Abstract
Background Methylation of deoxyribonucleic acid (DNA) is an epigenetic regulator of gene expression that changes with age, but its contribution to aging-related disorders, including high blood pressure (BP), is still largely unknown. We examined the relation of BP to the methylation of retrotransposon sequences of DNA and of selected candidate genes.
Methods This investigation included 789 elderly participants in the Normative Aging Study, ranging in age from 55 to 100 years, who had longitudinal measurements of DNA methylation. In these subjects’ DNA we measured the proportion of methylated sites in retrotransposable sequences and in pro-inflammatory genes, expressed as the percent of 5-methylated cytosines (%5mC) among all cytosines. From one to four methylation measurements were made for each subject between 1999 and 2009. We fit mixed-effects models, using repeated measures of BP as the outcome and DNA methylation as the explanatory variable, adjusting for confounding variables. We also fit a Bayesian mixed-effects structural equation model to account for heterogeneity in the effects of methylation sites within each gene.
Results An increase in inter-quartile range (IQR) in the methylation of Alu elements was associated with an increase of 0.97 mm Hg in diastolic blood pressure (DBP) (95% CI 0.32–1.57), but no such association was observed for long interspersed nuclear element-1 (LINE-1). We also found positive associations between DBP and methylation of the genes for toll-like receptor 2 (TLR2) and inducible nitric oxide synthase (iNOS), and a negative association between DBP and methylation of the gene for interferon-γ (IFN-γ). Associations between methylation and systolic blood pressure (SBP) were weaker than those between methylation and DBP. Bayesian mixed-effects structural equation model results were similar for both DBP and SBP models.
Conclusions The results of our study suggest that changes in DNA methylation of some pro-inflammatory genes and retrotransposable elements are related to small changes in BP.
Keywords: Epigenetics, DNA methylation, blood pressure, inflammation, Bayesian model
Introduction
Blood pressure (BP) is a well-established intermediary biomarker predictive of cardiovascular events including stroke, myocardial infarction, and mortality. Even moderate elevations in BP can increase the risk of these adverse health outcomes. High BP, or hypertension, affects approximately one-third of the US adult population1 and is a marker of accelerated arterial stiffening. A multitude of factors, including genetic, environmental, and lifestyle factors are suspected to influence hypertension.
The methylation of deoxyribonucleic acid (DNA) is an epigenetic process that helps regulate gene expression. In mammals, DNA methylation is predominantly found on cytosines at sites of CpG dinucleotides. As with other epigenetic markers, DNA methylation is mitotically stable yet modifiable in response to environmental factors.2 The methylation of DNA can be measured both in specific genes as well as in repetitive DNA sequences that are widespread throughout the genome.3,4
Retrotransposons are remnants of viral ribonucleic acid (RNA) incorporated into the human genome over evolutionary history5,6; these elements are highly methylated in order to suppress their expression.7 There is evidence that long interspersed nuclear element-1 (LINE-1) and Alu have current biological activity within the human genome.8,9 When transcribed, these retrotransposable elements can re-insert into DNA, causing mutation and genetic damage.10 Transcription of retrotransposons has been shown to contribute to transcriptional regulation of somatic genes11 as well as to cell growth and differentiation.12,13
Methylation of DNA plays an important role in regulation of the expression of individual genes. Methylation of promoter regions and, as more recently discovered, of CpG island shores upstream of the promoter region has been associated with transcriptional suppression of the corresponding gene.14,15 In addition, the differential methylation of promoter regions of certain genes has been found to be site-specific in cancer16 and in rheumatoid arthritis.17 There is also evidence that for some genes, the methylation of a particular CpG position may have a strong influence on transcriptional suppression, whereas methylation at other CpG positions has little influence.16–18 These site-specific findings indicate that there may be biologically relevant heterogeneity in DNA methylation levels across different sites within a promoter region. Hence, there may be a need for models that account for heterogeneity in the effects of methylation at different positions on the same gene.
The processes of inflammation and oxidative stress affect many inter-related outcomes of cardiovascular health.19 Higher levels of inflammatory biomarkers such as C-reactive protein (CRP) and cell adhesion molecules are independently and jointly associated with the risk of cardiovascular disease.20–23 In general, de-methylation of DNA in specified genes is expected to increase the expression and activity of the protein coded by the gene,24 and this has been demonstrated for some inflammatory genes.25–27 Thus, the methylation of pro-inflammatory genes may be related to markers of cardiovascular health by regulating inflammatory pathways that are key elements in the development of cardiovascular diseases.
Epigenetic regulation, including changes in DNA methylation, has recently been recognized as an important factor in the pathogenesis of atherosclerosis. The degree of loss of DNA methylation in patients with atherosclerosis is now well-established,28–30 with diminished genomic DNA methylation observed in a variety of tissues from patients with atherosclerotic disease, including smooth-muscle cells,31,32 atherosclerotic lesions,33 and peripheral blood leukocytes.34 However, the relationship between changes in DNA methylation and hypertension is still largely unknown.
We hypothesized that epigenetic changes in DNA methylation markers would be associated with changes in BP. In the study described here we examined the associations of DNA methylation in retrotransposable sequences (Alu, LINE-1) and in a panel of pro-inflammatory genes (the genes for coagulation factor 3 (F3), glucocorticoid receptor (GCR), inducible nitric oxide synthase (iNOS), intercellular adhesion molecule (ICAM-1), interferon-γ (IFN-γ), interleukin-6 (IL-6), and toll-like receptor-2 (TLR2), with diastolic blood pressure (DBP) and systolic blood pressure (SBP) in a cohort of elderly subjects in the greater Boston area. In a secondary analysis, we examined these associations using flexible Bayesian models that allowed for heterogeneous effects of different methylation positions on the same gene.
