Abstract
The UK's largest registry of adult twins, or TwinsUK Registry, started in 1992 and encompasses about 12 000 volunteer twins from all over the United Kingdom. More than 70% of the registered twins have filled at least one detailed health questionnaire and about half of them undergone a baseline comprehensive assessment and two follow-up clinical evaluations. The most recent follow-up visit, known as Healthy Ageing Twin Study (HATS), involved 3125 female twins aged >40 years with at least one previous clinical assessment to enable inspection of longitudinal changes in ageing traits and their genetic and environmental components. The study benefits from several state-of-the-art OMICs studies including genome-wide association, next-generation genome and transcriptome sequencing, and epigenetic and metabolomic profiles. This makes our cohort as one of the most deeply phenotyped and genotyped in the world. Several collaborative projects in the field of epidemiology of complex disorders are ongoing in our cohort and interested researchers are encouraged to get in contact for future collaborations.
How did the study come about?
The UK Adult Twin Registry (or TwinsUK Registry) is a cohort of volunteer adult twins from all over the United Kingdom. The Registry was started in 1992 with the primary aim of assessment of heritability of osteoarthritis and osteoporosis in women. The success of early studies led to rapid evolution of the registry and it now incorporates about 12 000 twins, both male and female aged 18–103 years, studied for a whole range of clinical and behavioural traits (Supplementary Appendix A1 available as Supplementary Data at IJE online). The main objective of the original study was to estimate heritabilities for ‘common’ diseases and traits and to discover associated genes. The details about the aims and design of the original twin registry, facilities and procedures for data collection, clinical and biological assessments, main findings, collaborations and working directions have been published in previous papers1,2 and described on the study website (www.twin-research.ac.uk).
Recently, the view of ageing as an extremely complex multifactorial process involving multiple organs and multiple molecular pathways has replaced the earlier search for a distinct unique cause such as a single gene or the decline of a key body system.3 Prospective observational twin studies provide an ideal setting for assessment of the genetic and environmental contributions to different ageing mechanisms. Since publication of the last cohort profile in 2006, the aim and focus of our study has been changed to ‘healthy ageing’. Age restriction and clinical data collection procedures in the Healthy Ageing Twin Study (HATS; new clinical visit of our cohort) have been modified to measure age-related longitudinal deteriorations in six different organ systems: cardiovascular, muscle, bone, respiration, vision and skin. ‘Healthy ageing’ is determined by assessment of the over-time changes in quantitative traits (rather than focusing on differences between aged population with/without various disorders) among HATS participants, who are female twins aged ≥40 years. Alongside detailed clinical measures, the cohort now benefits from several state-of-the-art technologies (the ‘OMICs’ studies) including data from genome-wide association, next-generation genome and transcriptome sequencing, and epigenetic and metabolomic profiles. This paper describes these advances and available opportunities in our cohort.
How is it funded?
Historically, much of the funding for the study has come from the Wellcome Trust project and programme grants and some core funding, with additional support from the UK Medical Research Council (MRC), British Heart Foundation, Chronic Disease Research Foundation, National Institute for Health Research (NIHR) Biomedical Research Centre, Pfizer and the European Union framework programme 7 (EU-FP7). In particular, the HATS project was funded by the Wellcome Trust and EU-FP7 and funding applications for further follow-up of the HATS cohort is ongoing. The Department of Twin Research and Genetic Epidemiology is based at St Thomas’ Hospital, Division of Genetics and Molecular Medicine at King’s College London. The team of over 50 staff includes genetic epidemiologists and statisticians, research nurses, laboratory and information technology teams, and administration team maintaining the cohort database and recruitment of studies. All studies have been approved by the local Research Ethics Committee.
What does it cover?
Our study examines the correlation between longitudinal age-related deteriorations in different organ systems, the heritability of biological ageing in different organs, the effects of genetic and environmental factors on biological ageing, and the value of putative biomarkers of ageing. The programme of work to achieve these goals can be summarized in five categories.
Longitudinal data collection: many of the twins in the cohort have at least two clinical visits with biological samples taken (blood, urine and DNA). Several new clinical measures were introduced in the follow-up visits to make HATS into a study of healthy ageing. These include grip strength, pain perception tests, ocular photos, hearing loss tests and naevi counts. Moreover, the key quantitative phenotypes on the six organ systems have been repeated in all clinical visits with similar protocols. Demographic, lifestyle and behavioural data are available from several postal questionnaires. These provide the means to assess the longitudinal changes over years of follow-up.
