Abstract
Well-controlled assembly of proteins on supramolecular templates of block copolymers can be extremely useful for high-throughput biodetection. We report the adsorption and assembly characteristics of a model antibody protein to various polystyrene-block-poly(4-vinylpyridine) templates whose distinctive nanoscale structures are obtained through time-regulated exposure to chloroform vapor. Strong adsorption preference of the protein to the polystyrene segment in the diblock copolymer templates leads to an easily predictable, controllable, rich set of nanoscale protein morphologies through self-assembly. We also demonstrate that the chemical identities of various subareas within individual nanostructures can be readily elucidated by investigating the corresponding protein adsorption behavior to each chemically distinct area of the template. In our approach, a rich set of intricate nanoscale morphologies of protein arrays that cannot be easily attained through other means can be generated straightforwardly via self-assembly of proteins on chemically treated diblock copolymer surfaces, without the use of clean room-based fabrication tools. Our approach provides much-needed flexibility and versatility for the use of block copolymer-based protein arrays in biodetection. The ease of fabrication in producing well-defined and self-assembled templates can contribute to a high degree of versatility and simplicity in acquiring intricate nanoscale geometry and spatial distribution of proteins in arrays. These advantages can be extremely beneficial both for fundamental research and biomedical detection, especially in the areas of solid-state based, high-throughput protein sensing.
Keywords: protein assembly, protein adsorption, protein array, diblock copolymer, polymeric nanotemplate
Introduction
Proteins assembled on supramolecular templates of block copolymers can be extremely useful to the area of proteomics and protein sensors due to the highly dense packing density and self-passivation capability demonstrated by various biomolecules on these substrates [14, 15, 18–22, 30]. Both the formation of the underlying nanoscale polymeric guides as well as the biomolecular arrangements on the substrates is driven by self-assembly. In the former case, the immiscibility and degree of polymerization determine the phase separation behavior of polymeric guides, whereas chemical and physical interaction parameters between polymers and proteins govern the specific protein arrangements on the polymer of preferred composition in the latter case. This bottom-up assembly process, in turn, yields well-organized protein arrays whose individual features are periodically arranged nanostructures. Since block copolymers produce characteristic domains with a repeat spacing on the order of tens of nanometers after their phase separation process [2, 6, 7, 10, 26, 31, 36], the spatial resolution of the individually addressable units in the resulting protein arrays is also on the order of nanometers. Creating nanoscale features through conventional lithography techniques can be costly and time-consuming as they require either specially engineered photomasks for a parallel fabrication process or the use of electron beam writing for a serial process. Even with such techniques, fabrication of intricate surface patterns below tens of nanometers cannot be easily and rapidly accomplished. These difficulties are circumvented in the case of spontaneous nanoscale organization through the self-assembly of block copolymers demonstrated in our previous studies [14, 15, 18–21, 30].
In addition to the capability of rapidly producing periodic nanoscale features through self-organization on a large scale, protein arrays created via diblock copolymer nanodomains can be effectively tuned by controlling the phase separation behavior of the underlying diblock copolymers. A variety of important parameters such as desired length scale in periodicity, spatial arrangement in repeated nanostructures, and geometric shape in individually addressable features can be modified. As an additional degree of freedom, a given diblock copolymer template initially produced by controlling the aforementioned variables can be further modified with a post-phase separation process. Recently, it has been shown that chemical treatment methods can be applied directly to the diblock copolymer and other polymers by exposing the surfaces to various chemical environments for modifying surface morphology [3, 11–13, 17, 24, 25, 29, 33, 34, 39, 40, 42]. Solvent annealing approaches effectively adjust the interfacial energies of diblock constituents through chemical selectivity towards one of the two polymeric components and, thus resulting in changes of the original size and shape of polymeric nanostructures. Nanostructures of useful block copolymers such as polystyrene-block-polymethylmethacrylate (PS-b-PMMA) and polystyrene-block-polyvinylpyridine (PS-b-PVP) are often investigated by various solvent annealing methods [3, 11–13, 17, 24, 25, 29, 33, 34, 39, 40, 42].
