Abstract
Copy number variation (CNV) is an important source of genomic diversity in humans, and influences disease susceptibility. The immunoglobulin-receptor genes FCGR3A and FCGR3B on chromosome 1q23.3 show CNV, and CNV of the FCGR3B gene is associated with glomerulonephritis in systemic lupus erythematosus and organ-specific autoimmunity. Large-scale case-control association studies of CNV require technologies that are amenable to high-throughput analysis with low error rates. Here we propose an integrated suite of five assays, four of them duplexed to reduce DNA usage, that assays for copy number variation at FCGR3A and FCGR3B, and genotype the polymorphic neutrophil antigen HNA1. We show how a maximum-likelihood approach to combining the results from these five assays allows estimation of statistical confidence for each individual copy number, and therefore an appropriate significance threshold to be set, controlling the error rate. This approach results in a high-throughput copy number genotyping system, with demonstrable precision and accuracy, that can be applied to large case-control cohort studies. We demonstrate Mendelian inheritance of this CNV, variation in frequency between Europeans and East Asians, and a lack of strong association between the CNV and flanking SNP genotypes, with important consequences for genome-wide association studies.
Keywords: Fc receptor, copy number variation, lupus, paralogue ratio test
Introduction
Copy number variation (CNV) is a common source of variation in the human genome (Iafrate, et al., 2004; Redon, et al., 2006; Sharp, et al., 2005; Tuzun, et al., 2005). Common CNV at various genes has been associated with different diseases, including psoriasis associated with CNV at the beta-defensin locus, and susceptibility to HIV-1 infection associated with CNV at the CCL3L1 locus (Gonzalez, et al., 2005; Hollox, et al., 2008). CNV of the RHD gene is responsible for determining Rh-negative blood group in Europeans (Colin, et al., 1991), and CNV of the HBA alpha-globin gene is responsible for some mild forms of alpha-thalassemia (Higgs, et al., 1980). A large proportion of multiallelic CNV appears to show recurrent mutation because certain diplotypes are not associated with flanking single-copy SNPs, and therefore individual copy-number alleles are not necessarily identical-by-descent (Locke, et al., 2006). Variation at these CNVs is therefore likely to be missed by current genomewide association studies that rely on SNPs within blocks of linkage disequilibrium to describe the majority of variation within that block. CNV therefore needs to be analysed directly in order to understand its evolution and phenotypic effect.
Fcγ receptors act as cellular receptors for the Fc region of IgG and IgE, and transmit their signal inside the cell either by tyrosine-based activation (ITAM) or inhibitory motifs (ITIM) (Nimmerjahn and Ravetch, 2008). In the latest build of the human genome assembly (hg18), two paralogous 82kb repeat regions on chromosome 1 contain the genes FCGR3A and FCGR3B, coding for two variants of the low-affinity activating receptor FcγRIII (CD16A and CD16B; OMIM 146740). They differ in their attachment to the cell surface and their expression pattern: FcγRIIIA has a transmembrane region and is expressed on natural killer cells, and FcγRIIIB is truncated by a mutation changing an arginine codon to a stop codon, is attached to the cell membrane by a glycophosphoinositol anchor and is expressed primarily on neutrophils (Ravetch and Perussia, 1989). Copy number variation of FCGR3B shows a gene dosage effect on protein levels of FcgammaIIIB in serum, and is correlated with neutrophil uptake of immune complexes (Koene, et al., 1996; Willcocks, et al., 2008). CNV of FCGR3B has been associated with glomerulonephritis, and systemic autoimmune diseases including systemic lupus erythematosus (Aitman, et al., 2006; Fanciulli, et al., 2007).
It is known that case-control association studies are particularly vulnerable to inaccuracies in the raw data, particularly systematic bias between case and control DNA which results in a false positive evidence for association (Clayton, et al., 2005). Therefore, accurate typing of copy number variation is a critical step in understanding the relevance of CNVs to human disease (McCarroll and Altshuler, 2007). Current methods are either based on hybridisation of large amounts of genomic DNA on arrays, or measurement of amplification products generated by polymerase chain reaction (PCR). The paralogue ratio test (PRT) is a PCR-based method that has been used to type the beta-defensin CNV and has been used in a replicated case-control association study that demonstrated association of high beta-defensin copy number with an increased susceptibility to psoriasis (Armour, et al., 2007; Hollox, et al., 2008). The first-pass error-rate of PRT at this locus was estimated to be around 8%, so it is clear that the assay has to be repeated several times or, perhaps more satisfactorily, must be combined with other assays to coordinate on a single correct diploid copy number for a given sample. An alternative way of measuring copy number is to amplify across a single nucleotide variant between copy number repeats (called multisite variants or MSVs (Fredman, et al., 2004)). A genotyping system that maintains variant dosage can be used to determine the relative dosage of the two variants: we have used in this paper restriction enzyme digest of PCR products followed by quantification of peaks after capillary electrophoresis (restriction enzyme digest variant ratio REDVR (Aldred, et al., 2005; Hollox, et al., 2008)). It is also possible to use short tandem repeat (STR) variation between copy number repeats to make inferences about copy number. This has been used to show that large expansions of the beta-defensin repeat region are not identical-by-descent and have occurred independently (Hollox, et al., 2003). By amplifying across a highly informative STR marker and counting peaks and dosage, an estimate of the likely copy number can be made (Hollox, et al., 2005).