Heritability of ageing: TwinsUK cohort, independently or in collaboration with other European twin registries (GenomEUtwin study),4,5 has estimated heritability of a large number of ageing-related clinical traits (please see http://www.twin-research.ac.uk/publications.html). Additionally, and as a separate phenotype, heritabilities for rate of changes in various quantitative traits attributed to ageing (e.g. bone loss over time as opposed to bone mineral density) can be estimated.
Genetic association studies: candidate gene analysis and genome-wide association studies (GWAS) have been conducted for several ageing traits (including longevity,6 telomere shortening or cellular senescence,7 oxidative stress8 and chronic inflammation9). There are ongoing collaborative projects for other genetic/epigenetic studies on the cohort (see below).
Environmental risk factors: lifestyle and demographic factors that can impact healthy ageing—such as exercise levels, smoking, alcohol intake, diet and nutrition, hormone replacement therapy, birth weight, sun exposure, occupation, socio-economic status, marital status, paternal/maternal age, and number of children —have been evaluated in the cohort.
Putative markers of biological ageing: some of the biomarkers of ageing, such as white cell telomere length, serum dehydroepiandrosterone sulphate, serum 25-hydroxy vitamin D, sensitive C-reactive protein and serum creatinine, have already been measured for our participants. Other clinical measures such as nuclear cataract, retinal vessel calibre and naevi counts have been added to the HATS clinical visit. Furthermore, the EuroBATS project (Biomarkers of Ageing using whole Transcriptome Sequencing) is a 3-year European (EU-FP7) project started in January 2011 with four other research partners. Incorporating novel RNA sequencing, telomere measurement and bioinformatics techniques, the hope is that EuroBATS will lead to discovery of novel biomarkers of ageing (please visit: http://ec.europa.eu/research/health/medical-research/human-development-and-ageing/projects/eurobats_en.html).
Who is in the sample, how often have they been followed up and what is attrition like?
The TwinsUK registry is a national twin volunteer population. Twins were recruited through a series of media campaigns asking for volunteers willing to participate in research investigating common diseases. Initially, only middle-aged women were recruited to the registry.1 From 1995 onwards, men and women >18 years of age were also invited to participate. As a result, 83% of the registry is female. The registry now contains 51% monozygotic (MZ) and 49% dizygotic (DZ) twins aged 18–103 years (Appendix A1 available as Supplementary Data at IJE online).
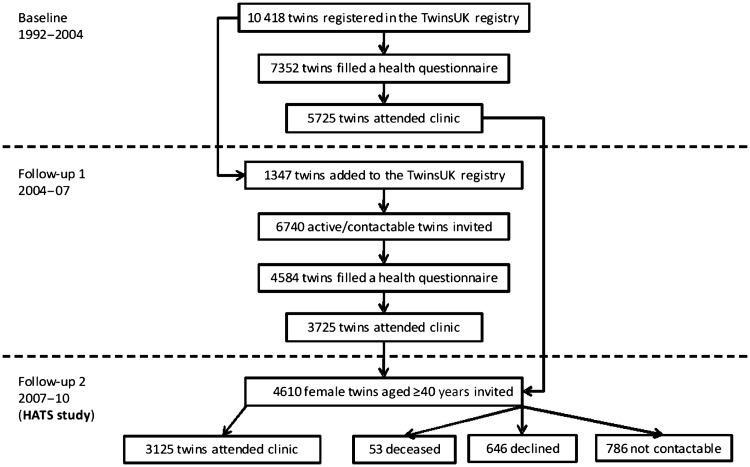
The flow of the participants through the study is depicted in Figure 1. Between 1992 and 2004, twins were invited for a comprehensive ‘baseline’ visit and several project-led studies, which evaluated different aspects of the bone and joint health in middle-aged and older women. Active twins in the registry were contacted in a yearly basis through different postal questionnaires or short follow-up visits. More than 7000 twins responded to annual questionnaires and 5725 of them (out of about 10 000 registered twins) attended a comprehensive visit. About half of them also returned for other visits related to different projects (about 700 twins had four or more visits). Attendants of the comprehensive visit have been compared with a singleton population-based cohort representative of the UK population (Chingford study).10 Apart from lower weight in MZ twins, all other age-matched characteristics of volunteer twins were found not to differ from singleton women of the same age.10
Figure 1.
