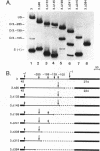
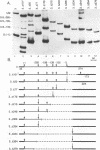
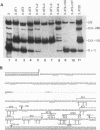
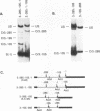
Abstract
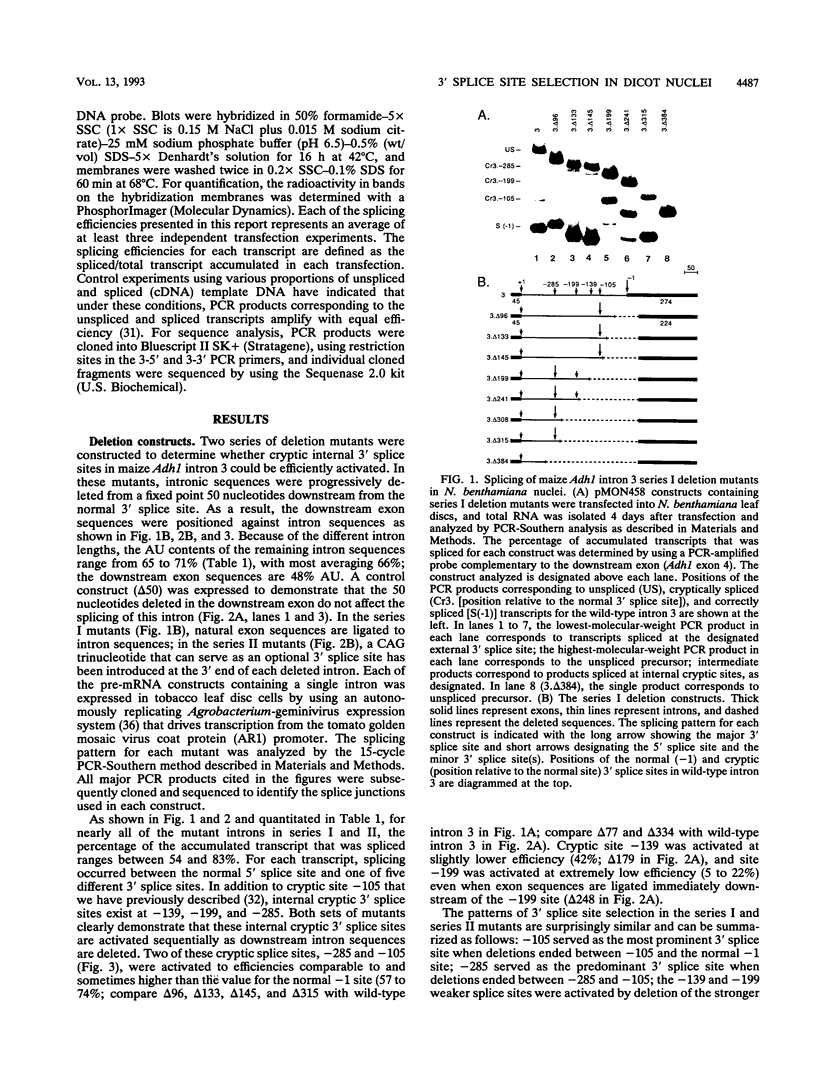
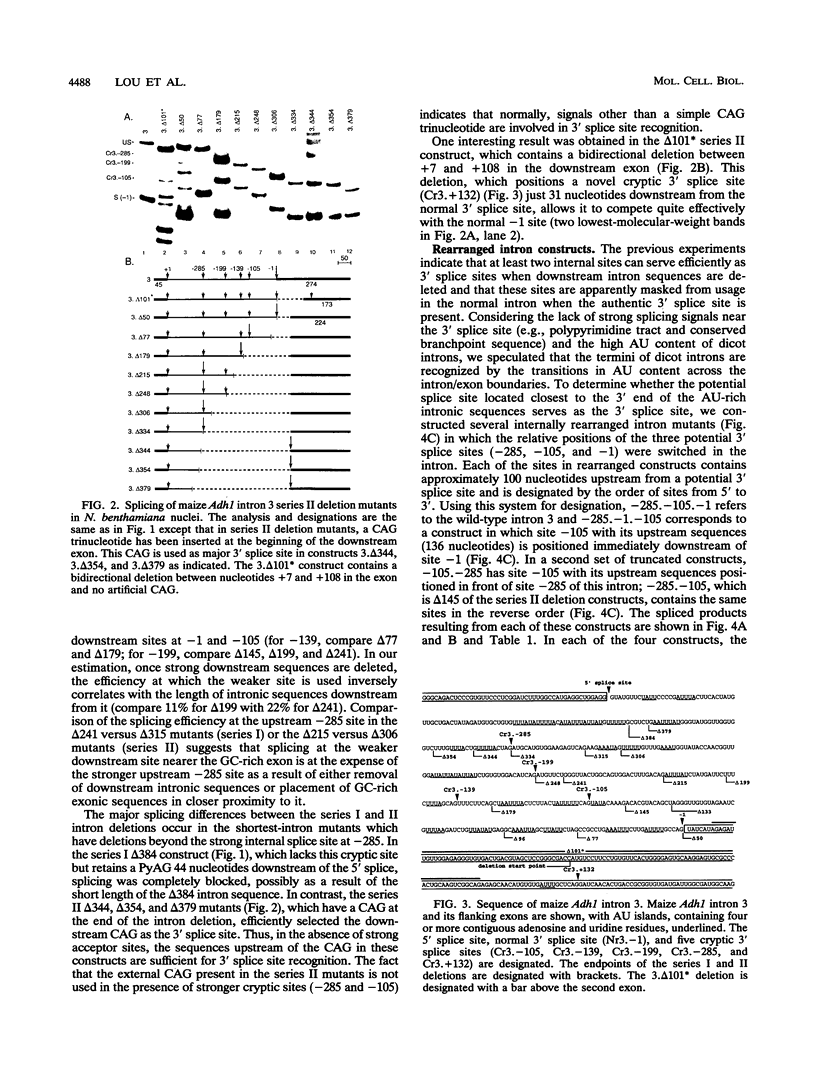
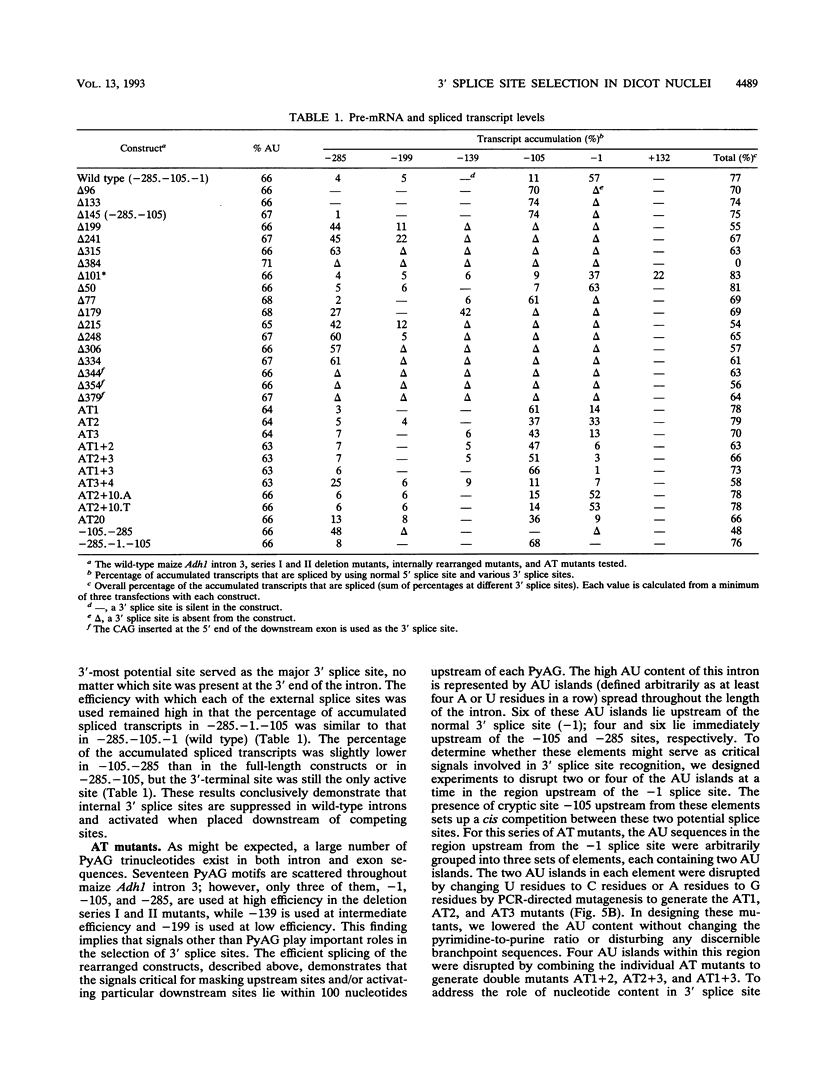
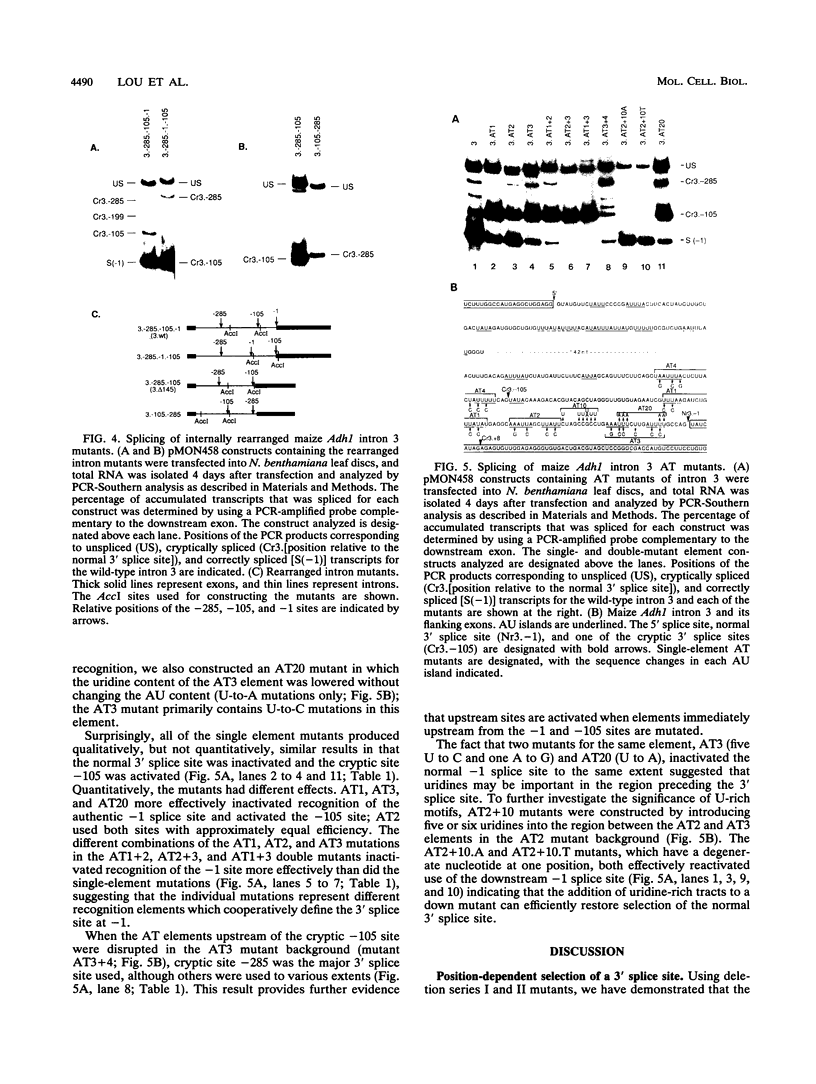
In contrast to mammalian and yeast systems, the mechanism for intron recognition and splice site selection in plant pre-mRNAs is poorly understood. Splice site sequences and putative branchpoint sequences are loosely conserved in plant introns compared with other eukaryotes. Perhaps to compensate for these variations, plant introns are significantly richer in adenosine and uridine residues than are their adjacent exons. To define elements critical for 3' splice site selection in dicotyledonous plant nuclei, pre-mRNA transcripts containing intron 3 of the maize Adh1 gene were expressed in Nicotiana benthamiana nuclei by using an autonomously replicating plant expression vector. Using a series of intron rearrangements which reposition the 3' intron-exon border, we demonstrate that the normal 3' splice site is defined in a position-dependent manner and that cryptic 3' splice sites within the intron are masked by the presence of a functional downstream 3' splice site. Disruption of the AU-rich elements upstream from the normal 3' splice site indicates that multiple AU elements between -66 and -6 cooperatively define the 3' boundary of the intron. These results are consistent with a model for plant intron recognition in which AU-rich elements spread throughout the length of the intron roughly define the intron boundaries by generating strong AU transition points. Functional 3' splice sites located downstream from these AU-rich sequences are preferentially selected over sites embedded within them.