Methods
Study Population
This study was conducted on a sub-sample of the Normative Aging Study (NAS), a multidisciplinary longitudinal study of aging established by the US Veterans Administration in 1963.35 Briefly, the NAS enrolled 2280 men from the Greater Boston area, ranging in age from 21–80 years, who were initially free of known chronic medical conditions as determined in a health screening examination. At the beginning of the study period in 1999, 30% of the original participants in the NAS had died and other subjects had moved out of the region after their retirement and were no longer being followed. All participants in the present study provided written informed consent, and the study was approved by the institutional review boards of all of the participating institutions.
Subjects who still live in the greater Boston area continue to visit the study center of the NAS at the Boston VA Hospital for medical examinations every 3–5 years. During these visits, extensive physical examination, laboratory, anthropometric, and questionnaire data are collected, including blood samples. Systolic blood pressure (SBP) and diastolic blood pressure (DBP) of subjects in the seated position are measured as the means of the measurements of each of these BP components in the left and right arm. Information about cigarette smoking, medical history, and medication use are obtained through a self-administered questionnaire. Each subject is interviewed to confirm the identity and purpose of medications being used by the subject. The incidence of new disease is also noted.
Samples of DNA in blood have been collected and stored since 1999 as part of the NAS. A total of 1722 blood samples from 789 subjects were collected between 1999 and 2009, and were successfully subjected to at least one measurement of DNA methylation. Subjects had from one to four DNA methylation measurements made at consecutive time points at intervals of approximately 3 years (a mean of 2.2 measurements). For some subjects, the extracted DNA was insufficient in quantity to be amplified for the performance of all methylation measurements, and in addition some assays occasionally failed. This should occur in a completely random manner because it is unrelated to the levels of methylation of a particular sequence of DNA.
DNA methylation
For the study of DNA methylation, DNA was extracted from the buffy coat of 7 ml of stored, frozen whole blood through the use of QiAmp DNA blood kits (QIAGEN, Valencia, CA, USA). The extracted DNA (500 ng; concentration: 50 ng/µl) was treated with the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s protocol. Final elution was done with 30 µl of M-Elution Buffer (Zymo Research)
Methylation of DNA was quantified with bisulfite treatment of DNA and simultaneous polymerase chain reaction (PCR) and by pyrosequencing, using previously described primers and conditions.36,37 A 50-µl PCR was done in 25 µl of GoTaq Green Master mix (Promega, Madison, WI, USA), 1 pmol biotinylated forward primer, 1 pmol reverse primer, 50 ng bisulfite-treated genomic DNA, and water. The degree of methylation was expressed as the percent of the sum total of methylated and unmethylated cytosines that consisted of 5-methylated cytosines (%5mC). Non-CpG cytosine residues were used as built-in controls to verify bisulfite conversion. Pyrosequencing-based assays produce individual measures of methylation at more than one CpG dinucleotide.
Candidate genes were identified through a literature review of genes involved in inflammatory pathways. The assays for methylated DNA were designed to cover the greatest possible number of CpG sites within the promoter region, taking into account the necessary length of the PCR amplicon, length of the target sequence, and primers that avoided CpGs. Methylation levels for LINE-1 and those for Alu were collected at three CpG sites. For the assessment of gene-specific methylation, the location of each CpG position and the promoter region is given in Supplementary Table 1. The primer sequences for each DNA methylation are listed in Supplementary Table 2.
Statistical analysis
In our initial analysis, we fit a mixed-effects model with a random intercept for each subject to account for correlation among repeated measurements of the same subject’s variables across different medical visits. This accounted for the longitudinal measurements of both BP and DNA methylation for a given subject. We performed analyses to evaluate the association between DNA methylation levels and BP. Potential confounding factors were chosen a priori and included age, body mass index (BMI), smoking (never, former, current), pack-years of smoking, diabetes mellitus or fasting blood glucose >126 mg/dl (yes/no), alcohol consumption (≥2 drinks per day: yes/no), race (white/others), education level, previous ischemic heart disease or stroke (yes/no), medication use, number of neutrophils in the white-blood-cell count, season of the year, and day of the week. The models in the study used time-varying covariates updated at each visit. Mixed models had the following form:
where the index i denotes the subject, the index j denotes the time, Yij is the health outcome (BP as SBP, or DBP), Cij is the vector of confounders, β0 is the overall intercept, ui is the separate random intercept for each subject, Mij is the mean methylation across CpG positions in a given gene, and βM is the slope representing the effect of DNA methylation in that gene. We assumed that the ui were mean zero normal random variables with common variance, yielding the simple compound symmetry variance structure. The errors represented by the error term eij were assumed to be independent and identically distributed (iid), with a mean of zero and common variance. The two variance components were estimated from the data and represent the inter- and the intrasubject variation.
We examined the DNA methylation measures as continuous covariates, and we modeled BP as a continuous outcome. The effect estimate is reported as the change in BP per change in interquartile range (IQR) of methylation for each gene. We report the effect estimate with 95% confidence intervals (CIs).
In our secondary analysis, we accounted for heterogeneity in the effect of DNA methylation at different CpG positions by using a mixed-effects structural equation model with a Bayesian framework. The use of mixed-effects structural equation models with application to environmental epidemiology and latent exposure modeling was described in detail by Sanchez et al. in 2005.38 The mixed-effects structural equation model allows the exposure, DNA methylation, to be represented as an unobserved latent variable, which we will refer to as the “latent methylation” for shorthand convenience. The latent methylation corresponds to the overall methylation level of a particular gene, and the model treats the measured levels of DNA methylation taken at particular CpG positions in the gene as surrogates of that overall methylation level.
The mixed-effects structural equation model consists of a joint model with three components: (i) a linear mixed model for the health endpoint (BP), given the latent methylation of a gene and the confounders; (ii) a linear mixed model for the latent methylation status given the confounders; and (iii) a factor-analytic structure which assumes that the latent methylation underlies K measured methylation positions. More formally, the set of regression equations is
| (1) |
| (2) |
| (3) |
where index i denotes the subject, index j denotes the time, and index k denotes the CpG position. For subject i on day j, the health response (SBP or DBP) is denoted by Yij, the vector of confounders is Cij, the methylation level measured at position k is denoted by Xijk, and the latent methylation is 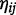 . The coefficients ui and bi represent subject-specific intercepts for the health-endpoint and latent-methylation models, respectively, and are assumed to be mean zero normal random variables with common variance estimated from the data. The error terms eij, ωij, δijk are all assumed to be independent normal random errors.