Understanding protein adsorption behavior on various polymeric surfaces is crucial, as evidenced by the rising demands for highly miniaturized, small-volume detection platforms for examining proteins both in laboratory and clinical diagnosis settings [8, 9, 27, 28]. Such solid-phase assays involving proteins on polymeric array or plate surfaces have the advantage of requiring only a very small amount (a few μL or smaller) of assay reagents in most detection settings [14, 15]. Solid-phase approaches also enable a large number of biosamples to be assayed rapidly and simultaneously. The nanostructures resulting from polystyrene-block-poly(4-vinylpyridine) (PS-b-P4VP) are particularly useful as protein arrays. This is because the control of size, shape, and spacing of individually addressable units in phase-separated PS-b-P4VP can be achieved with two dimensional degrees of freedom as compared to those of PS-b-PMMA nanotemplates which provide only one dimensional degree of spatial periodicity. Increased versatility in template sizes (pattern repeat spacing) and shapes (pattern geometry) that are bottom-up assembled on the nanoscale can allow for the use of protein arrays in basic biology research and biomedical detection. These advancements may also have the potential to embark on a truly nanoscale fabrication of protein arrays which facilitate non diffraction-limited optical detection.
Motivated by the aforementioned importance, we report for the first time the adsorption behavior of a model protein on a newly identified set of the chemically modified PS-b-P4VP micellar nanotemplates. Detailed evolution processes and mechanisms of the solvent-induced, nanoscale morphologies of PS-b-P4VP will be discussed elsewhere. In this paper, we focus on elucidating the unique protein adsorption behavior of anti-bovine immunoglobulin G antibody (anti-IgG) on various PS-b-P4VP nanostructures. Our study can provide insight into the evolution and modification process of the underlying polymeric chains under solvent annealing retrospectively. This can be achieved with spatial resolution down to the nanometer range while providing information on the nanoscale subarea-specific chemical composition simultaneously. We demonstrate that the specific arrangement of the polymeric chains and their chemical identities can be easily ascertained for each nanostructure in the new set of PS-b-P4VP templates by analyzing preferential interactions between proteins and their favored polymeric block. Resolving the exact spatial distribution of particular polymeric chains in various substructures within each individual micelle cannot be readily attained using other methods. This difficulty is due to the lack of straightforward experimental techniques that can simultaneously assess the complex individual nanostructures with topological and chemical resolution on the required sub-nanometer and nanometer level. In addition, the level of morphological sophistication and the nanometer size regime observed in the new set of solvent-induced PS-b-P4VP nanostructures cannot be created in a straight forward manner by using conventional fabrication techniques. The bottom-up method used in our approach to yield highly structured and efficiently assembled proteins eliminates the need for employing time-consuming and costly steps for producing sophisticated nanotemplates via techniques such as electron beam writing and sub-wavelength photolithography [4, 6, 7, 16, 17]. In addition to providing valuable information on nanoscale spatial maps and the corresponding chemical compositions of the intricate polymeric structures, our efforts can be highly beneficial for realizing the rich potential of high-density, diblock copolymer-assisted, protein array-based detection platforms in the areas of nanofabrication and nanobiotechnology.
Materials and Methods
Asymmetric PS-b-P4VP diblock copolymer with an average molecular weight of 68,500 Da is obtained from Polymer Source Inc. (Montreal, Canada). Silicon substrates, obtained from Silicon Inc. (Boise, Idaho), are cleaned with ethanol, acetone, and toluene and spun dry before coating ultrathin PS-b-P4VP films. The diblock contains 70% of PS by weight with a polydispersity of 1.14. Upon dissolution of 0.5% (w/v) PS-b-P4VP in toluene, micelles consisting of a P4VP core and a PS corona readily forms in toluene, a preferential solvent for PS. The amphiphilic PS-b-P4VP diblock copolymer with the specified volume composition is known to form micelles when dissolved in toluene [1, 5, 32, 41]. This micellar solution is then spun on silicon substrates at a speed of 3500~4000 rpm for 1min, in order to produce an ultrathin film of PS-b-P4VP with a thickness of 21 nm. The film thickness is determined by carrying out ellipsometry measurements. Upon spin-casting, hexagonal arrays of PS-b-P4VP micelles spontaneously form on the silicon substrate with an average repeat spacing of 50 nm. Subsequent chemical treatment of the original PS-b-P4VP micellar nanostructures is carried out under the vapor of chloroform, CHCl3, prepared in a small sealed chamber. Solvent-annealed samples are then imaged at various time intervals and the morphologies of the polymeric templates at each temporal stage are recorded by atomic force microscopy (AFM). AFM measurements are carried out using Digital Instruments Multimode Nanoscope IIIa in tapping mode at a scan speed of 1 Hz or lower. Silicon tips with a resonant frequency of 71.5 kHz and a spring constant of 5 N/m are used in our ambient AFM measurements. Homopolymers of PS and P4VP are purchased from Polymer Source, Inc. and dissolved in toluene and a 1:1 (v/v) mixture of acetone and ethanol, respectively, to make a 0.5% (w/v) solution. The homopolymer solution is spun-casted on silicon substrates using the same spin coating condition described above for the diblock copolymer. Dichlorotriazinylaminofluorescein-conjugated, anti-bovine immunoglobulin G antibody (anti-IgG) is received from Rockland Immunochemicals (Gilbertsville, PA) and the lyophilized powder of the proteins is reconstituted in a PBS buffer (10 mM mixture of Na2HPO4 and NaH2PO4, 140 mM NaCl, 3 mM KCl, pH 7.4) to varying concentrations ranging from 0.1 μg/ml to 1 μg/ml. A total of 8 μl of desired protein solution is then deposited onto various chemically treated, ultrathin PS-b-PVP micelle films for 2 min at room temperature. The sample surface is then thoroughly rinsed with PBS buffer followed by blow-drying under a gentle stream of compressed air.