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Bennett M., Piñol-Roma S., Staknis D., Dreyfuss G., Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol Cell Biol. 1992 Jul;12(7):3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W. A catalogue of splice junction and putative branch point sequences from plant introns. Nucleic Acids Res. 1986 Dec 22;14(24):9549–9559. doi: 10.1093/nar/14.24.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Feix G., Frendewey D. Accurate in vitro splicing of two pre-mRNA plant introns in a HeLa cell nuclear extract. EMBO J. 1986 Nov;5(11):2749–2758. doi: 10.1002/j.1460-2075.1986.tb04563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989 Oct 11;17(19):7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C., Taylor F. M., Martindale D. W. Nuclear pre-mRNA introns: analysis and comparison of intron sequences from Tetrahymena thermophila and other eukaryotes. Nucleic Acids Res. 1990 Sep 11;18(17):5133–5141. doi: 10.1093/nar/18.17.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. S., Gerlach W. L., Pryor A. J., Bennetzen J. L., Inglis A., Llewellyn D., Sachs M. M., Ferl R. J., Peacock W. J. Molecular analysis of the alcohol dehydrogenase (Adh1) gene of maize. Nucleic Acids Res. 1984 May 11;12(9):3983–4000. doi: 10.1093/nar/12.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu Rev Cell Biol. 1986;2:459–498. doi: 10.1146/annurev.cb.02.110186.002331. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Freyer G. A., O'Brien J. P., Hurwitz J. Alterations in the polypyrimidine sequence affect the in vitro splicing reactions catalyzed by HeLa cell-free preparations. J Biol Chem. 1989 Sep 5;264(25):14631–14637. [PubMed] [Google Scholar]