. The coefficients ui and bi represent subject-specific intercepts for the health-endpoint and latent-methylation models, respectively, and are assumed to be mean zero normal random variables with common variance estimated from the data. The error terms eij, ωij, δijk are all assumed to be independent normal random errors.
The latent methylation is related to each position k through its particular intercept λ0,k and slope λ1,k. For identifiability, we imposed the standard constraint in which λ0,K = 0 and λ1,K = 1, treating an arbitrary CpG position, K, as the reference for that gene. We used a Bayesian framework to flexibly model the distributional assumptions, and we fit the model using the WinBUGS software package (Medical Research Council Biostatistics Unit, Cambridge, UK) with a Markov Chain Monte Carlo (MCMC) sampling scheme. The priors for the effect of methylation were normal (1,10), allowing for a range of potential relationships between the CpG positions while also reflecting the prior knowledge that the methylation positions are related and moderately correlated. For all other model parameters, we chose non-informative prior distributions. We used uniform (0.01,100) priors for the variances of each methylation variable, since uniform priors have been shown to be much more stable for variance components than inverse-gamma priors.39 We report the median effect estimate and 95% credible intervals for each effect.
Results
The demographic characteristics of the NAS population across all visits used in the analysis are shown in Table 1. Subjects were elderly, with a baseline age ranging from 55 years to 100 years and a mean age of 74 years. Most subjects were overweight (median BMI 27.5 kg/m2). The majority of subjects were former smokers, and very few subjects were current smokers (3.5%). The average level of DNA methylation varied widely among genes, with the highest average methylation level in the gene for IFN-γ (84.4%) and the lowest average methylation level in the gene for F3 (2.5%). In our cohort, the measures of Alu and LINE-1 were not correlated (corr = 0.005). As discussed in the next section, this low correlation between the degree of methylation of LINE-1 and Alu is consistent with the findings of other research with non-cancerous cells. For the gene-specific DNA methylation measures, the adjacent CpG sites within each gene had moderate to high correlations ranging from 0.3 to 0.8. Over the 10-year study period, the percent change in methylation ranged from 2% to 20%.
Table 1.
Characteristics of the study population in the Normative Aging Study
| All visits |
||
|---|---|---|
| Variable | N = 1722 | |
| Age, years (SD) | 74.1 | (6.7) |
| Body mass index, Kg/m2 (SD) | 28.1 | (4.1) |
| Systolic blood pressure, mmHg (SD) | 128.5 | (17.5) |
| Diastolic blood pressure, mmHg (SD) | 73.4 | (10.3) |
| Race, n (%) | ||
| White | 1687 | (98.0) |
| Black/Hispanic | 35 | (2.0) |
| Smoking status, n (%) | ||
| Never smoker | 515 | (29.9) |
| Former smoker | 1147 | (66.6) |
| Current smoker | 60 | (3.5) |
| Pack years of smoking,a (SD) | 28.9 | (25.6) |
| Alcohol consumption (≥2 drinks per day), n (%) | 309 | (17.9) |
| Diabetes, n (%) | 329 | (19.1) |
| Ischaemic heart disease, n (%) | 548 | (31.8) |
| Stroke, n (%) | 130 | (7.5) |
| Blood count | ||
| White blood cells, cells/mm3 (SD) | 6541 | (3004) |
| Neutrophils, cells/mm3 (SD) | 3997 | (1301) |
| Lymphocytes, cells/mm3 (SD) | 1701 | (2192) |
| Monocytes, cells/mm3 (SD) | 545 | (192) |
| Eosinophils, cells/mm3 (SD) | 216 | (156) |
| Basophils, cells/mm3 (SD) | 37 | (36) |
| Day of the visit, n (%) | ||
| Monday | 67 | (3.9) |
| Tuesday | 436 | (25.3) |
| Wednesday | 968 | (56.2) |
| Thursday | 251 | (14.6) |
| Season of the visit, n (%) | ||
| Spring | 363 | (23.4) |
| Summer | 433 | (28.0) |
| Fall | 499 | (32.2) |
| Winter | 254 | (16.4) |
| DNA methylation, %5mCb (SD) | ||
| LINE-1, mean (SD) | 77.1 | (1.7) |
| ALU, mean | 25.9 | (1.3) |
| ICAM-1, mean | 4.3 | (1.7) |
| iNOS, mean | 68.3 | (7.5) |
| IFN-γ, mean | 84.8 | (5.2) |
| IL-6, mean | 43.3 | (10.5) |
| TLR2, mean | 3.0 | (1.3) |
| F3, mean | 2.4 | (1.3) |
| GCR, mean | 46.7 | (6.1) |
aMean and SD among ever smokers
bPercentage of 5-methyl cytosine
Table 2 presents the relationships between BP and DNA methylation in our initial analysis done with linear mixed-effects models with adjustment for confounding variables. Both SBP and DBP were positively associated with the degree of methylation of the gene for Alu. We also found positive associations between DBP and the level of methylation of the genes for TLR2 and iNOS, but the associations with SBP were weaker, and the 95% CIs included zero. We found that the methylation of the gene for IFN-γ was negatively associated with both SBP and DBP. The methylation of the gene for LINE-1 was also inversely associated with DBP, yet the association with SBP was weaker, with the 95% CI including zero. No clear associations were observed between SBP or DBP and the levels of methylation of the genes for ICAM-1, GCR, or F3.
Table 2.