Results and Discussion
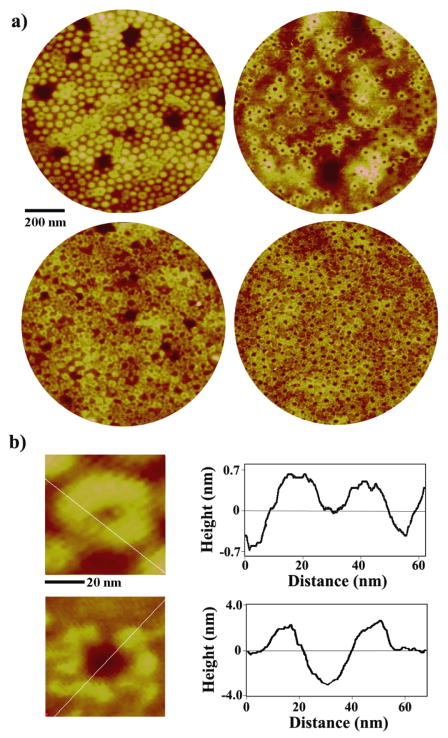
The vapor treatment of the micellar PS-b-P4VP template leads to time-dependent topology which can serve effectively as self-assembling guides for other inorganic or biological nanomaterials. Various, time-specific, surface features can be generated at large-scale via self-assembly using this method [38]. Figure 1 displays the diverse polymeric morphologies of PS-b-P4VP obtained by the aforementioned method that was used specifically in this protein adsorption study. The unique topological characteristics of the polymeric templates can be precisely controlled by varying solvent annealing time, where the series of PS-b-P4VP templates shown in Figure 1 is referred hereto as a) original spheres, b) holes, c) reformed spheres, d) embedded spheres, e) enlarged spheres and f) cylinder precursors, respectively.
Figure 1.
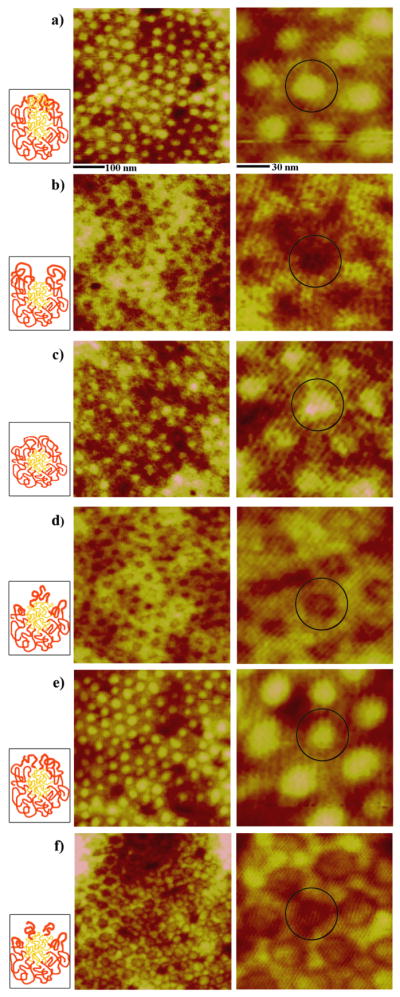
AFM panels of the unique PS-b-P4VP nanostructures monitored during time-dependent exposure to CHCl3 vapor; a) original spheres, b) holes, c) reformed spheres, d) embedded spheres, e) enlarged spheres and f) cylinder precursors. AFM images clearly demonstrate the periodic two-dimensional arrays consisting of the well-defined nanostructures at each time period of annealing for a) 0 h, b) 3 h, c) 9 h, d) 16 h, e) 25 h, and f) 29 h. The scan sizes of the images shown on the left and right column are 500 × 500 nm and 140 × 140 nm, respectively. A dark circle is inserted in the zoomed-in AFM panels in order to highlight each, independently addressable, PS-b-P4VP unit in its hexagonally packed configuration. As a guide, illustrations showing possible polymeric chain distributions in each micelle are inserted next to each AFM panel for each time-dependent PS-b-P4VP nanostructure. Red and yellow chains in the cartoons represent PS and P4VP, respectively.