Recruitment of TwinsUK and HATS study participants
Between April 2004 and May 2007, all the 6740 active twins on the registry (not deceased, withdrawn or uncontactable) were invited for a 1-day clinical visit (follow-up visit 1 in Figure 1). The study aimed to estimate heritabilities and discover genes for common diseases and complex traits.2 A sum of 3725 twins attended this visit. Additionally, 1299 twins posted their blood DNA samples (collected via their General Practitioners). The response rate was 55% for the clinical visit and 74% for the DNA sample collection. The age range of participants was 18–82 years (mean 52.5 ± 13 years) and 3299 of the clinic attendants (89%) were female. The data collected in this visit has led to over 80 collaborative and data sharing projects and has contributed to about 170 peer-reviewed publications in the past 3 years (www.twin-research.ac.uk/publications.html).
The second follow-up visit, also known as the HATS visit, started in August 2007 (Figure 1). Only women aged ≥40 years with at least one previous clinical visit were invited (n = 4610) and 3125 women (mean age 59.6 ± 9 years) attended the clinic (response rate = 68%). From 1485 twins who did not attend, 53 were deceased, 646 declined to participate and the remaining 786 could not be contacted. 1498 (48%) of participants were MZ twins as confirmed by genotyping (e.g. GWAS) or ‘peas in a pod’ questionnaire.11 Six hundred of the participants in this visit had four or more previous clinical visits. Follow-up time between first and last visits ranged between 6.1 and 17.4 years (mean 11.2 ± 2 years).
General characteristics of the participants in different stages of the study are summarized in Table 1. Compared with the baseline and first follow-up visits, participants in the HATS visit appeared to have higher socio-economic status, lower self-rated health status and be more health aware given their level of alcohol intake and smoking. Table 2 compares the characteristics of respondents and non-respondents to the HATS invitation based on data from their last clinical visit. These data show that the non-respondents to this visit were generally younger and of lower socio-economic status. No significant clinical differences were observed between attendants and non-attendants of this visit (Table 2), suggesting that the participants in this visit could be considered as representative of the original population in the study.
Table 1.
Characteristics of participants from the TwinsUK registry who participated in clinical assessments within different stages of the cohort
| Baseline visits (1992–2004) | Follow-up 1 (2004–07) | Follow-up 2 (2007–10) | |
|---|---|---|---|
| N | 5725 | 3725 | 3125 |
| Age (years) | 46.8 ± 12.6 | 52.5 ± 13.4 | 59.6 ± 9.3 |
| Sex (male/female) | 412/5313 | 401/3324 | 0/3125 |
| Zygosity (DZ/MZ/UZ) | 3662/2027/36 | 1587/2137/1 | 1627/1498/0 |
| Ever married (%) | 81.1 | 88.5 | 90.1 |
| Age leaving full-time education (years) | 17.3 ± 3.6 | 17.7 ± 3.6 | 17.4 ± 3.6 |
| Low socio-economic statusa (%) | 21.4 | 19.8 | 17.5 |
| Self-rated health status (fair/poor) (%) | 7.2 | 8.0 | 12.3 |
| Age at menarche (years) | 13.0 ± 1.6 | 12.9 ± 1.5 | 12.9 ± 1.5 |
| Current smoker (%) | 18.4 | 15.1 | 9.3 |
| Past smoker (%) | 28.4 | 27.9 | 37.3 |
| Alcohol intake (units/week) | 4 (0–9) | 4 (1–10) | 2 (0–5) |
UZ: unknown zygosity.
All age values are mean ± standard deviation. Values for alcohol intake are median (inter-quartile range) due to non-normality.
aThis is estimated for all twins in the registry based on their registered address and the English Indices of multiple deprivation (developed by the UK Department for Communities and Local Government: http://www.communities.gov.uk/). The lowest 2 quintiles of the general population of UK are considered as low socio-economic status here.
Table 2.
Comparison of the characteristics of twins who attended the second follow-up clinical visit with eligible twins who did not attend
| Follow-up 2 Respondents | Follow-up 2 Non-Respondents | P-value | |
|---|---|---|---|
| N | 3125 | 1485 | |
| Age (years) | 59.6 ± 9.3 | 57.4 ± 11.7 | <0.001 |
| Zygosity (DZ/MZ) | 1627/1498 | 874/604 | <0.001 |
| Age leaving full-time education (years) | 17.4 ± 3.6 | 17.3 ± 3.8 | 0.4 |
| Low socio-economic status (%) | 17.5 | 26.4 | <0.001 |
| Self-rated health status (fair/poor) (%) | 7.4 | 7.2 | 0.8 |
| Age at menarche (years) | 12.9 ± 1.6 | 13.1 ± 1.6 | 0.1 |
| Ever smoked (%) | 42.2 | 45.4 | 0.2 |
| Alcohol drinker (%) | 7.5 | 6.8 | 0.5 |
| Body mass index (kg/m2) | 26.4 ± 4.8 | 26.3 ± 5.2 | 0.6 |
| Grip strength (kg) | 28.9 ± 6.8 | 28.9 ± 7.1 | 0.9 |
| Diastolic blood pressure (mmHg) | 76.1 ± 10.2 | 76.0 ± 10.7 | 0.9 |
| Forced Expiratory Volume in 1 s (l) | 2.64 ± 0.55 | 2.68 ± 0.59 | 0.07 |
| Total hip bone mineral density (g/cm2) | 0.92 ± 0.12 | 0.93 ± 0.13 | 0.1 |
| Lumbar spine bone mineral density (g/cm2) | 0.99 ± 0.15 | 1.00 ± 0.15 | 0.1 |
Values are mean ± standard deviation.