Associations of blood pressure with mean DNA methylation of each element per IQR change in mean methylation, estimated by a mixed-effects model
| Diastolic blood pressure |
Systolic blood pressure |
||||
|---|---|---|---|---|---|
| Subjects N | Effecta | (95% Confidence Interval) | Effecta | (95% Confidence Interval) | |
| Methylation, %5mC | |||||
| Alu, mean | 760 | 0.81 | (0.36, 1.26) | 1.40 | (0.57, 2.23) |
| LINE-1, mean | 754 | –0.70 | (–1.20, –0.20) | –0.69 | (–1.59, 0.22) |
| ICAM-1, mean | 684 | 0.07 | (–0.52, 0.66) | 0.57 | (–0.51, 1.64) |
| iNOS, mean | 640 | 0.91 | (0.24, 1.57) | 0.42 | (–0.80, 1.63) |
| IFN-γ mean | 756 | –1.10 | (–1.68, –0.52) | –1.70 | (–2.76, –0.63) |
| IL-6, mean | 752 | 0.49 | (–0.10, 1.08) | 0.49 | (–0.60, 1.58) |
| TLR2, mean | 700 | 1.16 | (0.53, 1.78) | 1.07 | (–0.06, 2.21) |
| F3, mean | 732 | –0.31 | (–0.83, 0.21) | –0.50 | (–1.45, 0.46) |
| GCR, mean | 734 | –0.31 | (–0.80, 0.18) | –0.13 | (–1.02, 0.77) |
All models included confounding variables: age, body mass index, smoking (never, former, current), pack-years of smoking, diabetes mellitus (yes/no), consumption of alcohol (yes/no), race (white/others), ischaemic heart disease or stroke (yes/no), number of neutrophils in white blood count, season, and day of week. Alu, Alu transposable element; F3, coagulation factor 3; GCR, glucocorticoid receptor; ICAM-1, intercellular adhesion molecule-1; IL-6, interleukin-6; iNOS, inducible nitric oxide synthase; LINE-1, long interspersed nuclear element-1; TLR2, toll-like receptor 2.
aEffect estimate and 95% confidence interval (CI) expressing the change in blood pressure (mm Hg) associated with a change in inter-quartile range of the mean methylation for that element.
Table 3 presents the relationships between BP and DNA methylation in our mixed-effects structural equation model analysis. Comparing the results in Table 3 with those in Table 2 shows that most of the estimated effects of DNA methylation had the same direction and a similar magnitude per inter-quartile range (IQR) of methylation, and that most of the CIs in the mixed-effects structural equation model analysis were wider. The width of the CI reflects the additional modeling of the variation in the DNA methylation measures. In fact, the CIs for the mixed model may be underestimated because the measurement error in DNA methylation is not taken into account when the mean is used.
Table 3.
Associations of blood pressure with mean DNA methylation of each element per IQR change in latent methylation, estimated by a Bayesian Structural Equation Model
| Diastolic blood pressure |
Systolic blood pressure |
||||
|---|---|---|---|---|---|
| Subjects N | Effecta | (95% Credible Interval) | Effecta | (95% Credible Interval) | |
| Methylation, %5mC | 760 | ||||
| Alu, latent | 754 | 0.97 | (0.32, 1.57) | 1.51 | (0.36, 2.61) |
| LINE-1, latent | 684 | –0.32 | (–1.02, 0.40) | 0.82 | (–0.17, 1.89) |
| ICAM-1, latent | 640 | 0.23 | (–0.31, 0.73) | 0.87 | (–0.14, 1.83) |
| iNOS, latent | 756 | 1.28 | (0.46, 2.09) | 1.15 | (–0.28, 2.74) |
| IFN-γ, latent | 752 | –0.94 | (–1.57, –0.21) | –1.22 | (–2.38, –0.03) |
| IL-6, latent | 700 | 0.35 | (-0.06, 0.73) | 0.50 | (-0.25, 1.22) |
| TLR2, latent | 732 | 1.79 | (0.93, 2.64) | 1.73 | (0.20, 3.27) |
| F3, latent | 734 | –0.41 | (–1.14, 0.25) | –0.70 | (–1.97, 0.56) |
All models included confounding variables: age, body mass index, smoking (never, former, current), pack-years of smoking, diabetes mellitus (yes/no), alcohol (yes/no), race (white/others), ischaemic heart disease or stroke (yes/no), number of neutrophils in white blood count, season, and day of week. Alu, Alu transposable element; F3, coagulation factor 3; GCR, glucocorticoid receptor; ICAM-1, intercellular adhesion molecule-1; IL-6, interleukin-6; iNOS, inducible nitric oxide synthase; LINE-1, long interspersed nuclear element-1; TLR2, toll-like receptor 2.
aEffect estimate and 95% credible interval expressing the change in blood pressure (mm Hg) associated with a change in inter-quartile range of the latent methylation for that element.
Table 4 presents the factor loadings from the mixed-effects structural equation model analysis for each gene, which reflect the estimated relationships between the latent DNA methylation and the methylation at each measured CpG position. The factor loadings are interpreted by their relative sizes rather than by their absolute sizes because the reference is set to 1 and is chosen arbitrarily. A loading that is larger relative to the other loadings indicates that the CpG position with that loading is more closely related to the latent methylation variable than are the other CpG positions. We note that the estimated factor loadings in Table 4 are nearly identical for DBP and SBP, which is expected because they measure the relationship of the methylation levels to each other, with only a small amount of Bayesian “feedback” from the outcome and other covariates.
Table 4.