Here were show that integration of these three methods can yield accurate and precise copy number diplotypes at the FCGR3 locus. We also show that careful selection of variant ratio assays can yield important extra information on the sequence variation between copy number variable repeats. For example, we determine relative copy number of FCGR3A and FCGR3B, and confirm that both FCGR3A and FCGR3B are polymorphic in copy number (Breunis, et al., 2008). We also deduce the relative copy numbers of the HNA1a and HNA1b polymorphism in FCGR3B, which affects binding and phagocytosis of IgG-opsonised particles, and has been linked to periodontitis (Bux, 2008; de Souza and Colombo, 2006; Kobayashi, et al., 2000; Salmon, et al., 1990; Sugita, et al., 2001). We use the method to type a cohort of UK individuals, CEPH families, and HapMap samples, deduce variant frequencies and estimate how much copy number variation and HNA1 genotype could be tagged by flanking single-copy SNP genotypes.
Materials and Methods
DNA samples
Human genomic DNA samples were from the European Collection of Animal Cell Cultures (Control Box 1), Coriell Cell Repositories (HapMap plates 1,2,3) (International, 2005), and the Centre d’Etude des Polymorphisme Humain (CEPH families) and were derived from lymphoblastoid cell lines. A small number of genomic DNAs derived from peripheral blood leukocytes from volunteers were also used, with appropriate consent. Throughout this manuscript, YRI = Yoruba from Ibadan, Nigeria, CHB = Chinese from Beijing, JPT = Japanese from Tokyo, CEU = CEPH Europeans from Utah (mostly of Scandanavian or English origin).
Pulsed Field Gel Electrophoresis and Southern Blotting
Fresh peripheral blood leukocytes were isolated from whole blood by density centrifugation in Histopaque-1077 (Sigma-Aldrich, Poole, UK) and washed in 1×phosphate-buffered saline solution. DNA in agarose blocks was prepared from these cells (~5×107/ml agarose) by an in-block lysis method. DNA in 100μl agarose blocks (approximately 3.3μg) was digested with 40units SbfI (New England Biolabs, Beverly, MA) in 50mM potassium acetate, 20mM Tris-acetate, 10mM magnesium acetate, 1mM dithiothreitol (pH7.9@25°C) for 20 hours at 37°C. The DNA blocks were then loaded on a gel (1% low-EEO agarose (Sigma-Aldrich), in 1×TBE buffer) and run at 6V/cm, switch-time 50-90s, 120° switch angle at 14°C for 22 hours using a CHEF-III pulsed field gel apparatus (Biorad, Hercules, CA).
Transfer of the DNA on to uncharged nylon membrane, fixation by ultraviolet light and hybridisation by 32P-radiolabelled DNA probe was performed using standard laboratory methods. Probes containing no high-copy repeats were generated by PCR, and were a mixture of a 2kb amplicon (generated using primers 5′-GGCTTGTCCTAGGAGCTCAA-3′, 5′-TGAAAGCTTCCCATCCTGTT-3′), a 1.1kb amplicon (5′-TCCATATGGGGATTCTTGGA-3′, GCTTGGTTCAGGGAAATTCA) and a 1.7kb amplicon (5′- GCGCTTAAAGCTTACAAAACAGA-3′, 5′-TCCCAGTTTCTAGACCATAGCA-3′).
Paralogue ratio test
We amplified 5-10ng of genomic DNA in a final volume of 10μl, with 0.5μM primer F, 0.5μM FAM or HEX labelled primer R, in a buffer containing final concentrations of 50mM Tris-HCl (pH8.8@25°C), 12.5mM ammonium sulphate, 1.4mM magnesium chloride, 125μg/ml BSA, 7.5mM 2-mercaptoethanol and 200μM each dNTP (sodium salt), with 0.5U Taq DNA polymerase. Products were amplified using 30 cycles of 95°C for 30 seconds, 56°C for 30 seconds and 70°C for 30 seconds, followed by a single chase phase of 56°C for 1 minute/70°C for 20 minutes to reduce levels of single-stranded DNA products.
Two amplifications were carried out for each sample, with one primer fluorescently label with FAM or with HEX to allow detection and co-electrophoresis on a capillary sequencer. 1μl of a 10-20% dilution of each PCR product was added to 10μl deionised formamide, and analysed by electrophoresis on an ABI 3100 or 3130xl Genetic Analyser, with an injection time of 30 seconds.
Peak areas corresponding to the 67bp PCR product from chromosome 1 and the 72bp PCR product from chromosome 18 were recorded for both FAM- and HEX-labelled products using GeneScan software (Applied Biosystems, Warrington, UK). The ratio 67bp/72bp was compared between FAM- and HEX-labelled products, and results were accepted if the coefficient of variation (standard deviation divided by the mean) was less than 0.15 - this criterion led to the rejection of about 10% of tests. Mean ratios were used in conjunction with reference standards to calibrate each experiment, and the resulting (least-squares) linear regression used to infer the copy numbers for unknown samples. If accepted, the mean of the FAM and HEX ratios was used in further analysis.