Values are based on the last previous visit of participants.
What has been measured?
The structure of the baseline comprehensive and two follow-up visits were quite similar. Twins firstly filled self-completed questionnaires, asking about a range of lifestyle and behavioural traits, quality of life (Short Form-36 questionnaire), and recent clinical outcomes. During the clinical visit, fasting venous blood and urine were collected and participants went through a series of clinical measurements (Table 3). Twins are all now registered using their National Health Service (NHS) numbers with the Office for National Statistics of England for future follow-up regarding their cancer and mortality status.
Table 3.
Data available from different visits
| Baseline | Follow-up 1 | Follow-up 2 | |
|---|---|---|---|
| 1992–2004 | 2004–07 | 2007–10 | |
| Questionnaires | |||
| Demographics and lifestyle | ✓ | ✓ | ✓ |
| Quality of life (SF-36) | ✓ | ✓ | ✓ |
| Physical activity | ✓ | ✓ | ✓ |
| Medication history | ✓ | ✓ | ✓ |
| Gynaecological history (for women) | ✓ | ✓ | ✓ |
| Food frequency questionnaire | ✓ | ✓ | ✓ |
| Cognitive function (PRMQ and NART) | ✓ | – | – |
| Social deprivation class | ✓ | ✓ | ✓ |
| Biological samples | |||
| Fasting blood and urine samples | ✓ | ✓ | ✓ |
| DNA extraction and purification | ✓ | ✓ | ✓ |
| Lymphoid cell line culture | ✓ | ✓ | ✓ |
| Liver function tests | ✓ | ✓ | ✓ |
| Lipid and metabolic profiles | ✓ | ✓ | ✓ |
| Clinical assessments | |||
| Anthropometrics | ✓ | ✓ | ✓ |
| Blood pressure | ✓ | ✓ | ✓ |
| Electrocardiography | ✓ | ✓ | ✓ |
| Lung function tests | ✓ | ✓ | ✓ |
| Hip and spine DXA | ✓ | ✓ | ✓ |
| Whole-body DXA (fat and lean mass) | ✓ | ✓ | ✓ |
| Heel quantitative ultrasound | ✓ | – | – |
| Leg extension power test | ✓ | – | – |
| Grip strength | – | ✓ | ✓ |
| Visual acuity and refractive errors | ✓ | ✓ | ✓ |
| Intraocular pressure and optic disc photo | – | – | ✓ |
| Cataract (lens opacity) | ✓ | – | ✓ |
| Macular degeneration and fundus photo | ✓ | – | ✓ |
| Facial features (2- and 3-dimensional images) | ✓ | ✓ | ✓ |
| Pain perception tests | – | ✓ | ✓ |
| Distorted tune test | – | ✓ | ✓ |
| Hearing loss and audiometry | – | – | ✓ |
| Naevi counts | – | ✓ | ✓ |
| Genetic assessments | |||
| GWAS | ✓ | ✓ | ✓ |
| Whole-genome next-generation sequencing | – | ✓ | ✓ |
| Epigenetic analysis (MeDIP sequencing) | – | ✓ | ✓ |
| Gene expression (RNA sequencing) | – | – | ✓ |
| Specialized tests (not performed on all participants) | |||
| Echocardiography | ✓ | – | – |
| Sphygmocardiography (Arterial distensibility) | ✓ | – | – |
| Carotid ultrasound | – | ✓ | – |
| Hands, knees, spine and pelvis X-rays | ✓ | ✓ | – |
| Spinal MRI scans | ✓ | ✓ | – |
| Abdominal CT scans | ✓ | ✓ | – |
| Glucose tolerance test | ✓ | – | – |
| ActiHeart monitoring study | – | – | ✓ |
| Retinal macular pigment measures | – | ✓ | – |
| Skin, fat and muscle biopsy | – | – | ✓ |
| Telomere length analysis | – | ✓ | – |
| Metabolomic analysis | – | ✓ | – |
SF-36: Short-Form health survey; PRMQ: prospective and retrospective memory questionnaire; NART: National Adult Reading Test; CT: computed tomography; MeDIP: Methylated DNA ImmunoPrecipitation; MRI: magnetic resonance imaging.