Anti-IgG is chosen as a model protein system for assembly on PS-b-P4VP because it is widely used in a variety of protein detection settings to monitor antigen-antibody complex formation, enzymatic activity, and affinity-based purification. In addition, the size of the individual protein is smaller than the size of the nanostructures found in PS-b-P4VP thin films and the spherical shape of the protein matches closely with the ones of the polymer nanotemplates.
Periodic arrays of self-assembled proteins are readily formed on various PS-b-P4VP templates that are pretreated with the selective vapor of CHCl3. The highly discriminating nature of proteins against the P4VP polymeric block drives the extremely selective spatial organization of the proteins to PS-rich areas only. Hydrophobic interaction plays an important role as the driving force of the selective adsorption of the anti-IgG proteins to the PS phase. In a buffer environment, the interaction of anti-IgG proteins to water is unfavorable similar to that of another globular protein, bovine serum albumin [35]. The enthalpy is small and negative, and there is a large contribution from the negative entropy. When a hydrophobic solid surface is present, the entropic effect becomes largely positive as the protein surface near the solid substrate can exclude water molecules which, in turn, yields negative Gibbs free energy. The presence of a hydrophilic surface such as P4VP leads to an opposite effect.
In this process of protein adsorption, surface regions consisting of the P4VP block are left completely free of adsorbed proteins, serving as naturally generated areas of a passivated surface. The exclusive adsorption of proteins on the chemically competing nanoscale templates can eliminate the need for ‘surface passivation’ often required for producing protein arrays. The strong tendency of many proteins to adsorb randomly on various surfaces can hinder sensitivity and specificity of protein arrays and, therefore, treatments such as passivating certain surface areas of protein arrays chemically or biologically have been conventionally used in order to avoid this problem. In our approach, the chemically heterogeneous nature of the block copolymer nanotemplates serves as a ‘built-in’ self-passivating arrays where the facilitation and prevention of protein adsorption can be controlled precisely on the nanoscale by varying the size and shape of the polymeric nanotemplates.
Our AFM results demonstrate that the same type of diblock, PS-b-P4VP, can be successfully modified and utilized to create an assortment of highly intricate, nanoscale protein patterns which are otherwise difficult to produce. Figures 2 and 3 display 0.1 μg/ml of anti-IgG molecules deposited on the series of chemically treated PS-b-P4VP micellar templates. Proteins tend to decorate the outer edges of the micelles while leaving the centers of the spheres devoid of proteins. Our previous studies on similar diblock copolymer templates confirm that proteins such as immunoglobulin G and serum albumins show extremely high affinity for PS domains [18–21, 30].
Figure 2.
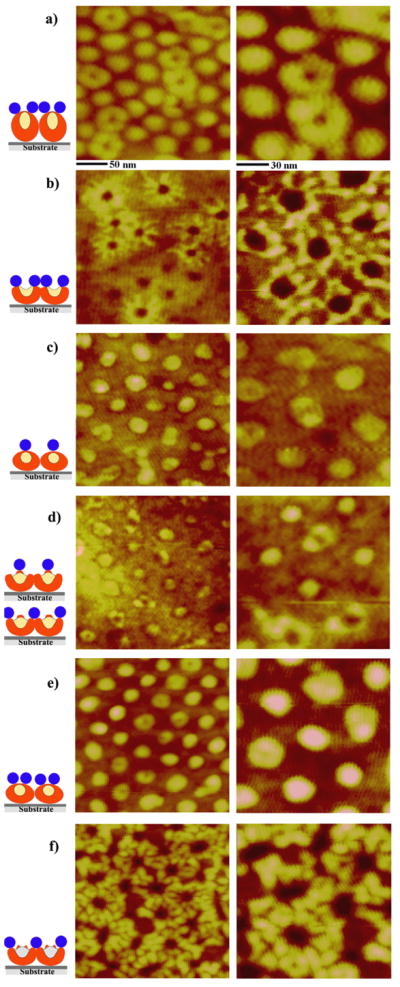
AFM images displaying the protein adsorption characteristics on each of the six polymeric nanotemplates indicated in Figure 1. The scan sizes of the topography images shown on the left and right column are 250 × 250 nm and 140 × 140 nm, respectively. The concentration of protein solution deposited onto various PS-b-P4VP templates is 0.1 μg/ml. This concentration is well below the saturation coverage, i.e. not all available PS areas on the polymeric film surface are covered by anti-IgG at this concentration. As a guide to understand the complex structures after protein adsorption, side-view illustrations of the AFM data are added. The proteins adsorbed on various surface locations of each polymer nanostructure are depicted in the illustrations as red (PS), yellow (P4VP), and blue (anti-IgG) areas.