Restriction enzyme digest variant ratios
The two REDVR assays used the distinguishing variant between FCGR3A and FCGR3B (C733T Arginine>Stop) and one of the five polymorphisms that distinguish the human neutrophil antigen HNA1a and HNA1b on FCGR3B (C147T) (Ory, et al., 1989a; Ory, et al., 1989b). We amplified in duplex two regions, using the primers 38L(5′FAM- AAGACTGAGCCACCAAGCAT-3′), 38R(5′-CTCCCTGGCACTTCAGAGTC-3′), 234L(5′-TTTTGCAGTGGACACAGGAC-3′) and 234R (5′HEX- GGGTTGCAAATCCAGAGAAA-3′) each at 0.5μM, and the conditions described above, except with an annealing temperature of 53°C. 2μl of PCR product were digested with 10 units of TaqαI restriction enzyme (New England Biolabs) in 50mM Tris-Cl (pH 7.9@25°C) 100mM NaCl 10mM MgCl2 1mM Dithiothreitol in a final volume of 10μl for at least 4 hours at 65°C. Peak areas were recorded for both FAM- and HEX-labelled products using GeneScan software (Applied Biosystems). As for PRT, mean ratios were used in conjunction with reference standards to calibrate each experiment, and the resulting (least-squares) linear regression used to infer the copy numbers for unknown samples.
Short tandem repeat analysis
We amplified a short tandem repeat using primers MSAT1L (5′FAM-TYTATCTGTGATTTTCAGCAG-3′) and MSAT1R (5′-AAAGAGAAAATGGCCAACAA-3′) at 0.5μM each. A short indel variation was co-amplified in the same mix, using primers IDL (5′HEX-TAAGTGCATGGGCTCTGTTG-3′) and IDR (5′TTTCTTCCCATCCCTGTTGA-3′) at 0.375μM each; this produced a product at 243bp and 246bp. The PCR used the same conditions as the paralogue ratio test described above, except amplification was for 26 cycles, with 2μl analysed by capillary electrophoresis. Slippage peaks were observed in MSAT1 amplification due to Taq DNA polymerase replication errors during amplification. In cases where the slippage peak did not overlap with a real peak, the area under the peak was added to the 2bp larger real peak. In some cases real variant peaks were 2bp apart, and therefore a slippage peak could not be distinguished from a real peak. We corrected for this by noting that, where visible, the amount of slippage peak generated was directly proportional to variant length, and so applied a length-dependant correction factor to real variant peaks coinciding with slippage peaks.
Following slippage correction, we obtained a value for copy number by dividing the area of each peak by the area of the peak showing the second smallest area, if there were more than two peaks, and the third smallest area if there were more than four peaks. In most cases, the number reflected an integer copy number, but occasionally, depending on informativeness, this could give a fraction (e.g. 2.5 reflecting a copy number of 5).
Combining copy number estimates
We combined copy number estimates from the five different measurements (2 REDVR, 1 PRT, 2 STR) to give an overall estimate of copy number diplotype for the FCGR3 repeat region, assuming no heterogeneity between repeats and integer copy numbers. We used a maximum-likelihood (ML) approach, calculating the probability that the observed data point with error following a Gaussian normal distribution reflected each integer value in turn. For example, the relative likelihoods of a PRT observed value of 3.8 reflecting integer values of 1 to 9 were calculated. Then the relative likelihoods of a REDVR observed ratio of 1.03 were calculated for integers of 1 to 9, and so on for all five methods. The highest probabilities for each method for each observed value were multiplied to give a ML estimate of copy number for that particular sample. The standard deviation of the Gaussian error distribution was estimated by multiplying the observed value (e.g. 3.8 for the PRT example above) by the coefficient of variation either used as a threshold in PRT, or from the maximum coefficient of variation from experimental repeat measurements in the case of REDVR and STR analyses. This approach reflects the increase in expected measurement error with increased copy number (Armour, et al., 2007).
An estimate of the degree of confidence in each diplotype was calculated by - 2(lnb-lna) where a is the probability of the most likely copy number diplotype, and b is the combined probability of all other copy number diplotypes (from 1 to 9). This statistic has a X2 distribution, so p-values for each copy number diplotype call can be determined using X2 tables.
For the data presented here, we assumed an equal probability of each copy number prior to the ML estimation of copy numbers (flat priors), because of the limited previous information about the copy number distribution in the different populations. However, we have also incorporated the ability to introduce estimates of copy number into our copy number estimation algorithm as Bayesian priors.
Results
PRT and REDVR
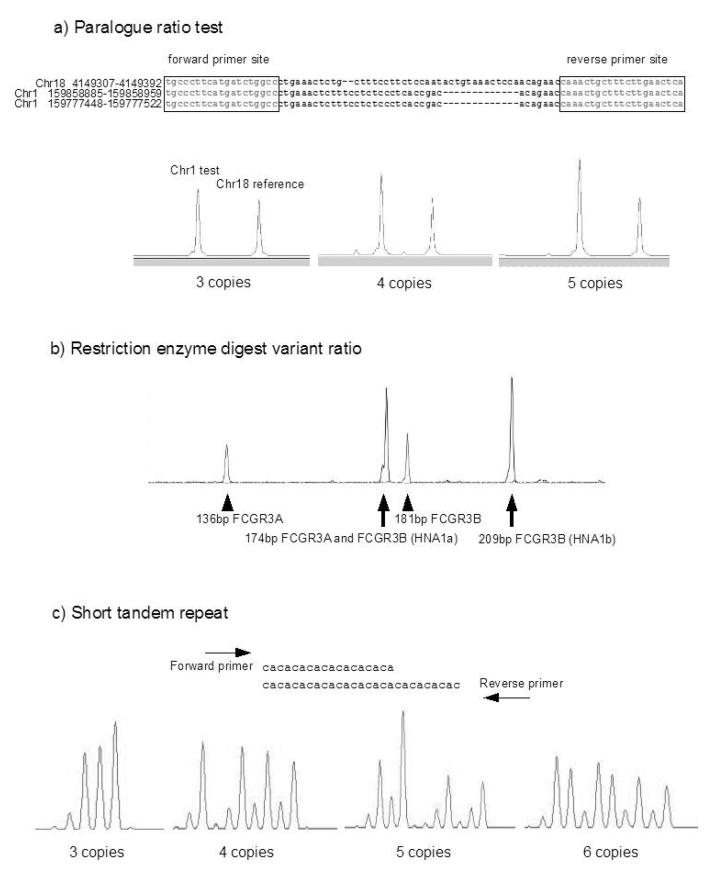
PRT relies on co-amplification of the test and reference locus using the same primer pair, with test and reference distinguished by a small deletion. Using a fluorescently labelled primer, the PCR products are subsequently differentiated and quantified using capillary electrophoresis. We analysed the genome sequence using the Self-Chain track on the UCSC genome browser for the FCGR3 repeat unit (82 kb) and identified a region that could be amplified by a primer pair that also amplified a unique reference locus on chromosome 18. The test and reference PCR products could be distinguished by a small difference in size (Figure 2a).