Clinical data
As summarized in Table 3, various clinical measures such as systolic and diastolic blood pressures, electrocardiographs, lung function tests, visual and hearing tests, biochemical assessments (including lipid profiles, diabetes tests and serum metabolites) and medication history is available from different visits. Musculoskeletal traits are the major strengths of our study particularly with longitudinal dual-energy X-ray absorptiometry (DXA) assessments (in total, about 14 000 DXA measures on 7400 twins with 3800 having two or more measurements). Incident clinical endpoints (e.g. cardiovascular events, stroke, fractures, osteoarthritis and different cancers) have been assessed over the course of study.
GWAS
In total, 5710 twins have undergone a genome-wide scan of either 317 000 single nucleotide polymorphism (SNP) markers (Illumina HumanHap300 Bead Chip) or 610 000 SNPs (Illumina HumanHap610 Quad Chip). From these twins, 2840 participated in the first follow-up and 2545 in the second follow-up visits. The data have been fully imputed using IMPUTE version 2 software,12 quality checked, and used in many international consortia for different phenotypes (Table 4). There are several ongoing consortia for meta-analysis of other traits such as lean mass and reproductive health (CHARGE and GEFOS consortia), vitamin D and related traits (SUNLIGHT consortium), molecular phenotypes (MolPAGE consortium), external visible traits (VisiGEN consortium), liver function and other traits (ENGAGE consortium).
Table 4.
Names of phenotypes and consortia that TwinsUK cohort has contributed to date
| Phenotype | Consortium |
|---|---|
| Adiponectin concentrations | GIANT27 |
| Age at menarche | GIANT28 |
| Blood coagulation | CHARGE29 and EuroCLOT30 |
| Blood lipids | ENGAGE31,32 |
| Blood pressure | Global bpGEN-CHARGE33 |
| Body mass index | PROCARDIS34 and GIANT35 |
| Cardiac repolarization | WTCCC-QTGEN-QTSCD36 |
| Primary glaucoma | Glaucoma37 |
| Haemoglobin concentrations | CHARGE-HaemGEN38 |
| Height | GIANT39 |
| Melanoma | GenomMEL40 |
| Myopia and refractive errors | Myopia41 |
| Obesity and fat distribution | PROCARDIS-GIANT42 |
| Osteoarthritis | arcOGEN43 and TreatOA44 |
| Osteoporosis | GEFOS45 |
| Respiratory function | SpiroMeta-CHARGE46 |
| Smoking behaviour | ENGAGE47 |
| Type 2 diabetes | DIAGRAM-GIANT-Global BPgen48 |
| Uric acid concentrations | EUROSPAN-ENGAGE- PROCARDIS49 |
Next-generation sequencing
One of the main ongoing collaborations of our department is with the Wellcome Trust Sanger Institute for the UK10K study. Using the state-of-the-art next-generation sequencing methods,13 UK10K aims to find rare genetic variants associated with health and disease. The study involves whole-genome sequencing of 4000 healthy people with well-documented physical characteristics. Two thousands twins from our cohort have been genotyped (6× depth) in this study. The sequence and clinical data will be publically available soon (www.uk10k.org).
Epigenetics
The EpiTwin study, in collaboration with the Beijing Genomics Institute, is the largest-ever epigenetic project. This study aims to capture the subtle epigenetic signatures that mark the differences between 5000 twins. This will be achieved by Methylated DNA ImmunoPrecipitation (MeDIP) sequencing, examining ‘DNA methylation’ patterns of 20 million sites across genome of each twin and comparing them with the patterns in the co-twin. Finding the crucial differences between twins will reveal the key genes that are being turned on and off, and so too the causes of diseases. Obesity, diabetes, allergies, heart disease, osteoporosis and longevity are the initial targets of the study (http://www.epitwin.eu).
Gene expression
The MuTHER (Multiple Tissue Human Expression Resource) project is a Wellcome Trust funded study designed to understand the mechanisms involved in common trait susceptibility via gene expression across multiple tissues. The project aims to develop a resource for assessment of the relationships between genome sequence variation, methylation status, mRNA expression and disease phenotypes.14 Lymphocytes and fat, muscle and skin biopsies have been obtained from 855 twins from the HATS visit. The genome-wide expression profiling of all samples is near complete and GWAS data and DNA methylation data will also be available shortly. Interested researchers can contact us via: muther@kcl.ac.uk.
