3% TritionX-100, 20mM EGTA, 1mM MgSO4. Fix solution should be prepared fresh every time.
10XPBS: 1.37M NaCl, 27mM KCl, 100mM Na2HPO4, 20mM KH2PO4. Dissolve and adjust pH to 7.4 with concentrated HCl. Bring the volume up to 1 L with dH2O and sterilize. Store at room temperature.
10% Triton X-100: 100% Trition X-100 diluted in sterilized water. 3. PBST (500 ml): 1X PBS, 0.3% TritonX-100. Store at room temperature.
10X Glycine: 1M Glycine, 2% sodium azide, 1XPBS.
Block solution: 1X PBST, 0.1% Normal goat serum, 1X Glycine. Prepare fresh and keep on ice.
70% Glycerol: 100% Glycerol diluted in sterilized water.
Prolong Gold Anti-fade mounting medium (Invitrogen) 8. Schneider’s Insect Medium (Sigma-Aldrich)
2.2 Equipment
22 × 22 mm coverslips, #1 thickness
24 × 40 mm coverslips
Dissection dishes
Fine-tipped forceps (2 pairs)
Fine micro knife with thickness about 0.15 mm
Microfuge tubes (0.5 μl)
Microscope slides
Nutator or rocker
Pipettes & sterile tips
3. Methods
3.1. Dissection of Larval Brains
Fill wells of dissection dishes with 200–400 μl cold Schneider’s medium.
Dissect larvae by rolling them onto their dorsal side so the denticle belts are facing up.
Using a pair of forceps, gently grasp the larva just posterior of the midpoint. With the second pair of forceps, grasp the anterior end of the larva with one tip pushing mouth hook inwards and the other tip outside on the cuticle.
Carefully tear the cuticle at the tip of second pair of forceps while slowly drawing the body away from the mouthpart. The brains will remain attached to the head and be clearly visible among the gut and salivary glands. Remove excess tissues, but leave the brains attached to the mouth hooks. (see Note 1–2)
Place the brains in a 0.5 ml tube containing cold Schneider’s medium. (see Note 3)
3.2. Fixation and Staining
-
1
Remove Schneider’s medium from the tube containing the brains.
-
2
Add 500 μl fixative to the brains and incubate with rocking for 23 minutes at room temperature.
-
3
Quickly wash the brains for three times in ~500 μl of PBST.
-
4
Incubate the brains in primary antibodies diluted in PBST for 3 hours at the room temperature or overnight at 4°C. (see Note 4–7)
-
5
Quickly wash the brains three times in PBST.
-
6
Incubate the brains in secondary antibodies overnight at 4°C. Secondary antibodies are typically diluted 1:200–500 in PBST. Wrap the tube with foil to keep the brains protected from light after this point. (see Note 4–7)
-
8
Quickly wash the brains three times in PBST.
-
9
Equilibrate the brains in Prolong Gold at the room temperature. Samples can be stored in the dark at room temperature or 4°C.
3.3. Mounting Samples
3.3.1. Taking a Z-stack from posterior to anterior side
Adhere two 22 × 22 mm coverslips to a slide with a small amount of 70% glycerol, leaving a ~5 mm space between them. (see Note 8)
Transfer the brains to a slide using a pipette with the tip cut off.
Remove all excess tissues including discs from each brain with forceps.
Orient the brains so that the ventral side is down. Arrange the brains in an array for easy tracking during confocaling. (see Note 9)
Place a 24 × 40 mm coverslip over the samples and back-fill the space between the slide and coverslip by pipetting a small amount of mounting medium along the edge of the coverslip.
3.3.2. Taking a Z-stack from lateral to medial side
Adhere two 22 × 22 mm coverslips to a slide with a small amount of 70% glycerol, leaving a ~5 mm space between them.
Transfer one brain to the slide using a pipette with the tip cut off. (see Note 10)
Remove all excess tissues including the discs from each brain with forceps.
Using one pair of forceps to hold the brain and cut the brain lobes apart from the ventral nerve cord using a micro knife. (see Note 11)
Place a 24 × 40 mm coverslip over the samples and back-fill the space between the slide and coverslip by pipetting a small amount of mounting medium along the edge of the coverslip. Do not seal the coverslip with nail polish. (see Note 12)
Figure 1. Neuroblast lineages in the fly larval central brain.
(A) A cartoon summarizes the expression profile of molecular markers in the type I neuroblast and type II neuroblast lineage. Imm INP: immature INP. (B–C) A third instar larval brain was stained with antibodies against Dpn, Ase and PntP1. The cell cortex was marked by phalloidin. White arrows indicate type I neuroblasts whereas arrowheads indicate type II neuroblasts and asterics indicate immature INPs. The scale bar is 50μm.
Figure 2. Neuroepithelial cells and neuroblasts in the fly larval optic lobe.
(A) A cartoon summarizes the expression profile of molecular markers in neuroepithelial cells and neuroblasts. Non-diff NEC: non-differentiating neuroepithelial cells. Diff NEC: differentiating neuroepithelial cells. imm neurob: immature neuroblasts. Neurob: neuroblasts. (B) A third instar larval optic lobe was stained with antibodies against PatJ, E(spl)mγ-GFP and Dpn. Arrows indicate non-differentiating neuroepithelial cells whereas arrowheads indicate differentiating neuroepithelial cells and asterics indicate immature neuroblasts. The scale bar is 50μm.