Figure 2. Copy number measurement approaches.
a) Paralogue Ratio Test (PRT)
Shown are the three sequences present in the genome assembly that are amplified using the primers marked by a box. The different sizes of the products allow the chromosome 1 product to be distinguished from the chromosome 18 product. Three examples of an electrophoretogram of PRT products are shown.
b) Restriction enzyme digest variant ratio (REDVR)
A sample electrophoretogram of TaqI digestion products showing an approxiamate 1:1 ratio of both FCGR3A:FCGR3B and FCGR3A/FCGR3B(HNA1a):FCGR3B(HNA1b).
c) Short tandem repeat (STR)
Illustrating the principle of amplification across an STR, with example electrophoretograms of MSAT1 products from four different copy number samples. Note that the five copy sample produces four differing sized peaks, but one has twice the area of the other three, implying a total of (1+2+1+1)=5 copies.
A multiplex REDVR assay was used to amplify across the nucleotide change that changes an arginine in FCGR3A to a stop codon in FCGR3B, and a nucleotide that distinguished HNA1a from HNA1b. The A/B variant and the HNA1a/HNA1b variant can be distinguished by digestion using TaqαI (Figure 2b).
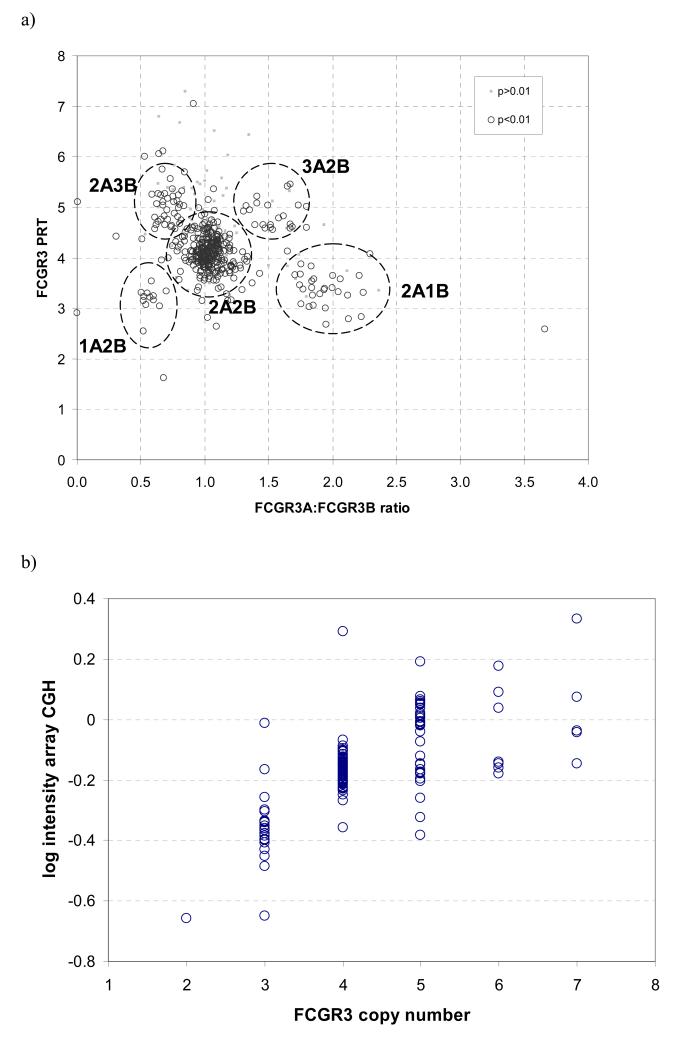
Seven samples were confirmed by PFGE and Southern blotting (two 3 copy, three 4 copy, one 5 copy), showing bands of size consistent with total loss or gain of one 82kb repeat unit (supplementary figure S1). In both PRT and REDVR, inter-experiment calibration was corrected by running these known controls together with several other controls showing high agreement for total copy number between methods. We typed a small panel of samples in duplicate, each from different DNA batches and determined an empirical error rate of the PRT/REDVR method of 8.6% (95%CI 0-18%). A scatterplot of PRT values against values from one REDVR assay shows clustering of results (figure 3a).
