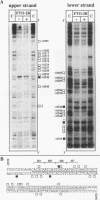
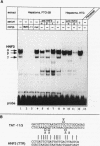
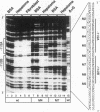
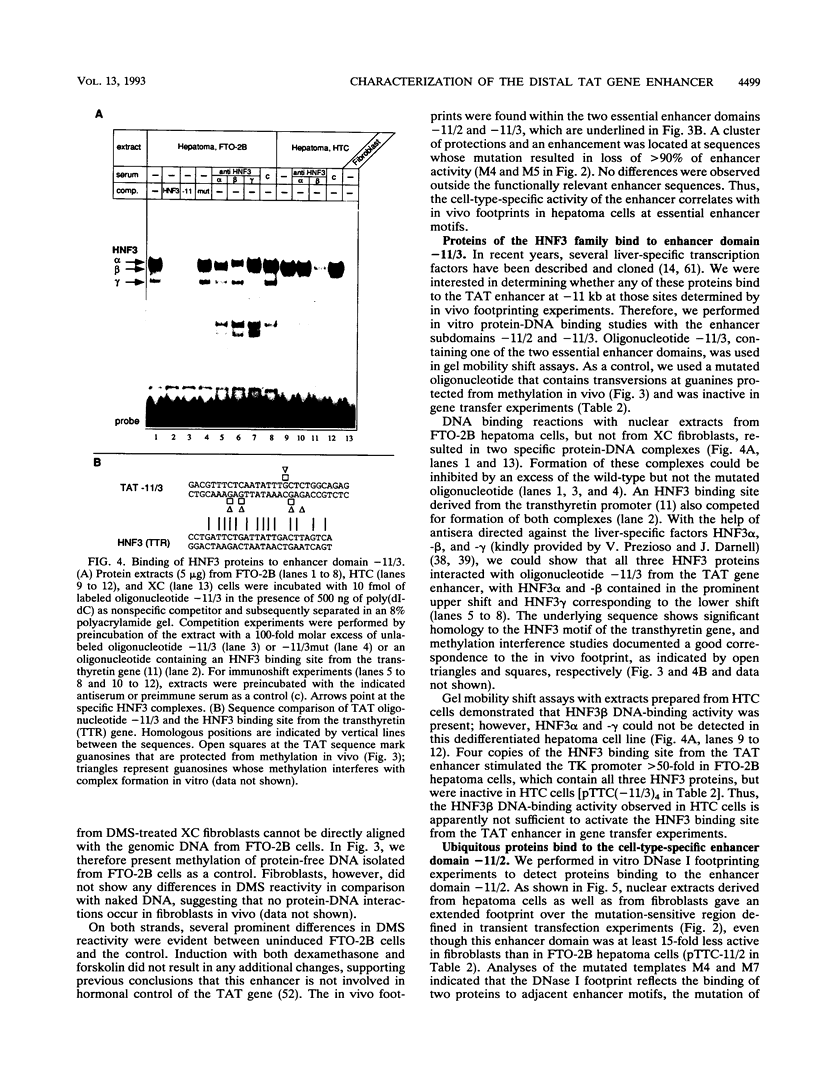
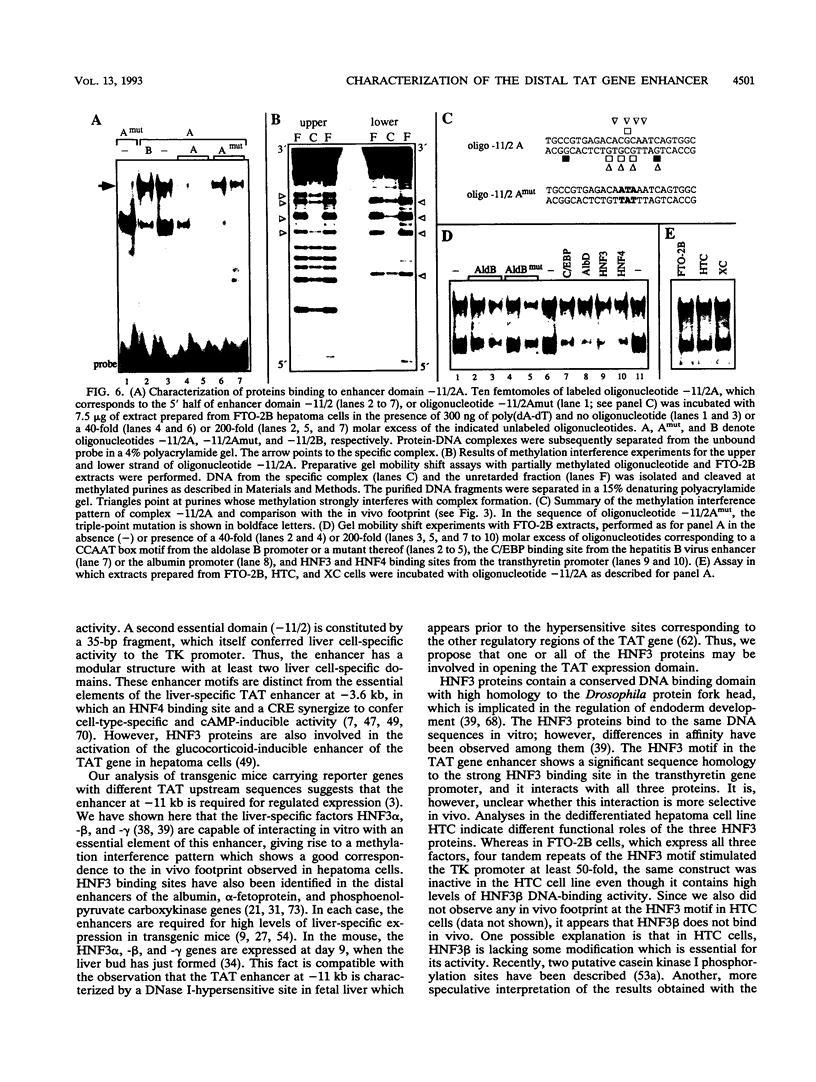
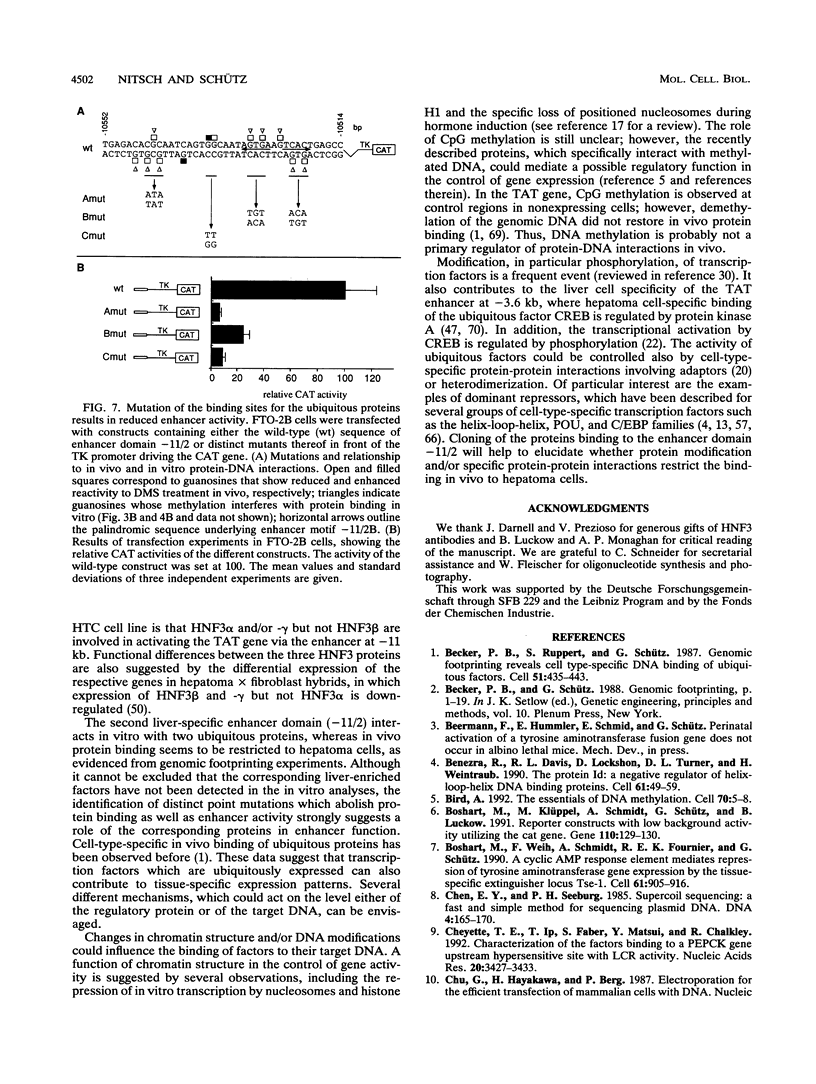
Abstract
Tyrosine aminotransferase gene expression is confined to parenchymal cells of the liver, is inducible by glucocorticoids and glucagon, and is repressed by insulin. Three enhancers control this tissue-specific and hormone-dependent activity, one of which, located at -11 kb, is implicated in establishing an active expression domain. We have studied in detail this important regulatory element and have identified a 221-bp fragment containing critical enhancer sequences which stimulated the heterologous thymidine kinase promoter more than 100-fold in hepatoma cells. Within this region, we have characterized two essential liver-specific enhancer domains, one of which was bound by proteins of the hepatocyte nuclear factor 3 (HNF3) family. Analyses with the dedifferentiated hepatoma cell line HTC suggested that HNF3 alpha and/or -gamma, but not HNF3 beta, are involved in activating the tyrosine aminotransferase gene via the -11-kb enhancer. Genomic footprinting and in vitro protein-DNA binding studies documented cell-type-specific binding of ubiquitous factors to the second essential enhancer domain, which by itself stimulated the thymidine kinase promoter preferentially in hepatoma cells. These results will allow further characterization of the role of these enhancer sequences in developmental activation of the tyrosine aminotransferase gene.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990 Apr 6;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Boshart M., Klüppel M., Schmidt A., Schütz G., Luckow B. Reporter constructs with low background activity utilizing the cat gene. Gene. 1992 Jan 2;110(1):129–130. doi: 10.1016/0378-1119(92)90456-y. [DOI] [PubMed] [Google Scholar]
- Boshart M., Weih F., Schmidt A., Fournier R. E., Schütz G. A cyclic AMP response element mediates repression of tyrosine aminotransferase gene transcription by the tissue-specific extinguisher locus Tse-1. Cell. 1990 Jun 1;61(5):905–916. doi: 10.1016/0092-8674(90)90201-o. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Cheyette T. E., Ip T., Faber S., Matsui Y., Chalkley R. Characterization of the factors binding to a PEPCK gene upstream hypersensitive site with LCR activity. Nucleic Acids Res. 1992 Jul 11;20(13):3427–3433. doi: 10.1093/nar/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Grayson D. R., Darnell J. E., Jr Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989 Apr;9(4):1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone V., Cortese R. Transcriptional regulation of liver-specific gene expression. Curr Opin Cell Biol. 1991 Dec;3(6):960–965. doi: 10.1016/0955-0674(91)90114-e. [DOI] [PubMed] [Google Scholar]