- Fu X. Y., Ge H., Manley J. L. The role of the polypyrimidine stretch at the SV40 early pre-mRNA 3' splice site in alternative splicing. EMBO J. 1988 Mar;7(3):809–817. doi: 10.1002/j.1460-2075.1988.tb02879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Blanco M. A., Jamison S. F., Sharp P. A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989 Dec;3(12A):1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- Ge H., Manley J. L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990 Jul 13;62(1):25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J. 1991 Sep;10(9):2635–2644. doi: 10.1002/j.1460-2075.1991.tb07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Goux-Pelletan M., Libri D., d'Aubenton-Carafa Y., Fiszman M., Brody E., Marie J. In vitro splicing of mutually exclusive exons from the chicken beta-tropomyosin gene: role of the branch point location and very long pyrimidine stretch. EMBO J. 1990 Jan;9(1):241–249. doi: 10.1002/j.1460-2075.1990.tb08101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Elmer J. S., Rogers S. G. Transient expression of heterologous RNAs using tomato golden mosaic virus. Nucleic Acids Res. 1988 Nov 25;16(22):10511–10528. doi: 10.1093/nar/16.22.10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley B. A., Schuler M. A. Plant intron sequences: evidence for distinct groups of introns. Nucleic Acids Res. 1988 Jul 25;16(14B):7159–7176. doi: 10.1093/nar/16.14.