Figure 3.
(a) Anti-IgG assembled on the PS-b-PVP nanotemplates of original spheres (left) and holes (right), showing the protein adsorption behavior on the ultrathin film on a large scale. AFM panels of 1 μm in diameter are obtained after depositing 0.1 μg/ml (top row) and 0.5 μg/ml (bottom row) protein solutions. (b) Zoomed-in, 60 × 60 nm, AFM topography of anti-IgG proteins decorating original micellar (top) and hole (bottom) templates. Height profiles along the inserted white lines are displayed on the right to clearly show the morphology of anti-IgG upon adsorption to each template.
If a micelle in the film consists of a P4VP core and a PS corona as expected in solution where PS is uniformly encapsulating the inner P4VP core, the adsorption tendency of the proteins should show no inclination for a particular sub-region within a micelle and proteins should evenly distribute themselves on the entire surface of the micelle. However, our AFM results from anti-IgG adsorption repeatedly confirm that at both low (less than monolayer coverage) and high (saturation coverage) concentrations, proteins evade the center surface areas of the micelles during adsorption, see Figures 2 and 3. This observation, in conjunction with the known adsorption tendency of the protein, demonstrates that the center of the sphere is P4VP rich at the air/polymer interface. This phenomenon suggests that PS-b-P4VP micelles tend to adjust their isotropic spherical configuration when prepared on a solid substrate. Due to the presence of the underlying silicon surface, each micelle is likely to be flattened in the vertical direction and take the form of a collapsed micelle. Unlike its concentric core/corona micellar counterpart of PS-b-P4VP found in solution, our data indicate that the center region of the collapsed micelle prepared on a substrate is devoid of PS blocks in its corona structure near the surface and that the PS chains are likely to have moved down toward the substrate. As a result of this rearrangement of PS chains, some P4VP fragments close to the surface of the micelle, which would have otherwise been buried in the inner core, are now exposed to the air/polymer interface.
Another interesting behavior of protein adsorption occurs at low protein coverage resulting from 0.1 μg/ml deposition. In these instances, proteins occupy less than a monolayer of the available PS-b-P4VP surface sites. Anti-IgG assembly favors fully saturating a subset of micelles with proteins rather than partially filling in all available micelles with a few proteins per micelle, see Figure 2(a). Each saturated micellar template hosts four to five individual protein molecules around its center. When a group of saturated micelles is nearby, a network of loops is observed where each loop consists of four to five beads of protein molecules. The maximum number of proteins that can decorate each micelle is determined by the size of the protein and the micellar template. No proteins are found at the center areas of the loops nor at the shared areas between neighboring loops. The relatively high presence of P4VP chains at the air/polymer interface is attributed to the lack of adsorbed proteins at the center of the loops, whereas steric hindrance from proteins forming the loops is responsible for limiting protein adsorption in the neighboring areas.
Because the sizes of the polymeric templates and proteins are well-defined, it is possible to predict and control the maximum number of proteins that can be gathered on each template. The size of the micellar templates can be controlled in order to fine-tune the adsorption density of proteins per given surface area of the substrate. A micellar template of approximately 50 nm in diameter will host three globular proteins with a diameter of ~15 nm on average if the proteins tend to occupy the top of the micellar template. The same polymeric template will be able to accommodate approximately six protein molecules of the same size around the perimeter of the micelle if protein adsorption favors the outer periphery of the template.
Quantitative analysis and comparison of detection signal from multiple locations on the templates are often necessary in protein array measurements. However, the task of obtaining well-controlled density of proteins on arrays is difficult to achieve using conventional protein printing methods. They require either additional fabrication steps for array production or user-calibration processes for the normalization of background signal between all spots in the arrays prior to the intended protein measurements. The unique ability of the protein arrays prepared by our method in controlling the exact protein density on surfaces can overcome the drawbacks of conventional arrays and, therefore, is extremely beneficial for quantitatively comparable protein detection.