- Descombes P., Chojkier M., Lichtsteiner S., Falvey E., Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 1990 Sep;4(9):1541–1551. doi: 10.1101/gad.4.9.1541. [DOI] [PubMed] [Google Scholar]
- Descombes P., Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991 Nov 1;67(3):569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Fromm M., Taylor L. P., Walbot V. Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5824–5828. doi: 10.1073/pnas.82.17.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C. K., Devary O. V., Rosenfeld M. G. Multiple cell type-specific proteins differentially regulate target sequence recognition by the alpha retinoic acid receptor. Cell. 1990 Nov 16;63(4):729–738. doi: 10.1016/0092-8674(90)90139-6. [DOI] [PubMed] [Google Scholar]
- Godbout R., Ingram R. S., Tilghman S. M. Fine-structure mapping of the three mouse alpha-fetoprotein gene enhancers. Mol Cell Biol. 1988 Mar;8(3):1169–1178. doi: 10.1128/mcb.8.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez G. A., Montminy M. R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989 Nov 17;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange T., Roux J., Rigaud G., Pictet R. Cell-type specific activity of two glucocorticoid responsive units of rat tyrosine aminotransferase gene is associated with multiple binding sites for C/EBP and a novel liver-specific nuclear factor. Nucleic Acids Res. 1991 Jan 11;19(1):131–139. doi: 10.1093/nar/19.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer R. E., Krumlauf R., Camper S. A., Brinster R. L., Tilghman S. M. Diversity of alpha-fetoprotein gene expression in mice is generated by a combination of separate enhancer elements. Science. 1987 Jan 2;235(4784):53–58. doi: 10.1126/science.2432657. [DOI] [PubMed] [Google Scholar]
- Hashimoto S., Schmid W., Schütz G. Transcriptional activation of the rat liver tyrosine aminotransferase gene by cAMP. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6637–6641. doi: 10.1073/pnas.81.21.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T., Karin M. The regulation of transcription by phosphorylation. Cell. 1992 Aug 7;70(3):375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Poon D., Stone D., Granner D. K., Chalkley R. Interaction of a liver-specific factor with an enhancer 4.8 kilobases upstream of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1990 Jul;10(7):3770–3781. doi: 10.1128/mcb.10.7.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen H. M., Strähle U., Gloss B., Stewart F., Schmid W., Boshart M., Miksicek R., Schütz G. Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell. 1987 Apr 10;49(1):29–38. doi: 10.1016/0092-8674(87)90752-5. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Killary A. M., Fournier R. E. A genetic analysis of extinction: trans-dominant loci regulate expression of liver-specific traits in hepatoma hybrid cells. Cell. 1984 Sep;38(2):523–534. doi: 10.1016/0092-8674(84)90507-5. [DOI] [PubMed] [Google Scholar]
- Killary A. M., Lugo T. G., Fournier R. E. Isolation of thymidine kinase-deficient rat hepatoma cells by selection with bromodeoxyuridine, Hoechst 33258, and visible light. Biochem Genet. 1984 Apr;22(3-4):201–213. doi: 10.1007/BF00484224. [DOI] [PubMed] [Google Scholar]
- Kuo C. J., Conley P. B., Chen L., Sladek F. M., Darnell J. E., Jr, Crabtree G. R. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992 Jan 30;355(6359):457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- Lai E., Prezioso V. R., Smith E., Litvin O., Costa R. H., Darnell J. E., Jr HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990 Aug;4(8):1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- Lai E., Prezioso V. R., Tao W. F., Chen W. S., Darnell J. E., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991 Mar;5(3):416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., McKnight S. L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988 Jul;2(7):786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Lee W., Mitchell P., Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987 Jun 19;49(6):741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mueller C. R., Maire P., Schibler U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell. 1990 Apr 20;61(2):279–291. doi: 10.1016/0092-8674(90)90808-r. [DOI] [PubMed] [Google Scholar]
- Müller G., Ruppert S., Schmid E., Schütz G. Functional analysis of alternatively spliced tyrosinase gene transcripts. EMBO J. 1988 Sep;7(9):2723–2730. doi: 10.1002/j.1460-2075.1988.tb03126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols M., Weih F., Schmid W., DeVack C., Kowenz-Leutz E., Luckow B., Boshart M., Schütz G. Phosphorylation of CREB affects its binding to high and low affinity sites: implications for cAMP induced gene transcription. EMBO J. 1992 Sep;11(9):3337–3346. doi: 10.1002/j.1460-2075.1992.tb05412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch D., Boshart M., Schütz G. Extinction of tyrosine aminotransferase gene activity in somatic cell hybrids involves modification and loss of several essential transcriptional activators. Genes Dev. 1993 Feb;7(2):308–319. doi: 10.1101/gad.7.2.308. [DOI] [PubMed] [Google Scholar]