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. In vitro processing of a plant pre-mRNA in a HeLa cell nuclear extract. Nucleic Acids Res. 1986 Oct 10;14(19):7513–7528. doi: 10.1093/nar/14.19.7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Roscigno R. F., Mulligan G. J., Finn L. A., Weber K. S. Identification of two distinct intron elements involved in alternative splicing of beta-tropomyosin pre-mRNA. Genes Dev. 1990 Jan;4(1):98–110. doi: 10.1101/gad.4.1.98. [DOI] [PubMed] [Google Scholar]
- Keith B., Chua N. H. Monocot and dicot pre-mRNAs are processed with different efficiencies in transgenic tobacco. EMBO J. 1986 Oct;5(10):2419–2425. doi: 10.1002/j.1460-2075.1986.tb04516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5' splice site selection by activating proximal sites. Cell. 1990 Jul 13;62(1):35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Lou H., McCullough A. J., Schuler M. A. Expression of maize Adh1 intron mutants in tobacco nuclei. Plant J. 1993 Mar;3(3):393–403. doi: 10.1046/j.1365-313x.1993.t01-22-00999.x. [DOI] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992 Jan 24;68(2):365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayrand S. H., Pederson T. Crosslinking of hnRNP proteins to pre-mRNA requires U1 and U2 snRNPs. Nucleic Acids Res. 1990 Jun 11;18(11):3307–3318. doi: 10.1093/nar/18.11.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough A. J., Lou H., Schuler M. A. Factors affecting authentic 5' splice site selection in plant nuclei. Mol Cell Biol. 1993 Mar;13(3):1323–1331. doi: 10.1128/mcb.13.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough A. J., Lou H., Schuler M. A. In vivo analysis of plant pre-mRNA splicing using an autonomously replicating vector. Nucleic Acids Res. 1991 Jun 11;19(11):3001–3009. doi: 10.1093/nar/19.11.