Figure 3. Analysis of CNV typing methods.
a) Paralogue ratio test (PRT) versus restriction enzyme digest variant ratio (REDVR). PRT values represent absolute copy numbers, REDVR values are ratios of FCGR3A:FCGR3B. For example, there are two clusters for around PRT values of 3, one at (0.5,3) representing 1 FCGR3A : 2 FCGR3B, and one at (2,3) representing 2 FCGR3A : 1 FCGR3B. Values that result in a significance level p>0.01 for the combined copy number are shown in grey. All 485 samples are shown.
b) Combined copy number measurements versus arrayCGH
Maximum-likelihood copy number estimates of 187 HapMap samples are used, with a significance threshold of p<0.01. Array CGH results are from whole-genome BAC array, using BAC 1tp-8H4 (Redon et al. 2006).
Short tandem repeat (STR) analysis
We devised a duplex PCR to co-amplify a (CA)n repeat, called MSAT1, and an (ATG)n repeat, called ID1. Analysis on the cohort of UK individuals revealed 13 length variants for MSAT1 (167bp-192bp) and two repeat length variants (243bp and 246bp). Repeat testing of a panel of 78 UK samples showed a MSAT1 error rate of 5.8% (95%CI 0%-11%) and an ID1 error rate of 8.1% (95%CI 0%-16%). In contrast to PRT, STR methods seem much more robust to variation between experiments and do not require calibration against a set of known copy number controls.
Consensus copy number
Of 485 samples, 453 (93%) were informative for four or all five measurements of copy number. The theoretical error rate can be set as the significance rate threshold; so for example, a p value of 0.01 corresponds to an error rate of 1%. The error rate was determined empirically by comparing independent duplicate tests of a small panel of samples. The empirical error rate is consistent with the theoretical error rate: for example at a significance value of p<0.1 (predicted error rate 10%), the empirical error rate is 12.5% (95%CI 6.8%-18%). Because the theoretical error rate is estimated using high values of experimental error (see Methods), we expect that the true error rate will be at the lower end of these confidence limits. Therefore the threshold set for all further analyses (p<0.01, predicted error rate 1%), is likely to reflect or slightly overestimate the actual error rate.
The estimation of consensus copy number depends on two assumptions: firstly, that integer copy number variation underlies the observed data, and, secondly, that all five methods are measuring the same copy number and that there is no copy number heterogeneity between repeats. All the evidence from examining raw data suggests that copy number varies as integers (2,3,4,5 etc), because of clustering around expected integer classes (figure 3a), and this is observed at other similar multiallelic loci such as the beta-defensins (Armour, et al., 2007; Hollox, et al., 2008). Differences in copy number estimates on one sample from the five different methods could reflect either measurement error or real copy number heterogeneity between repeats. These differences are reflected in a low confidence p-value for a given copy number for that particular sample, and can be excluded by selecting an appropriate p-value threshold. Given the observed errors estimated by duplicate measurements, and the large number of samples analysed, most of the apparent disagreements in copy number estimates between methods are likely to be due to experimental error. However, retesting of several discrepant samples showed that in a minority of these samples the discrepant measurements are reproducible. This may reflect small repeat-specific deletions, or, perhaps more likely, reflect rare single nucleotide variation underneath oligonucleotide primers that reduce or abolish amplification from one repeat. Whatever the cause, samples which show such large discrepancies are rare and will show low confidence values for copy number: in the UK cohort of 86 samples only two repeatedly showed discrepancies (with p values for overall copy number of 0.97 and 0.53).
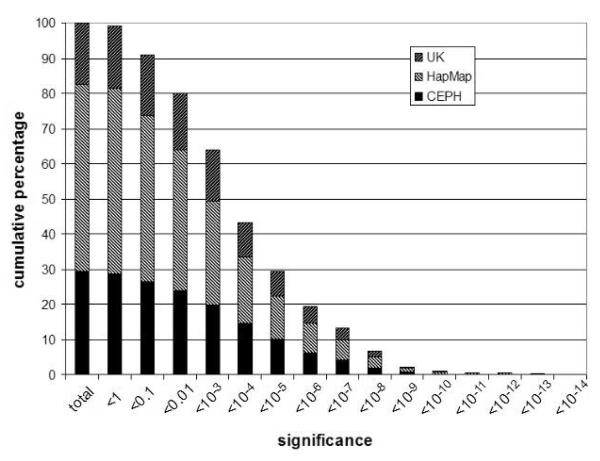
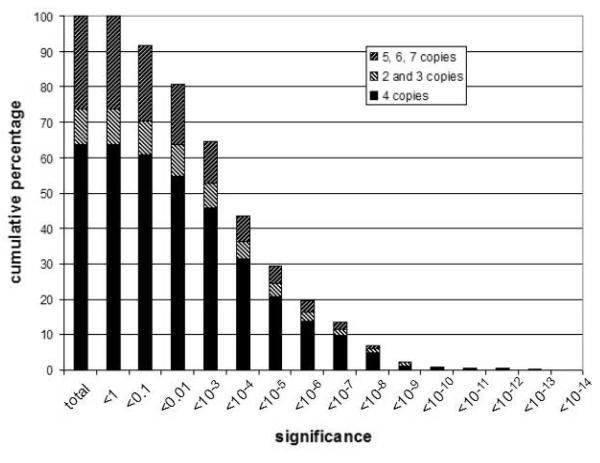
We analysed the significance of individual FCGR3 copy numbers stratified by DNA source (figure 4a) and by copy number (figure 4b). This shows that the combined approach is not severely affected by DNA source, but that there is a small effect of copy number itself: individuals with 5, 6 and 7 copies tend to have measurements with weaker statistical support. This is consistent with previous single method observations, and reflects the fact that the difference between 2 and 3 copies is 50% relative dosage, and the difference between 5 and 6 copies is 20% relative dosage. Full results from all 485 samples are provided (Supplementary table S1).