Specialized tests
Several specialized scans and assays (e.g. spinal Magnetic Resonance Imaging scans and echocardiography) are available in different subsets of twins (Table 3). Two important ones are ‘telomere length’ and ‘metabolomic’ data. In 3256 participants with GWAS data, telomere length has been measured using the Southern blot method on DNA extracted from peripheral leukocytes. These data have contributed to detection of several genes implicated to affect biological age15,16 and the collaboration is ongoing. For 1270 twins with GWAS data, fasting serum concentrations of 163 metabolites, covering a biologically relevant panel of amino acids, sugars, acylcarnitines and phospholipids, have been measured using electrospray ionization tandem mass spectrometry (Biocrates AbsoluteIDQ targeted metabolomics technology). Variation in all possible metabolite concentration ratios (163 × 162 = 26 406 traits), can influence different phenotypes (e.g. osteoarthritis)17 or be genetically determined.18 The same sample has also been assayed for non-targeted metabolomics using ultrahigh performance liquid-phase chromatography and gas chromatography separation coupled with tandem mass spectrometry.
What has it found?
TwinsUK contributions to science have evolved with the advances, in both methodology and technology, over the past two decades. In the early stages, linkage studies in twins found several replicated loci for different ageing traits such as bone mineral density19 and telomere shortening.20 Heritability of many complex traits, from obvious clinical endpoints (e.g. blood pressure21 and cataract)22 to non-clinical personality-related traits (e.g. sexual functioning23 or entrepreneurship),24 have been estimated in this cohort. Nearly all traits measured to date show a heritable component. Interestingly, many diseases previously believed to be dull ‘wear and tear’ diseases (such as degenerative disc disease or osteoarthritis) have a major genetic component. TwinsUK has particularly contributed to the clinical and genetic epidemiology of osteoporosis and osteoarthritis. We have collaborated with several GWAS consortia that have found polymorphisms associated with several important clinical conditions (Table 4). Recently, we showed that ∼50–60% of biological variations in metabolite concentrations in plasma and urine are related to familial and environmental factors.25 GWAS of approximately 37 000 metabolomic traits also identified several genes involved in metabolic individuality in humans.26 Unusually high effect sizes for some of these traits promise significant advances in future functional studies. Work on several projects like UK10K, EpiTwin, MuTHER and EuroBATS is ongoing by several groups in our department. For a comprehensive list of publications, interested researchers can refer to http://www.twin-research.ac.uk/publications.html.
What are the main strengths?
The twin-based nature of this cohort, frequent data collection and detailed characterization of participants enables researchers to dissect the contribution of genetic and environmental risk factors for complex diseases and domains of healthy ageing. All measurements have been made by a trained research team working with strict study protocols for about two decades. Blood, urine and DNA samples from all visits are stored for future measurements. Follow-up for cause-specific mortality and cancer is ongoing for all participants. We are moving towards new methods of active engagement with twins (e.g. email lists and social networking websites). The current plan is to start the third follow-up visit in 2012 and interested researchers are encouraged to contact us for inclusion of their phenotypes in this visit.
What are the main weaknesses?
The UK Adult Twin Registry is based on volunteer subjects, which might be considered as non-representative of the total population. Twins may also differ from singleton populations in several in utero or early-life exposures and this is a potential limitation of all twin studies. We have compared our twins with a population-based study sample and found no important clinical differences.10 The status of participants in the last clinical visit (Table 2) also shows no sign of attrition bias. Given the variable structure of the baseline visits (before 2004), and the fact that TwinsUK study was not founded as a usual longitudinal cohort, there are missing values for some of the study measures for different sub-samples. Currently, a specialized group inside our department is working on cleaning and harmonizing data. At this time, our study population might be considered somewhat young for an ageing study with mean age of 61 years and only ∼10% aged >75 years. We prefer to describe this as an ‘early ageing’ cohort. Hence, most of our health outcomes are quantitative traits rather than discrete endpoints (such as end-stage diseases). The advantage is, however, that we can study them prospectively and useful data will continue to emerge from future evaluations.
Can I get hold of the data? Where can I find out more?