Relationship between individual methylation positions and the latent methylation variable for each gene, with factor loading estimates and credible intervals from the Bayesian structural equation model
| Diastolic blood pressure |
Systolic blood pressure |
|||
|---|---|---|---|---|
| Gene | Effect | (95% Credible Interval) | Effect | (95% Credible Interval) |
| Alu | ||||
| Position 1 | 1 | reference | 1 | reference |
| Position 2 | 2.08 | (1.96, 2.18) | 2.08 | (1.96, 2.18) |
| Position 3 | 1.28 | (1.18, 1.38) | 1.28 | (1.18, 1.38) |
| LINE | ||||
| Position 1 | 1 | reference | 1 | reference |
| Position 2 | 1.01 | (0.94, 1.08) | 1.01 | (0.94, 1.08) |
| Position 3 | 0.92 | (0.85, 0.99) | 0.92 | (0.85, 0.98) |
| ICAM-1 | ||||
| Position 1 | 1 | reference | 1 | reference |
| Position 2 | 0.98 | (0.90, 1.05) | 0.98 | (0.90, 1.05) |
| Position 3 | 0.34 | (0.29, 0.39) | 0.34 | (0.28, 0.39) |
| iNOS | ||||
| Position 1 | 1 | reference | 1 | reference |
| Position 2 | 1.33 | (1.21, 1.45) | 1.32 | (1.20, 1.44) |
| IFN-γ | ||||
| Position 1 | 1 | reference | 1 | reference |
| Position 2 | 0.87 | (0.83, 0.91) | 0.87 | (0.83, 0.92) |
| IL-6 | ||||
| Position 1 | 1 | reference | 1 | reference |
| Position 2 | 0.85 | (0.80, 0.90) | 0.85 | (0.80, 0.90) |
| TLR2 | ||||
| Position 1 | 1 | reference | 1 | reference |
| Position 2 | 0.95 | (0.81, 1.12) | 0.94 | (0.79, 1.11) |
| Position 3 | 0.99 | (0.83, 1.17) | 0.98 | (0.82, 1.15) |
| F3 | ||||
| Position 1 | 1 | reference | 1 | reference |
| Position 2 | 1.38 | (1.13, 1.66) | 1.38 | (1.13, 1.66) |
| Position 3 | 1.53 | (1.27, 1.84) | 1.52 | (1.27, 1.83) |
| Position 4 | 1.88 | (1.61, 2.26) | 1.88 | (1.61, 2.25) |
| Position 5 | 1.63 | (1.34, 1.99) | 1.62 | (1.34, 1.98) |
Results for the relationship between each position and latent methylation correspond to the effects of latent methylation for the models in Table 3.
Abbreviations: Alu, Alu transposable element; F3, coagulation factor 3; GCR, glucocorticoid receptor; ICAM, intercellular adhesion molecule; IL-6, interleukin-6; iNOS, inducible nitric oxide synthase; LINE, long interspersed nuclear element-1; TLR2, toll-like receptor 2
What is most interesting is the relative weight for each CpG position, which indicates the potentially different importance of different positions. For Alu, both position 2 and position 3 are estimated to better reflect the latent methylation variable for Alu than is position 1, and the strength of the relationship with position 2 is estimated to be approximately twice that of position 1. In contrast, all three CpG positions for LINE-1 are estimated to nearly equally reflect the LINE-1 latent methylation variable. For ICAM-1, positions 1 and 2 are estimated to equally reflect the latent methylation variable, and the strength of the relationship is estimated to be approximately 3-fold greater than that of position 3. For IFN-γ, iNOS, IL-6, and F3, there are also some differences in the strength of the relationship of one position as compared with another, although the magnitudes of these differences are less than 2-fold. In contrast, the estimated effects of all three positions for TLR2 are nearly the same, indicating that no particular CpG position reflects latent methylation to a noticeably better degree than another.
As a sensitivity analysis, we considered the association between the change in BP and the changes in methylation levels. Results were similar but not as strong. Changes in the mean methylation of Alu showed a strong relationship with both SBP (P = 0.0085) and DBP (P = 0.0214). Also, changes in the mean methylation of IFN-γ showed a strong relationship with both SBP (P = 0.0145) and DBP (P = 0.0001). The associations between the other methylation markers were not as strong.
We also considered whether there were any deviations from linearity in the effects of methylation by using a spline fit for mean methylation. There was no evidence of a nonlinear relationship for most of the methylation markers. The only evidence of thresholding observed was for F3 and ICAM-1, where we saw an effect of approximately zero at methylation levels below 5% and a linear trend above 5% methylation. This could help explain why the effects appeared not as strong for ICAM-1 and F3 in our analyses.
Discussion
In this study we found associations between BP and DNA methylation levels in several of the pro-inflammatory genes, with the direction of the effects varying by gene. We also found a positive association between the methylation of Alu and BP, but not between the methylation of LINE-1 and BP. These findings of associations between DNA methylation levels and BP have a number of implications as well as potential limitations, which are acknowledged below.
With regard to gene-specific DNA methylation and cardiovascular disease, we measured the association between BP and the DNA methylation of seven candidate genes in inflammatory pathways and expressed in leukocytes.40 In our data, BP showed negative associations with methylation of the gene for IFN-γ and positive associations with methylation of the genes for iNOS and TLR2. No clear associations were observed between BP and the degree of methylation of the genes for ICAM-1, F3, or IL-6. Because few studies of gene-specific methylation currently exist, further research is warranted to better understand the observed results.
The activation of TLR2 is now thought to play an important signaling role in the development of cardiovascular diseases, particularly of atherosclerosis and potentially of myocardial inflammation and injury.41,42 The associations we found between BP and methylation of the gene for TLR2 are consistent with developing research in this area. Expression of the enzyme iNOS produces nitric oxide during inflammation and is thought to affect vascular reactivity and to contribute to the development of inflammatory cardiovascular diseases such as atherosclerosis, with expression of iNOS present in the vascular smooth-muscle cells of atherosclerotic lesions in animal studies.43 Our finding of an association between methylation of the gene for iNOS and DBP is consistent with this research, although the association between methylation of the gene for iNOS and SBP was much weaker. Interferon-γ has also been closely linked with atherosclerosis, and the gene for IFN-γ is also highly expressed in atherosclerotic lesions, with its expression having been classified as proatherogenic in animal studies.44,45 Thus, on the assumption that decreased levels of methylation lead to increased expression of a gene, our findings of negative associations between methylation of the gene for IFN-γ and BP are consistent with this literature.