Figure 4. Cumulative distribution of significance levels for 485 copy number measurements.
a) showing distribution of significance levels in different DNA cohorts
b) showing distribution of significance levels of results of different copy number.
Comparison with array-CGH data
We compared the results from the Wellcome Trust Sanger Institute whole-genome tiling BAC array with our copy number estimates for the HapMap samples where paired data were available (figure 3b). Copy number estimates correspond to differential relative dosage assayed by array-CGH, although the correspondence weakens for higher copy numbers, so that the increased dosage of 6 and 7 copy individuals is not reflected in increased signal from array-CGH. This perhaps reflects a limit of dose-response of the BAC-array CGH method, at least for this particular BAC clone. Several 5 copy samples cluster clearly with the four copy samples according to array-CGH, and the reason for this discrepancy is not clear. Reexamination of our results from the five assays described above show that the estimate of five copies for these samples has very strong support.
Copy number distribution in populations
In all population cohorts studied, the most frequent copy number was 4, comprising 2 FCGR3A genes and 2 FCGR3B genes. Analysis of FCGR3 copy number distribution in Europeans, represented by the unrelated CEPH samples and the UK cohort, shows that deletion and duplication alleles are relatively rare: 3 and 5 copy individuals make up 6% and 13% of the population respectively (table 1a). These comprise almost equally of copy number variation of FCGR3A and FCGR3B, with duplication more frequent than deletion. The estimation of the frequency of individuals with one copy of FCGR3B differs from previously published data which suggested a frequency in healthy UK individuals of between 15 and 20% (Fanciulli, et al., 2007). In this study we estimate the frequency of FCGR3B 1 copy individuals to be around 4%. The duplication frequency has similarly been overestimated, with FCGR3B 3 copy individuals at a frequency of around 6% in the UK cohort used in this study, in contrast to 10-20% published elsewhere. The frequencies presented here are consistent with those previously obtained on a healthy Dutch population using multiplex ligation probe amplification (MLPA) (Breunis, et al., 2008).
Table 1. Population copy number frequencies.
YRI = Yoruba from Ibadan, Nigeria, CHB = Chinese from Beijing, JPT = Japanese from Tokyo. Significance threshold p<0.01
| a) FCGR3A | |||||
|---|---|---|---|---|---|
| copies | JPT | CHB | YRI | Ceph | UK |
| 1 | 0 | 1 | 1 | 3 | 1 |
| 2 | 28 | 26 | 33 | 98 | 73 |
| 3 | 3 | 5 | 1 | 9 | 5 |
| 4 | 1 | 0 | 0 | 0 | 0 |
| n | 32 | 32 | 35 | 110 | 79 |
| b) FCGR3B | |||||
|---|---|---|---|---|---|
| copies | JPT | CHB | YRI | Ceph | UK |
| 1 | 2 | 3 | 5 | 5 | 3 |
| 2 | 19 | 19 | 30 | 97 | 69 |
| 3 | 11 | 9 | 0 | 6 | 6 |
| 4 | 0 | 1 | 0 | 2 | 1 |
| n | 32 | 32 | 35 | 110 | 79 |
| c) Total FCGR3 | |||||
|---|---|---|---|---|---|
| copies | JPT | CHB | YRI | Ceph | UK |
| 2 | 0 | 1 | 0 | 0 | 0 |
| 3 | 2 | 2 | 6 | 8 | 4 |
| 4 | 19 | 15 | 28 | 86 | 63 |
| 5 | 7 | 12 | 1 | 14 | 11 |
| 6 | 3 | 2 | 0 | 1 | 1 |
| 7 | 1 | 0 | 0 | 1 | 0 |
| n | 32 | 32 | 35 | 110 | 79 |
In East Asian populations, represented by Japanese and Chinese, there is a similar frequency of FCGR3A deletion and duplication but a much higher frequency of FCGR3B duplication (tables 1b and 1c). There is no large difference in copy number distribution between the Nigerian Yoruba and the Europeans, although the sample size is small.
The FCGR3B polymorphism HNA1a/b differs in frequency between populations as reported previously (table 2)(Bux, 2008). The variant ratio results generated are consistent with the assumption that FCGR3A carries a C nucleotide at this position, is cut by TaqI and therefore there is no evidence of gene conversion of the T nucleotide variant between FCGR3B and FCGR3A. We typed three three-generation CEPH families using the five assays and demonstrated Mendelian inheritance of FCGR3 copy number repeats (Supplementary figure S2).