Exposing spherical PS-b-P4VP micelles in the original template to chloroform vapor for up to 3 h leads to a hole geometry due to the favored interaction between the P4VP block and chloroform, Figure 1(b). Figure 2(b) displays 0.1 μg/ml anti-IgG deposited on the hole template, where both bare holes as well as those decorated with proteins are observed at this low deposition concentration. It is evident that anti-IgG proteins deposited onto this hole template completely avoid the depressed regions in the polymer film where the dominant polymeric phase is the surface-exposed P4VP. This arrangement of the proteins onto the hole template is, in part, similar to the process observed on the original template. At the same time, the assembled proteins on holes exhibit an additional grouping geometry that is not seen on the original template. Strings of proteins are found to be placed into distinctive lines emanating from the protein loops around the holes. This assembly behavior results in a unique windmill-like pattern of proteins at this surface coverage as seen in Figure 2(b). The formation of these distinctive lines may indicate that there is a dominant morphological pathway for PS chains to fold away from the center of the original sphere during the chemically-induced rearrangement process of the polymeric chains into holes. Similar to the observation in the original templates, not all available hole templates are occupied by proteins at this concentration. Proteins also show the same tendency to complete assembling around a subset of hole templates, instead of incompletely distributing among all available hole templates with only a few protein molecules per hole. This behavior is seen clearly in Figure 3 which displays AFM panels acquired from a larger scan area. The original micelles in Figure 3(a) exhibit a repeat spacing and height profile of 50 nm and 2.43 nm, respectively. The width and height of a single anti-IgG protein adsorbed on top of the sphere are measured as 14.8 nm and 0.59 nm, respectively. The protein presents no net charge under the specified deposition condition. The Y-shaped IgG antibody is known to exhibit widths along the short and long axes and a height of 14.5 nm, 8.5 nm, 4.0 nm, respectively [23, 37]. The height of the observed IgG antibody is much smaller than expected due to the tapping of the Si tip onto the surface of the soft protein sample during AFM imaging. This observation is similar to previously reported AFM measurements [18–21]. However, a strikingly different outcome is revealed when carrying out a line analysis of the same protein on the hole template. Anti-IgG proteins in Figure 3(b) adsorbed on the hole nanostructures, showing an average repeat spacing of 49 nm and depth of 2.5 nm, are measured to have a greatly reduced width and an increased height of 9.2 nm and 1.9 nm, respectively. This observation indicates that the anti-IgG proteins near more hydrophilic chains of P4VP sit tighter on the adhesion area of the surface while pushing the protein domains upward away from the polymer.
Further exposure of the hole template to the chloroform vapor for up to 9 h yields reformed spheres, Figure 1(c). These structures are similar to the original templates but exhibit much shallower height variations within micelles. Anti-IgG proteins prefer to gather on the tops of spheres, Figure 2(c). However, no pronounced tendency to resist protein adsorption in the middle of the sphere is seen on the reformed spheres. This observation indicates that PS chains in the reformed spheres sweep back fully toward the center of the sphere when transitioning from holes to the reformed spheres. This process ends up burying the unfavored P4VP chains deep into the film. On average, each reformed sphere accommodates three protein molecules on its surface.
The adsorption behavior of proteins on embedded spheres obtained by vapor annealing of the template for up to 16 h shows two dominant adsorption pathways in the 0.1 μg/ml concentration regime, Figure 2(d). One adsorption route is for the protein molecules to engage in the center lobes of the embedded spheres. The other path is for the proteins to encircle around the outer edges of the embedded spheres. The surface-assembled proteins serve as a faithful indicator of the chemical phase difference of the underlying polymeric blocks of PS and P4VP. The protein molecules adsorb onto the embedded sphere at its center whose dominant surface phase is PS. At the same time, the nested sphere in the center is encircled by a circular band of depressed area where no proteins are found. The lack of protein adsorption in this area suggests that P4VP chains are either exposed to or lie very close to the air/polymer interface. In addition, the proteins tend to gather outside the depressed band regions, indicating that the surface areas outside of the circular bands are predominantly PS. Anti-IgG proteins do not show strong preference to take one of the two available adsorption paths at this concentration as each case is represented by approximately half of the templates. Assembled proteins on the enlarged spheres created by annealing the polymeric template for up to 25 h closely mimic the adsorption behavior observed on the reformed spheres. Proteins prefer to adsorb on top of the enlarged spheres at a deposition concentration of 0.1 μg/ml, Figure 2(e).