The mechanism by which IL-6 influences BP may involve complex interactions between multiple factors which could explain why we did not detect an association between BP and the methylation level of the gene for IL-6. Interleukin-6 regulates the expression of CRP, which is a well-established biomarker for cardiovascular disease risk.22,23 However, other studies of BP have not found it to be associated with BP and either expression of the mRNA for IL-646 or serum levels of IL-6.47 In addition, a review of animal studies has shown that the expression of IL-6 can be proatherogenic or antiatherogenic, depending on the experimental conditions.45 Thus, more research is needed to understand how transcription and expression of the gene for IL-6 may affect pathways to cardiovascular diseases. Studies of the effect on hypertension and coronary heart disease of variations in individual single nucleotide polymorphisms (SNPs) in the GCR gene have found no clear effects,48,49 and our finding of the lack of an association of BP with the methylation level of GCR is consistent with the literature on polymorphism.
With regard to global DNA methylation and cardiovascular disease, epigenetic regulation is now recognized as an important factor in the pathogenesis of atherosclerosis, with well-documented associations between the hypomethylation of global genomic DNA and atherosclerosis.28-30 These changes in DNA methylation may be mediated by homocysteine,50 or could be a secondary effect of changes in homocysteine. Studies with mice have demonstrated that the hypomethylation of global genomic DNA is a predictor of future atherosclerosis.51
We found that methylation of Alu was positively associated with DBP and SBP. This finding is consistent with the recent finding in a Chinese cohort that individuals with higher degrees of methylation of Alu in peripheral blood leukocytes were at higher risk of developing cardiovascular disease.52 Although earlier studies proposed that the methylation of Alu and LINE-1 reflected global DNA methylation,53,54 measures of LINE-1 and Alu methylation made on peripheral blood leukocytes, such as the ones we used in our study, should be seen as focused on distinct markers assessing two different types of retrotransposon. The correlation of the methylation levels of LINE-1 and Alu has been established only in cancer cells,54 in studies in which the pathogenesis of cancer has included a loss of methylation affecting both types of repetitive elements.55,56 No correlation between the methylation levels of Alu and LINE-1 exists in DNA taken from peripheral blood leukocytes.57,58 Recent studies have also revealed differences between LINE-1 and Alu elements in the mechanisms that regulate their methylation, and their responses to cellular stressors and environmental exposures may account for different methylation states of these elements.36,59-62 Thus, our findings of a lack of correlation between the levels of DNA methylation in LINE-1 and Alu elements, and the lack of a clear association of methylation of Alu with BP but not of methylation of LINE-1 with BP, are consistent with other findings of DNA methylation in peripheral blood leukocytes. Taken together, these findings suggest distinct roles for the methylation of LINE-1 and Alu as markers of cardiovascular risk.
Retrotransposons are now thought to have a functional role in the regulation of gene expression. LINE-1 elements encode enzymes that allow them to replicate and insert themselves into different genomic regions, altering gene expression and cell function.6,63 The insertion of Alu into genes can also regulate gene expression or function or both.6 Transposable elements have recently been shown to play a critical role in human development, tissue differentiation, and gene expression, in that these elements can be transcribed and translated into functional proteins.9,64 In this context, our finding of an association between BP and the DNA methylation of Alu elements could represent an alteration in gene expression that has a mechanistic role in cardiovascular health during aging.
A strength of our study is that we measured methylation at more than one CpG site on each gene and analyzed the overall effect of methylation in two different ways. The Bayesian mixed-effects structural equation model analysis accounted for the heterogeneity of the CpG position effects of DNA methylation. This flexibility of modeling is important when the extent of heterogeneity of an effect is unknown. The differences seen in many of the factor loadings in our analysis suggest that there may be differences in the relationships of methylation at different sites, although these differences were small and the overall effect of methylation was similar in both our mixed-effects and Bayesian mixed-effects structural equation model analyses. If all the factor loadings are equal, then each CpG position is equally related to the latent methylation of a gene or transposable element, and using the mean methylation instead of the latent variable may therefore not eliminate very much information.
In some cases we observed stronger effects for DBP than for SBP in association with the gene-specific degree of methylation. This difference in effect on BP may reflect differences in the process of aging. In general, BP increases with age, but DBP tends to rise until the age of 50 or 60 years and to then level off, whereas SBP continues to increase until 70 or 80 years of age.65 However, even small changes in SBP and DBP are associated with an increased risk of cardiovascular mortality. For example, a meta-analysis of 61 prospective cohort studies estimated that a DBP that was even 1 mm Hg lower usual was associated with about a 10% lower mortality from stroke and 7% lower mortality from ischaemic heart disease (IHD) and other vascular causes of death, and reported that the age-specific associations of BP and mortality remained strong even in older age groups (70–79 years and 80–89 years).66 Thus, even small changes in BP may be clinically relevant.
One limitation of this study is that the measurement of DNA methylation was done with DNA from peripheral blood leukocytes, which may not reflect DNA methylation in blood vessels and neural tissues. In addition, because the measures of DNA methylation used in the study were made with blood leukocytes, our results might have reflected shifts in the proportions of white blood-cell subsets caused by alterations related to impending disease onset. Although we tried to account for this problem by controlling for the number of neutrophils in the white blood count as a confounding variable, there is still the possibility of further residual confounding. In addition, the assays we used were designed to cover the greatest possible number of CpG sites within the promoter region of each of the genes that we studied, but it is possible that other, nearby CpG sites which were not examined could also be biologically relevant.
Another limitation of this study is the restricted demographics of the study population. All of the study subjects were elderly men, and most of them were white, which prevents us from generalizing our results to other populations. However, the elderly represent a particularly disease-susceptible population subgroup, growing in number and proportion, and so represent an important population for study. Future studies should address the role of methylation of retrotransposons among women, as well as in various age and ethnic groups.
Beyond being elderly, many of the subjects in our study were overweight or obese. Thus, our results may reflect relationships of methylation levels that are more relevant to metabolic pathways involved in obesity, inflammation, and metabolic syndrome. Hence, these results should be generalized only to a similar population that is mainly overweight. In healthy, non-overweight populations, the methylation of inflammatory genes may or may not be as important in determining BP.