Table 2. HNA1 genotypes in different populations.
| Genotype | Ratio 1a:1b | JPT | CHB | YRI | Ceph | UK |
|---|---|---|---|---|---|---|
| 1a | 1 | 2 | 2 | 2 | 2 | 3 |
| 1b | 0 | 0 | 1 | 3 | 3 | 3 |
| 1a1a | 1 | 10 | 6 | 9 | 4 | 5 |
| 1a1b | 0.5 | 12 | 8 | 13 | 42 | 26 |
| 1b1b | 0 | 1 | 5 | 7 | 51 | 37 |
| 1a1a1a | 1 | 2 | 1 | 0 | 0 | 0 |
| 1a1a1b | 0.66 | 0 | 4 | 0 | 1 | 2 |
| 1a1b1b | 0.33 | 3 | 4 | 0 | 4 | 0 |
| 1b1b1b | 0 | 0 | 0 | 0 | 0 | 2 |
| 1a1a1b1b | 0.5 | 0 | 1 | 0 | 2 | 1 |
| 1a1b1b1b | 0.25 | 0 | 0 | 0 | 1 | 0 |
| Mean population 1a:1b ratio |
0.66 | 0.55 | 0.50 | 0.27 | 0.30 |
Linkage disequilibrium with neighbouring SNPs
For the 200kb either side of the FCGR3 repeat region, we compared SNP genotypes generated by the International HapMap project (phase II) with our copy number estimates for the HapMap samples where paired data was available. Individual genotypes were coded as 1, 2 and 3 representing homozygote 1, heterozygote, and homozygote 2 respectively. These values were compared with total copy number, copy number of FCGR3A, copy number of FCGR3B, HNA1 genotype, and the square of Pearson’s correlation coefficient (r2) calculated for each comparison. The r2 values were plotted against the genomic region (Supplementary figure S3).
The maximum r2 values observed were 0.38 (rs10800032) and 0.31 (rs2340719) associating with HNA1 genotype and total copy number respectively in the Japanese population. Such values are too low to allow any predictive power of copy number from SNP genotype, and suggest that duplication and deletion at the FCGR3 locus has occurred multiple times on different SNP haplotypes. At such a low level, association of copy number with human disease will not have been detected by current SNP-based genome-wide association studies. The values are population specific: the association of HNA1 genotype with rs10800032 has r2 values of 0.06 in Europeans, 0.003 in Chinese and this SNP is monomorphic in Yoruba. The higher r2 values may reflect recent population-specific mutation events on certain SNP haplotypes.
Discussion
We have described an approach for CNV typing at the FCGR3 locus where statistical support for each copy number estimate can be assessed. In addition, the number of FCGR3A and FCGR3B genes, and the allelic status of the FCGR3B HNA1 polymorphism, is directly interrogated. Our methods are PCR-based, with between 20-40ng required for all five tests, in contrast to MLPA where at least 200ng is required for a test. For the MLPA, the probe discriminating A from B does not discriminate the actual functional Arg-Stop change, but other nucleotide variants up to 6.7kb away, which have unknown relationships to the A/B change (Breunis, et al., 2008). We suggest that our approach, where significance thresholds can be adjusted appropriately, is sufficiently robust for case-control association studies at this locus.
The East Asian populations tested have a higher frequency of FCGR3B duplication compared to Europeans. It is unclear whether this is due to variation in mutation rate, genetic drift or natural selection. Investigation of clinical consequences of FCGR3B duplication in these populations will help understand the difference in observed duplication frequency.
We show that copy number variation of the FCGR3 region is not strongly associated with any SNP in the flanking 400kb region. Similarly, at least one sequence variant, the HNA1 polymorphism on FCGR3B, has only weak association with flanking SNPs. Therefore both copy number and at least some sequence variation between copy number repeats will not be assayed in the current generation of whole-genome association studies. This is important because both copy number variation and sequence variation are likely to act separately or synergistically to influence phenotype. For example, copy number of FCGR3B can influence disease directly, presumably by a gene dosage effect (Aitman, et al., 2006). Also, copy number variation of the FCGR2C fusion gene, and sequence variation of that gene introducing a polymorphic truncation allele, can interact to affect susceptibility to idiopathic thrombocytopenic purpura (Breunis, et al., 2008). Direct assays for copy number combined with an appreciation of the allelic association of single nucleotide variants within copy number repeats are required to fully interrogate CNV regions for association with human disease.
Supplementary Material
Supplementary table S1 Full data for the 485 samples analysed in this paper, in comma separated value format (.csv).
Supplementary Figures S1-S3
Figure 1. Human FCGR3 genomic region on chromosome 1q23.3.
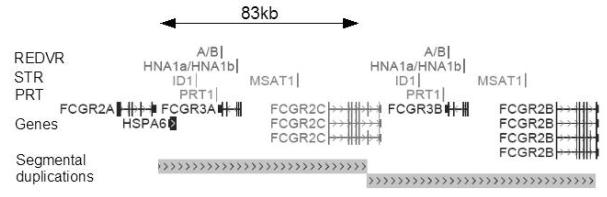
RefSeq genes are shown in blue, together with segmental duplication (98.3% identity between the two assembled repeats) as defined by Bailey et al. Location of the five copy number measures are shown (2 REDVR, 2 STR, 1 PRT). Based on the UCSC Genome Browser March 2006 (build 36.1, hg18).
Acknowledgements
EJH would like to thank colleagues in the Department of Genetics for support and access to the HapMap and CEPH samples. J-CD was part of the EU Erasmus Exchange programme with the University of Heidelberg, Germany.