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen M. P., Smith C. W., Patton J. G., Nadal-Ginard B. Alpha-tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. Genes Dev. 1991 Apr;5(4):642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988 Nov;2(11):1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Patterson B., Guthrie C. A U-rich tract enhances usage of an alternative 3' splice site in yeast. Cell. 1991 Jan 11;64(1):181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- Pautot V., Brzezinski R., Tepfer M. Expression of a mouse metallothionein gene in transgenic plant tissues. Gene. 1989 Apr 15;77(1):133–140. doi: 10.1016/0378-1119(89)90367-3. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Reed R. Protein composition of mammalian spliceosomes assembled in vitro. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8031–8035. doi: 10.1073/pnas.87.20.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985 Oct 24;317(6039):732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Porro E. B., Patton J. G., Nadal-Ginard B. Scanning from an independently specified branch point defines the 3' splice site of mammalian introns. Nature. 1989 Nov 16;342(6247):243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988 May;8(5):2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3' end of introns. EMBO J. 1988 Nov;7(11):3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Herrero J. J., Filipowicz W. Nuclear pre-mRNA processing in plants: distinct modes of 3'-splice-site selection in plants and animals. Mol Cell Biol. 1988 May;8(5):2042–2051. doi: 10.1128/mcb.8.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen V. L., Spritz R. A. Splicing of plant pre-mRNAs in animal systems and vice versa. Gene. 1987;56(2-3):253–265. doi: 10.1016/0378-1119(87)90142-9. [DOI] [PubMed] [Google Scholar]