In summary, we found that increases in the degree of methylation of Alu elements were associated with increases in BP. We also found positive associations between BP and the degree of methylation of the genes for TLR2 and iNOS, and negative associations of BP with methylation of the gene for IFN-γ. These findings support the hypothesis that the methylation of pro-inflammatory genes may play a role in the pathogenesis of cardiovascular diseases in the elderly. We also found that the associations of DNA methylation in retrotransposons vary for the elements LINE-1 and Alu, leaving to be determined the question of whether the associations in the case of Alu represent functional differences.
Funding
This research was supported by National Institute of Environmental Health Sciences grant R01 ES015172. The Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the US Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC), Boston, MA. This research was also supported by a VA Research Career Scientist award to D.S.
KEY MESSAGES.
This paper reports novel associations between DNA methylation and BP in a longitudinal cohort of elderly males.
These findings support the hypothesis that methylation of the DNA of pro-inflammatory genes may play a role in the pathogenesis of cardiovascular diseases in the elderly.
As in some other studies, the associations of DNA methylation in retrotransposons differ in the elements LINE-1 and Alu.
We compare two analytical approaches, and we discuss the potential benefits of a Bayesian mixed-effects structural equation model with a latent variable representing the overall degree of DNA methylation for incorporating heterogeneity in measures of DNA methylation of the same gene.
Supplementary Material
References
- 1.Fields LE, Burt VL, Cutler JA, et al. The burden of adult hypertension in the United States 1999 to 2000: a rising tide. Hypertension. 2004;44:398–404. doi: 10.1161/01.HYP.0000142248.54761.56. [DOI] [PubMed] [Google Scholar]
- 2.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21:243–51. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 4.Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 5.Carnell AN, Goodman JI. The long (LINEs) and the short (SINEs) of it: altered methylation as a precursor to toxicity. Toxicol Sci. 2003;75:229–35. doi: 10.1093/toxsci/kfg138. [DOI] [PubMed] [Google Scholar]
- 6.Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol. 2006;310:211–50. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- 8.Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet. 2005;117:411–27. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 9.Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annual review of genetics. 2001;35:501–38. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 11.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–74. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 12.Ergun S, Buschmann C, Heukeshoven J, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem. 2004;279: 27753–63. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 13.Sciamanna I, Landriscina M, Pittoggi C, et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene. 2005;24:3923–31. doi: 10.1038/sj.onc.1208562. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg AP, Tycko B. The history of cancer epigenetics. Nature Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pogribny IP, Pogribna M, Christman JK, James SJ. Single-site methylation within the p53 promoter region reduces gene expression in a reporter gene construct: possible in vivo relevance during tumorigenesis. Cancer Res. 2000;60:588–94. [PubMed] [Google Scholar]
- 17.Ishida K, Kobayashi T, Ito S, et al. Interleukin-6 gene promoter methylation in rheumatoid arthritis and chronic periodontitis. J Periodontol. 2012;83:917–25. doi: 10.1902/jop.2011.110356. [DOI] [PubMed] [Google Scholar]
- 18.Zou B, Chim CS, Zeng H, et al. Correlation between the single-site CpG methylation and expression silencing of the XAF1 gene in human gastric and colon cancers. Gastroenterology. 2006;131:1835–43. doi: 10.1053/j.gastro.2006.09.050. [DOI] [PubMed] [Google Scholar]
- 19.Pashkow FJ. Oxidative stress and inflammation in heart disease: do antioxidants have a role in treatment and/or prevention? Int J Inflam. 2011;2011:514623. doi: 10.4061/2011/514623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pradhan AD, Rifai N, Ridker PM. Soluble intercellular adhesion molecule-1, soluble vascular adhesion molecule-1, and the development of symptomatic peripheral arterial disease in men. Circulation. 2002;106:820–25. doi: 10.1161/01.cir.0000025636.03561.ee. [DOI] [PubMed] [Google Scholar]
- 21.Rana JS, Arsenault BJ, Despres JP, et al. Inflammatory biomarkers, physical activity, waist circumference, and risk of future coronary heart disease in healthy men and women. Eur Heart J. 2011;32:336–44. doi: 10.1093/eurheartj/ehp010. [DOI] [PubMed] [Google Scholar]
- 22.Ridker PM, Buring JE, Cook NR, Rifai N. C-reactive protein, the metabolic syndrome, and risk of incident cardiovascular events: an 8-year follow-up of 14 719 initially healthy American women. Circulation. 2003;107:391–97. doi: 10.1161/01.cir.0000055014.62083.05. [DOI] [PubMed] [Google Scholar]
- 23.Danesh J, Whincup P, Walker M, et al. Low grade inflammation and coronary heart disease: prospective study and updated meta-analyses. BMJ. 2000;321:199–204. doi: 10.1136/bmj.321.7255.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Methods Mol Biol. 2009;507:3–20. doi: 10.1007/978-1-59745-522-0_1. [DOI] [PubMed] [Google Scholar]
- 25.Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79:1514–19. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]
- 26.Bayarsaihan D. Epigenetic mechanisms in inflammation. J Dent Res. 2011;90:9–17. doi: 10.1177/0022034510378683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–93. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 28.Wierda RJ, Geutskens SB, Jukema JW, Quax PH, van den Elsen PJ. Epigenetics in atherosclerosis and inflammation. J Cell Mol Med. 2010;14:1225–40. doi: 10.1111/j.1582-4934.2010.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turunen MP, Aavik E, Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta. 2009;1790:886–91. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci. 2009;66:2249–61. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiltunen MO, Yla-Herttuala S. DNA methylation, smooth muscle cells, and atherogenesis. Arterioscler Thromb Vasc Biol. 2003;23:1750–53. doi: 10.1161/01.ATV.0000092871.30563.41. [DOI] [PubMed] [Google Scholar]