References
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439(7078):851–855. doi: 10.1038/nature04489. others. [DOI] [PubMed] [Google Scholar]
- Aldred PMR, Hollox EJ, Armour JAL. Copy number polymorphism and expression level variation of the human alpha-defensin genes DEFA1 and DEFA3. Human Molecular Genetics. 2005;14(14):2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- Armour JA, Palla R, Zeeuwen PL, Heijer MD, Schalkwijk J, Hollox EJ. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35(3):e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunis WB, van Mirre E, Bruin M, Geissler J, de Boer M, Peters M, Roos D, de Haas M, Koene HR, Kuijpers TW. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood. 2008;111(3):1029–38. doi: 10.1182/blood-2007-03-079913. [DOI] [PubMed] [Google Scholar]
- Bux J. Human neutrophil alloantigens. Vox Sang. 2008;94(4):277–85. doi: 10.1111/j.1423-0410.2007.01031.x. [DOI] [PubMed] [Google Scholar]
- Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM, Smink LJ, Lam AC, Ovington NR, Stevens HE. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37(11):1243–6. doi: 10.1038/ng1653. others. [DOI] [PubMed] [Google Scholar]
- Colin Y, Cherif-Zahar B, Le Van Kim C, Raynal V, Van Huffel V, Cartron JP. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78(10):2747–52. [PubMed] [Google Scholar]
- de Souza RC, Colombo AP. Distribution of FcgammaRIIa and FcgammaRIIIb genotypes in patients with generalized aggressive periodontitis. J Periodontol. 2006;77(7):1120–8. doi: 10.1902/jop.2006.050305. [DOI] [PubMed] [Google Scholar]
- Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L, Heward JM, Gough SC, de Smith A, Blakemore AI. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39(6):721–3. doi: 10.1038/ng2046. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredman D, White SJ, Potter S, Eichler EE, Den Dunnen JT, Brookes AJ. Complex SNP-related sequence variation in segmental genome duplications. Nat Genet. 2004;36(8):861–6. doi: 10.1038/ng1401. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307(5714):1434–1440. doi: 10.1126/science.1101160. others. [DOI] [PubMed] [Google Scholar]
- Higgs DR, Old JM, Pressley L, Clegg JB, Weatherall DJ. A novel alpha-globin gene arrangement in man. Nature. 1980;284(5757):632–5. doi: 10.1038/284632a0. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Armour JA, Barber JC. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am J Hum Genet. 2003;73(3):591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox EJ, Davies J, Griesenbach U, Burgess J, Alton EW, Armour JA. Beta-defensin genomic copy number is not a modifier locus for cystic fibrosis. J Negat Results Biomed. 2005;4:9. doi: 10.1186/1477-5751-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, van de Kerkhof PC, Traupe H, de Jongh G, den Heijer M. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40(1):23–5. doi: 10.1038/ng.2007.48. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- International HapMap Project A haplotype map of the human genome. Nature. 2005;437(7063):1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, van der Pol WL, van de Winkel JG, Hara K, Sugita N, Westerdaal NA, Yoshie H, Horigome T. Relevance of IgG receptor IIIb (CD16) polymorphism to handling of Porphyromonas gingivalis: implications for the pathogenesis of adult periodontitis. J Periodontal Res. 2000;35(2):65–73. doi: 10.1034/j.1600-0765.2000.035002065.x. [DOI] [PubMed] [Google Scholar]
- Koene HR, de Haas M, Kleijer M, Roos D, von dem Borne AE. NA-phenotype-dependent differences in neutrophil Fc gamma RIIIb expression cause differences in plasma levels of soluble Fc gamma RIII. Br J Haematol. 1996;93(1):235–41. doi: 10.1046/j.1365-2141.1996.4971038.x. [DOI] [PubMed] [Google Scholar]
- Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79(2):275–90. doi: 10.1086/505653. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39(7 Suppl):S37–42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8(1):34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- Ory PA, Clark MR, Kwoh EE, Clarkson SB, Goldstein IM. Sequences of complementary DNAs that encode the NA1 and NA2 forms of Fc receptor III on human neutrophils. J Clin Invest. 1989a;84(5):1688–91. doi: 10.1172/JCI114350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory PA, Goldstein IM, Kwoh EE, Clarkson SB. Characterization of polymorphic forms of Fc receptor III on human neutrophils. J Clin Invest. 1989b;83(5):1676–81. doi: 10.1172/JCI114067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravetch JV, Perussia B. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J Exp Med. 1989;170(2):481–97. doi: 10.1084/jem.170.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon JE, Edberg JC, Kimberly RP. Fc gamma receptor III on human neutrophils. Allelic variants have functionally distinct capacities. J Clin Invest. 1990;85(4):1287–95. doi: 10.1172/JCI114566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R. Segmental duplications and copy-number variation in the human genome. American Journal of Human Genetics. 2005;77(1):78–88. doi: 10.1086/431652. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita N, Kobayashi T, Ando Y, Yoshihara A, Yamamoto K, van de Winkel JG, Miyazaki H, Yoshie H. Increased frequency of FcgammaRIIIb-NA1 allele in periodontitis-resistant subjects in an elderly Japanese population. J Dent Res. 2001;80(3):914–8. doi: 10.1177/00220345010800031301. [DOI] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D. Fine-scale structural variation of the human genome. Nature Genetics. 2005;37(7):727–732. doi: 10.1038/ng1562. others. [DOI] [PubMed] [Google Scholar]
- Willcocks LC, Lyons PA, Clatworthy MR, Robinson JI, Yang W, Newland SA, Plagnol V, McGovern NN, Condliffe AM, Chilvers ER. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205(7):1573–1582. doi: 10.1084/jem.20072413. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table S1 Full data for the 485 samples analysed in this paper, in comma separated value format (.csv).
Supplementary Figures S1-S3