Invaluable data have been gathered within the TwinsUK and HATS study that may provide answers to hundreds of research questions about the healthy ageing process. These cannot all be analysed by our research group alone and we believe this is the best time for new collaborators to join our study. One of our missions is to collaborate with other groups and promote the wide use of our data. There is an embedded search engine in our website (www.twin-research.ac.uk/phenotypes.html) that researchers can enter their phenotype of interest and find the relevant assessments performed in different stages of our study. We invite colleagues to find out if TwinsUK data can help them answer their research questions. Contact information can be found at the study website: www.twin-research.ac.uk.
Supplementary Data
Supplementary Data are available at IJE online.
Acknowledgements
The authors acknowledge financial support from the Wellcome Trust, the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London/Arthritis Research Campaign/EC FP6 Programme Grant-512066 (LSHG-CT-2004); MOLPAGE/EC Framework 7 programme grant 200800; Treat OA/EC Framework 7 Health-2007-A; ENGAGE/EC Framework 7 Health-2007-2.4.5-4; GEFOS/EC FP6 MRTN-CT-2006-034021; MyEuropia Research Training Network/Chronic Disease Research Foundation (CDRF)/Pfizer Pharmaceuticals/National Health and Medical Research Council (NHMRC)/National Institute of Aging (NIA)/Guide Dogs for the blind Association(GDBA)/Biotechnology and biological Sciences Research Council (BBSRC). The authors acknowledge the funding and support of the National Eye Institute via an NIH/CIDR genotyping project (R01EY018246-01-1 PI: Terri Young). We also would like to thank all the twins who participated and supported this cohort and staff in the Department of Twin Research and Genetic Epidemiology, St Thomas’ Hospital, King’s College London.
Conflict of interest: None declared.
KEY MESSAGES.
Twin studies with longitudinal data provide an ideal setting for assessment of the genetic and environmental contributions to complex multifactorial processes like ageing.
The vision of the TwinsUK registry has been recently changed to focus on healthy ageing.
Clinical and genetic epidemiology of various quantitative traits associated with ageing process has been assessed longitudinally in our cohort.
Several state-of-the-art OMIC technologies have also been employed in our cohort.
Interested researchers are welcome to collaborate on analysis of available data and planning for the future stages of the study.
References
- 1.Spector TD, MacGregor AJ. The St. Thomas’ UK Adult Twin Registry. Twin Res. 2002;5:440–43. doi: 10.1375/136905202320906246. [DOI] [PubMed] [Google Scholar]
- 2.Spector TD, Williams FM. The UK Adult Twin Registry (TwinsUK) Twin Res Hum Genet. 2006;9:899–906. doi: 10.1375/183242706779462462. [DOI] [PubMed] [Google Scholar]
- 3.Vina J, Borras C, Miquel J. Theories of ageing. IUBMB Life. 2007;59:249–54. doi: 10.1080/15216540601178067. [DOI] [PubMed] [Google Scholar]
- 4.Perola M, Sammalisto S, Hiekkalinna T, et al. Combined genome scans for body stature in 6,602 European twins: evidence for common Caucasian loci. PLoS Genet. 2007;3:e97. doi: 10.1371/journal.pgen.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kettunen J, Perola M, Martin NG, et al. Multicenter dizygotic twin cohort study confirms two linkage susceptibility loci for body mass index at 3q29 and 7q36 and identifies three further potential novel loci. Int J Obes. 2009;33:1235–42. doi: 10.1038/ijo.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhai G, Teumer A, Stolk L, et al. Eight common genetic variants associated with Serum DHEAS levels suggest a key role in ageing mechanisms. PLoS Genet. 2011;7:e1002025. doi: 10.1371/journal.pgen.1002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangino M, Richards JB, Soranzo N, et al. A genome-wide association study identifies a novel locus on chromosome 18q12.2 influencing white cell telomere length. J Med Genet. 2009;46:451–54. doi: 10.1136/jmg.2008.064956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrew T, Aviv A, Falchi M, et al. Mapping genetic loci that determine leukocyte telomere length in a large sample of unselected female sibling pairs. Am J Hum Genet. 2006;78:480–86. doi: 10.1086/500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehghan A, Dupuis J, Barbalic M, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–38. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res. 2001;4:464–77. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- 11.Martin NG, Martin PG. The inheritance of scholastric abilities in a sample of twins. I. Ascertainments of the sample and diagnosis of zygosity. Ann Hum Genet. 1975;39:213–18. doi: 10.1111/j.1469-1809.1975.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 12.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 14.Nica AC, Parts L, Glass D, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Codd V, Mangino M, van der Harst P, et al. Common variants near TERC are associated with mean telomere length. Nat Genet. 2010;42:197–99. doi: 10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangino M, Richards JB, Soranzo N, et al. A genome-wide association study identifies a novel locus on chromosome 18q12.2 influencing white cell telomere length. J Med Genet. 2009;46:451–54. doi: 10.1136/jmg.2008.064956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhai G, Wang-Sattler R, Hart DJ, et al. Serum branched-chain amino acid to histidine ratio: a novel metabolomic biomarker of knee osteoarthritis. Ann Rheum Dis. 2010;69:1227–31. doi: 10.1136/ard.2009.120857. [DOI] [PubMed] [Google Scholar]