- Nitsch D., Stewart A. F., Boshart M., Mestril R., Weih F., Schütz G. Chromatin structures of the rat tyrosine aminotransferase gene relate to the function of its cis-acting elements. Mol Cell Biol. 1990 Jul;10(7):3334–3342. doi: 10.1128/mcb.10.7.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani L., Overdier D. G., Porcella A., Qian X., Lai E., Costa R. H. Hepatocyte nuclear factor 3 beta contains two transcriptional activation domains, one of which is novel and conserved with the Drosophila fork head protein. Mol Cell Biol. 1992 Sep;12(9):3723–3732. doi: 10.1128/mcb.12.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkert C. A., Ornitz D. M., Brinster R. L., Palmiter R. D. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1987 May;1(3):268–276. doi: 10.1101/gad.1.3.268. [DOI] [PubMed] [Google Scholar]
- Reik A., Schütz G., Stewart A. F. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J. 1991 Sep;10(9):2569–2576. doi: 10.1002/j.1460-2075.1991.tb07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud G., Roux J., Pictet R., Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991 Nov 29;67(5):977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- Ron D., Habener J. F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992 Mar;6(3):439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- SVOBODA J. Presence of chicken tumour virus in the sarcoma of the adult rat inoculated after birth with Rous sarcoma tissue. Nature. 1960 Jun 18;186:980–981. doi: 10.1038/186980b0. [DOI] [PubMed] [Google Scholar]
- Schmid E., Schmid W., Jantzen M., Mayer D., Jastorff B., Schütz G. Transcription activation of the tyrosine aminotransferase gene by glucocorticoids and cAMP in primary hepatocytes. Eur J Biochem. 1987 Jun 15;165(3):499–506. doi: 10.1111/j.1432-1033.1987.tb11467.x. [DOI] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Müller M. M., Schaffner W. Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989 Aug 11;17(15):6419–6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart U. K. Regulation of gene expression in rat hepatocytes and hepatoma cells by insulin: quantitation of messenger ribonucleic acid's coding for tyrosine aminotransferase, tryptophan oxygenase, and phosphoenolpyruvate carboxykinase. Endocrinology. 1986 Oct;119(4):1741–1749. doi: 10.1210/endo-119-4-1741. [DOI] [PubMed] [Google Scholar]
- Sladek F. M., Darnell J. E. Mechanisms of liver-specific gene expression. Curr Opin Genet Dev. 1992 Apr;2(2):256–259. doi: 10.1016/s0959-437x(05)80282-5. [DOI] [PubMed] [Google Scholar]