The next chloroform annealing stage to 29 h yields cylinder precursor templates, whose individual nanostructures can be viewed as two concentric rings of alternating PS (brighter rings in the AFM topography) and P4VP (darker depressed areas between the two rings in the AFM topography), Figure 1(f). At the same protein concentration of 0.1 μg/ml, proteins gather on top of the two concentric rings as shown in Figure 2(f). A striking adsorption behavior of the proteins is monitored on these cylinder precursors; the proteins tend to assemble in much higher density than what is observed from prior templates. This tendency is confirmed with repeated measurements although the exact reason for the increase in protein density on cylinder precursors is not yet clear. From our earlier study that was carried out on a series of polystyrene and polymethylmethacrylate homopolymers, blends, and diblock copolymers, it was demonstrated that the chemical heterogeneity of template surfaces plays a very important role in protein adsorption, especially with regards to achieving high protein density [20]. Similar effects may be seen in our current study where the density of adsorbed proteins is highest in the polymeric nanotemplates whose degree of chemical heterogeneity on surfaces is the largest: i.e. when using identical protein deposition conditions, protein density is the highest in the cylinder precursor templates amongst the various vapor-annealed PS-b-PVP templates. When comparing the most plausible polymeric chain compositions between the six templates used in our protein experiment, the heterogeneity in chemical composition of a given surface area is expected to be the highest in cylinder precursors. This may explain why the unusually high protein density is achieved on cylinder precursor templates.
At a higher protein concentration of 0.5 μg/ml, the proteins engage all available template nanostructures and reach close to a monolayer-forming surface coverage of the proteins. We define monolayer coverage as the surface coverage where all available and preferred surface adsorption sites of PS are taken by proteins. Protein molecules adsorbed on the original micelles before any vapor annealing favor the PS domains on the spheres while avoiding the center surfaces of the micellar spheres at this concentration. This observation is identical to the protein adsorption behavior monitored at the lower concentration of 0.1 μg/ml. The resulting network of adsorbed proteins is formed by the complex link between protein bands on all available original micelles, where each band consists of four to five individual protein molecules, see Figure 4(a). Protein assembly on the hole template after deposition of the 0.5 μg/ml solution also leads to the involvement of all existing nanotemplates. Every available PS area encircling the holes is engaged with proteins. The wind-mill patterns of the assembled proteins originating from each hole are monitored as in the previous case of the lower concentration and, at this higher concentration, they are intricately interlaced with each other, Figure 4(b). On reformed spheres, proteins deposited at a concentration of 0.5 μg/ml fill all available areas on the tops of the spheres and, on some places of the nanotemplates, begin to spread out into the regions between the reformed spheres. This result is displayed in the AFM panel of Figure 4(c) as merged patches of various sizes and shapes, derived from the amalgamation of protein domains from neighboring templates of reformed spheres. No strong resistance against protein adsorption is seen in the center region of the reformed sphere, a phenomenon similar to that of the assembly on reformed spheres at the lower concentration. Proteins deposited on the embedded spheres from the 0.5 μg/ml solution primarily adsorb on the outside perimeter of the nanostructure. Five to six protein molecules circle around each embedded sphere, Figure 4(d). Unlike the assembly behavior of the lower concentration of deposited protein, no embedded spheres show proteins positioned on the nested sphere (center lobe) of the template and proteins preferentially adsorb to PS domains located outside the embedded spheres at this increased concentration. Proteins on enlarged spheres closely resemble the adsorption behavior of protein on the reformed spheres at the same concentration. At a deposition concentration of 0.5 μg/ml, proteins occupy all available sites on the tops of the enlarged spheres and they additionally start to fill up surface regions between enlarged spheres, see Figure 4(e). Assembled proteins on cylinder precursors at the same solution concentration, Figure 4(f), pack all areas encircling the nanostructures as well as neighboring areas between nanostructures. This supramolecular assembly behavior leads to the formation of two concentric protein bands that are connected together into large-scale protein networks, as shown in the zoomed-in AFM topography panel. The distinctively assembled protein structures described in this paper are observed consistently on the template substrate whose typical size was kept as 0.5 × 0.5 cm2 in our experiments.
Figure 4.
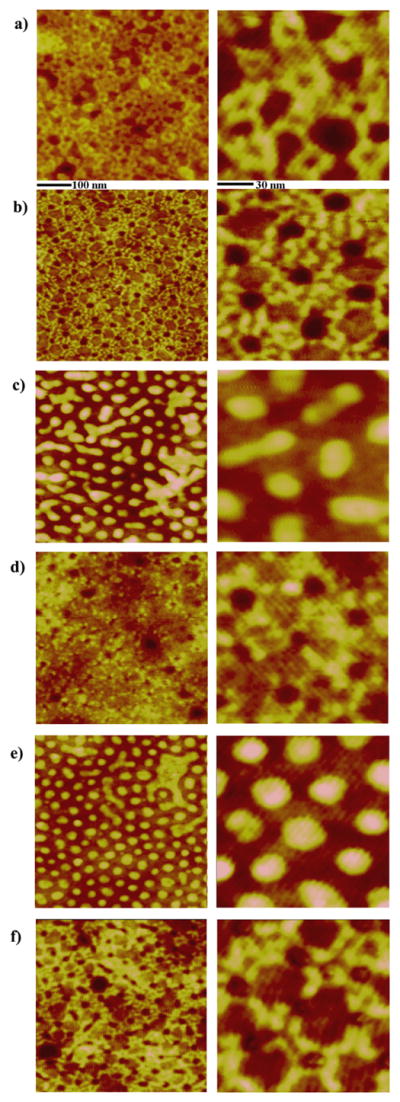
AFM images displaying protein adsorption characteristics on each of the six polymeric PS-b- P4VP nanotemplates. The concentration of protein solution used is 0.5 μg/ml, which is approximately at the saturation coverage. The scan sizes of AFM panels displayed in the left and right columns are 500 × 500 nm and 140 × 140 nm, respectively.