- 18.Illig T, Gieger C, Zhai G, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–41. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keen RW, Snieder H, Molloy H, et al. Evidence of association and linkage disequilibrium between a novel polymorphism in the transforming growth factor beta 1 gene and hip bone mineral density: a study of female twins. Rheumatology. 2001;40:48–54. doi: 10.1093/rheumatology/40.1.48. [DOI] [PubMed] [Google Scholar]
- 20.Andrew T, Aviv A, Falchi M, et al. Mapping genetic loci that determine leukocyte telomere length in a large sample of unselected female sibling pairs. Am J Hum Genet. 2006;78:480–86. doi: 10.1086/500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snieder H, Hayward CS, Perks U, Kelly RP, Kelly PJ, Spector TD. Heritability of central systolic pressure augmentation: a twin study. Hypertension. 2000;35:574–79. doi: 10.1161/01.hyp.35.2.574. [DOI] [PubMed] [Google Scholar]
- 22.Hammond CJ, Snieder H, Spector TD, Gilbert CE. Genetic and environmental factors in age-related nuclear cataracts in monozygotic and dizygotic twins. N Engl J Med. 2000;342:1786–90. doi: 10.1056/NEJM200006153422404. [DOI] [PubMed] [Google Scholar]
- 23.Dunn KM, Cherkas LF, Spector TD. Genetic influences on variation in female orgasmic function: a twin study. Biol Lett. 2005;1:260–63. doi: 10.1098/rsbl.2005.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolaou N, Shane S, Cherkas L, Hunkin J, Spector TD. Is the tendency to engage in entrepreneurship genetic? Manage Sci. 2008;54:167–79. [Google Scholar]
- 25.Nicholson G, Rantalainen M, Maher AD, et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Mol Syst Biol. 2011;7:525. doi: 10.1038/msb.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suhre K, Shin SY, Petersen AK, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards JB, Waterworth D, O'Rahilly S, et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 2009;5:e1000768. doi: 10.1371/journal.pgen.1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elks CE, Perry JR, Sulem P, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–85. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith NL, Chen MH, Dehghan A, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010;121:1382–92. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams FM, Carter AM, Kato B, et al. Identification of quantitative trait loci for fibrin clot phenotypes: the EuroCLOT study. Arterioscler Thromb Vasc Biol. 2009;29:600–605. doi: 10.1161/ATVBAHA.108.178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aulchenko YS, Ripatti S, Lindqvist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nolte IM, Wallace C, Newhouse SJ, et al. Common genetic variation near the phospholamban gene is associated with cardiac repolarisation: meta-analysis of three genome-wide association studies. PLoS One. 2009;4:e6138. doi: 10.1371/journal.pone.0006138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42:906–909. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganesh SK, Zakai NA, van Rooij FJ, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41:1191–98. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lango AH, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–38. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duffy DL, Iles MM, Glass D, et al. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am J Hum Genet. 2010;87:6–16. doi: 10.1016/j.ajhg.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hysi PG, Young TL, Mackey DA, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42:902–905. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindgren CM, Heid IM, Randall JC, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panoutsopoulou K, Southam L, Elliott KS, et al. Insights into the genetic architecture of osteoarthritis from stage 1 of the arcOGEN study. Ann Rheum Dis. 2011;70:864–67. doi: 10.1136/ard.2010.141473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhai G, van Meurs JB, Livshits G, et al. A genome-wide association study suggests that a locus within the ataxin 2 binding protein 1 gene is associated with hand osteoarthritis: the Treat-OA consortium. J Med Genet. 2009;46:614–16. doi: 10.1136/jmg.2009.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richards JB, Kavvoura FK, Rivadeneira F, et al. Collaborative meta-analysis: associations of 150 candidate genes with osteoporosis and osteoporotic fracture. Ann Intern Med. 2009;151:528–37. doi: 10.7326/0003-4819-151-8-200910200-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Repapi E, Sayers I, Wain LV, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolz M, Johnson T, Sanna S, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]