- Thompson E. B., Tomkins G. M., Curran J. F. Induction of tyrosine alpha-ketoglutarate transaminase by steroid hormones in a newly established tissue culture cell line. Proc Natl Acad Sci U S A. 1966 Jul;56(1):296–303. doi: 10.1073/pnas.56.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J. M., Schibler U. Tissue-specific expression of the gene encoding hepatocyte nuclear factor 1 may involve hepatocyte nuclear factor 4. Genes Dev. 1991 Dec;5(12A):2225–2234. doi: 10.1101/gad.5.12a.2225. [DOI] [PubMed] [Google Scholar]
- Treacy M. N., He X., Rosenfeld M. G. I-POU: a POU-domain protein that inhibits neuron-specific gene activation. Nature. 1991 Apr 18;350(6319):577–584. doi: 10.1038/350577a0. [DOI] [PubMed] [Google Scholar]
- Tsutsumi K., Ito K., Ishikawa K. Developmental appearance of transcription factors that regulate liver-specific expression of the aldolase B gene. Mol Cell Biol. 1989 Nov;9(11):4923–4931. doi: 10.1128/mcb.9.11.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Jäckle H. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell. 1990 Nov 2;63(3):455–456. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- Weih F., Nitsch D., Reik A., Schütz G., Becker P. B. Analysis of CpG methylation and genomic footprinting at the tyrosine aminotransferase gene: DNA methylation alone is not sufficient to prevent protein binding in vivo. EMBO J. 1991 Sep;10(9):2559–2567. doi: 10.1002/j.1460-2075.1991.tb07796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih F., Stewart A. F., Boshart M., Nitsch D., Schütz G. In vivo monitoring of a cAMP-stimulated DNA-binding activity. Genes Dev. 1990 Aug;4(8):1437–1449. doi: 10.1101/gad.4.8.1437. [DOI] [PubMed] [Google Scholar]
- Weih F., Stewart A. F., Schütz G. A novel and rapid method to generate single stranded DNA probes for genomic footprinting. Nucleic Acids Res. 1988 Feb 25;16(4):1628–1628. doi: 10.1093/nar/16.4.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildeman A. G., Sassone-Corsi P., Grundström T., Zenke M., Chambon P. Stimulation of in vitro transcription from the SV40 early promoter by the enhancer involves a specific trans-acting factor. EMBO J. 1984 Dec 20;3(13):3129–3133. doi: 10.1002/j.1460-2075.1984.tb02269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Liu J. K., DiPersio C. M. Site-directed mutagenesis reveals a liver transcription factor essential for the albumin transcriptional enhancer. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5469–5473. doi: 10.1073/pnas.87.14.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff E. B. Transcription factors: a new family gathers at the cAMP response site. Trends Genet. 1990 Mar;6(3):69–72. doi: 10.1016/0168-9525(90)90081-g. [DOI] [PubMed] [Google Scholar]
- de Wet J. R., Wood K. V., DeLuca M., Helinski D. R., Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987 Feb;7(2):725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]