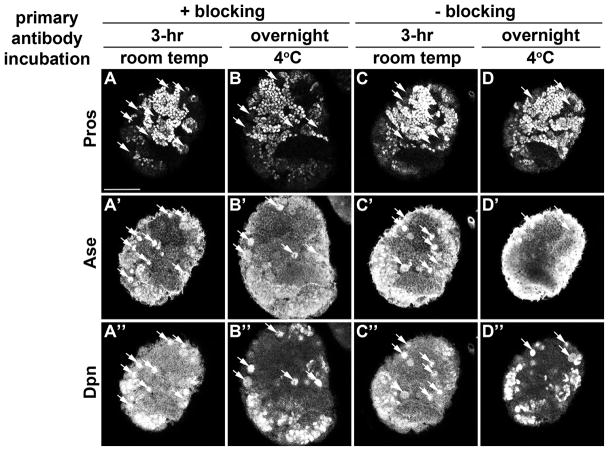
Figure 3. Comparative analyses of four distinct primary antibody staining protocols.
(A–D) Third instar larval brains were stained with Dpn, Pros and Ase. Arrows indicate neuroblasts. The scale bar is 50μm.
Footnotes
Leaving the brains connected to the mouth hooks will help the brains sink to the bottom of the tube during washes and their dark color will make it easier to see the brains while pipetting solutions in the tubes.
Leaving the ventral nerve cord intact will aid in preferred orientation of the brain on the slide: the brain can rest steadily on its ventral surface.
Fix the brain within 20 minutes following dissection to prevent protein degradation.
Four conditions are compared in Figure 3. Skipping the blocking step has no effect on the quality of the staining.
Conditions for primary antibody incubation are dependent on the specific antibody being used. In general, primary antibody incubation at room temp for 3 hours and secondary antibody incubation at 4°C overnight give a good balance between quality and efficiency for the majority of the antibodies. However, some primary antibodies work significantly better when incubated at the room temperature for 3 hours than at 4°C overnight or the reverse. Thus different staining conditions should be tested when the staining quality is not satisfactory.
Due to the thickness of the brain, it is recommended to incubate secondary antibodies at 4°C overnight for complete penetration, especially for anti-rabbit secondary antibodies.
Phalloidin (Invitrogen) is a high-affinity filamentous actin probe conjugated with specific fluorophores. It is useful to mark the cell cortex as shown in Figure X. To do this, incubate the samples with phalloidin diluted 1:100 in PBST for 30 minutes at room temperature after step 5 of the fixation and staining protocol.
Building a bridge using cover slips provides the space to maintain larval brains without destroying their shape.
Orienting the brain with the ventral surface down will give a better confocal quality since this leaves the brain at a higher position than the ventral nerve cord. The 24 × 40 mm coverslip will apply pressure on the brain and result in a larger angle between nerve cord and the brain lobe and leave posterior side up. This is especially important for examining type II neuroblast lineages for them being located at posterior side.
It is preferred to mount one brain per slide, as it is hard to keep track of multiple lobes when rolling the brains under coverslip.
Despite being discarded, an intact ventral nerve cord helps in positioning the brain during cutting, thus resulting in a clean cut and a smoother lobe surface. This will aid in rolling the brain to the desired orientation during confocaling.
Put the slide on the confocal microscope stage and find the brain lobes under low magnification. Slowly and gently slide the coverslip to roll the brain lobe to the desired orientation determined by visualizing markers like PatJ and E(spl)mγ-GFP under an epifluorescent scope.
References
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Develop. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neurobiol. 2008;68:1185–95. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila. Dev Cell. 2008;14:535–46. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–87. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens N, Brand AH, Doe C. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Chell JM, Brand AH. Insights into neural stem cell biology from flies. Philos Trans R Soc Lond B Biol Sci. 2008;363:39–56. doi: 10.1098/rstb.2006.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010;137:2981–2987. doi: 10.1242/dev.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–97. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Ngo KT, Wang J, Junker M, Kriz S, Vo G, Asem B, Olson JM, Banerjee U, Hartenstein V. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Dev Biol. 2010;346:284–95. doi: 10.1016/j.ydbio.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BVVG, Rauskolb C, Irvine KD. Influence of Fat-Hippo and Notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137:2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R, Cheng LY, Gould AP. Regulating neural proliferation in the Drosophila CNS. Curr Opin Neurobiol. 2010;20:50–7. doi: 10.1016/j.conb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Wang W, Liu W, Wang Y, Zhou L, Tang X, Luo H. Notch signaling regulates neuroepithelial stem cell maintenance and neuroblast formation in Drosophila optic lobe development. Dev Biol. 2011;350:414–28. doi: 10.1016/j.ydbio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Weng M, Lee CY. Keeping neural progenitor cells on a short leash during Drosophila neurogenesis. Curr Opin Neurobiol. 2011;21:36–42. doi: 10.1016/j.conb.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T, Sugie A, Umetsu D, Tabata T. Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development. 2010;137:3193–3203. doi: 10.1242/dev.048058. [DOI] [PubMed] [Google Scholar]
- Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–80. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]