- 32.Yideng J, Jianzhong Z, Ying H, et al. Homocysteine-mediated expression of SAHH, DNMTs, MBD2, and DNA hypomethylation potential pathogenic mechanism in VSMCs. DNA Cell Biol. 2007;26:603–11. doi: 10.1089/dna.2007.0584. [DOI] [PubMed] [Google Scholar]
- 33.Hiltunen MO, Turunen MP, Hakkinen TP, et al. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med. 2002;7:5–11. doi: 10.1191/1358863x02vm418oa. [DOI] [PubMed] [Google Scholar]
- 34.Castro R, Rivera I, Struys EA, et al. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–96. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 35.Bell B, Rose CL, Damon A. The Veterans Administration longitudinal study of healthy aging. Gerontologist. 1966;6:179–84. doi: 10.1093/geront/6.4.179. [DOI] [PubMed] [Google Scholar]
- 36.Baccarelli A, Wright RO, Bollati V, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–78. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bollati V, Schwartz J, Wright R, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–39. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez B, Budtz-Jorgensen E, Ryan L, Hu H. Structural equation models: a review with applications to environmental epidemiology. JAMA. 2005;100:1443–55. [Google Scholar]
- 39.Gelman A. Prior distributions for variance parameters in hierarchical models. Bayesian Analysis. 2006;1:515–33. [Google Scholar]
- 40.Thierry-Mieg D, Thierry-Mieg J. AceView: integrative annotation of cDNA-supported genes in human, mouse, rat, worm and Arabidopsis. http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly (28 December 2012, date last accessed) [Google Scholar]
- 41.Frantz S, Ertl G, Bauersachs J. Mechanisms of disease: toll-like receptors in cardiovascular disease. Nat Clin Pract Cardiovasc Med. 2007;4:444–54. doi: 10.1038/ncpcardio0938. [DOI] [PubMed] [Google Scholar]
- 42.Chao W. Toll-like receptor signaling: a critical modulator of cell survival and ischemic injury in the heart. Am J Physiol Heart Circ Physiol. 2009;296:H1–H12. doi: 10.1152/ajpheart.00995.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu V, Huang PL. Cardiovascular roles of nitric oxide: a review of insights from nitric oxide synthase gene disrupted mice. Cardiovasc Res. 2008;77:18–29. doi: 10.1016/j.cardiores.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLaren JE, Ramji DP. Interferon gamma: a master regulator of atherosclerosis. Cytokine Growth Factor Rev. 2009;20:125–35. doi: 10.1016/j.cytogfr.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Kleemann R, Zadelaar S, Kooistra T. Cytokines and atherosclerosis: a comprehensive review of studies in mice. Cardiovasc Res. 2008;79:360–76. doi: 10.1093/cvr/cvn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura T, Ebihara I, Takahashi T, et al. Increased interleukin 6 mRNA expression by peripheral blood T cells from patients with IgA nephropathy. Autoimmunity. 1993;15:171–79. doi: 10.3109/08916939309019924. [DOI] [PubMed] [Google Scholar]
- 47.Wannamethee SG, Whincup PH, Rumley A, Lowe GD. Inter-relationships of interleukin-6, cardiovascular risk factors and the metabolic syndrome among older men. J Thromb Haemost. 2007;5:1637–43. doi: 10.1111/j.1538-7836.2007.02643.x. [DOI] [PubMed] [Google Scholar]
- 48.Kuningas M, Mooijaart SP, Slagboom PE, Westendorp RG, van Heemst D. Genetic variants in the glucocorticoid receptor gene (NR3C1) and cardiovascular disease risk. The Leiden 85-plus Study. Biogerontology. 2006;7:231–38. doi: 10.1007/s10522-006-9021-2. [DOI] [PubMed] [Google Scholar]
- 49.Rosmond R, Holm G. A 5-year follow-up study of 3 polymorphisms in the human glucocorticoid receptor gene in relation to obesity, hypertension, and diabetes. J Cardiometab Syndr. 2008;3:132–35. doi: 10.1111/j.1559-4572.2008.00008.x. [DOI] [PubMed] [Google Scholar]
- 50.Newman PE. Can reduced folic acid and vitamin B12 levels cause deficient DNA methylation producing mutations which initiate atherosclerosis? Med Hypotheses. 1999;53:421–24. doi: 10.1054/mehy.1998.0794. [DOI] [PubMed] [Google Scholar]
- 51.Lund G, Andersson L, Lauria M, et al. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004;279:29147–54. doi: 10.1074/jbc.M403618200. [DOI] [PubMed] [Google Scholar]
- 52.Kim M, Long TI, Arakawa K, et al. DNA methylation as a biomarker for cardiovascular disease risk. PloS one. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang AS, Estecio MR, Doshi K, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32: e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bollati V, Fabris S, Pegoraro V, et al. Differential repetitive DNA methylation in multiple myeloma molecular subgroups. Carcinogenesis. 2009;30:1330–35. doi: 10.1093/carcin/bgp149. [DOI] [PubMed] [Google Scholar]
- 56.Szpakowski S, Sun X, Lage JM, et al. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009;448:151–67. doi: 10.1016/j.gene.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi JY, James SR, Link PA, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–97. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou L, Wang H, Sartori S, et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer. 2010;127:1866–74. doi: 10.1002/ijc.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276: 135–41. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 60.Tarantini L, Bonzini M, Apostoli P, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environmental health perspectives. 2009;117:217–22. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pavanello S, Bollati V, Pesatori AC, et al. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer. 2009;125:1692–97. doi: 10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- 62.Wright RO, Schwartz J, Wright RJ, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790–95. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–11. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–84. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 65.Burt VL, Whelton P, Roccella EJ, et al. Prevalence of hypertension in the US adult population. Results from the Third National Health and Nutrition Examination Survey, 1988-1991. Hypertension. 1995;25:305–13. doi: 10.1161/01.hyp.25.3.305. [DOI] [PubMed] [Google Scholar]
- 66.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–13. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