The characteristic adsorption behavior of proteins on each polymeric nanotemplate displayed in Figures 2 through 4 occurs throughout the sample where the assembly of the proteins takes place instantaneously at large scale. The reproducibility of the unique protein assembly behavior is confirmed by repeated measurements on at least four sets of CHCl3-annealed template series and the height/width data of the adsorbed proteins are obtained from analyzing at least 50 unit structures per template. High magnification images are provided in Figures 2 through 4 in order to show both the configurations of the proteins and the underlying polymeric nanotemplates clearly. Fluorescence images were then taken with a Zeiss A2M microscope in order to assess the functionality of adsorbed proteins on PS-b-P4VP templates, Figures 5a and 5b. Due to the optical diffraction limit, fluorescence signal from individual nanoscale templates cannot be resolved. However, the strong fluorescence signal combined with the AFM data indicates that a large portion of the protein molecules keep their bioactivity upon their surface adsorption onto the polymer film. More systematic and quantitative measurements are currently underway in order to investigate the degree of denaturation and biofunctionality of the proteins upon surface adsorption to all the aforementioned PS-b-P4VP nanotemplates. When ultrathin homopolymer films of PS and P4VP are used instead of the diblock copolymer, the strong adsorption preference of the protein to PS is unambiguously determined by the significant differences in protein density on the two homopolymer films. The phase AFM panels displayed in Figures 5c and 5d show that DTAF-antiIgG density is much higher on the PS homopolymer film than on the P4VP homopolymer surface, even though identical biodeposition conditions are used to prepare the two samples.
Figure 5.
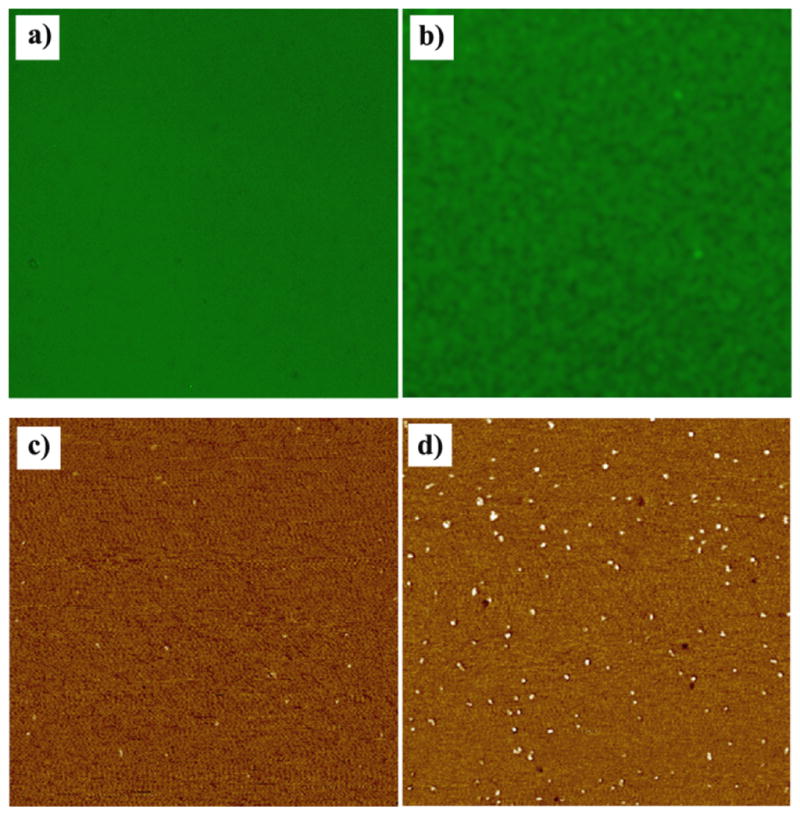
150 × 150 μm fluorescence panels obtained from self-assembled proteins on PS-b-P4VP templates and 1.5 × 1.5 μm AFM phase panels collected from DTAF-antiIgG molecules treated identically on the two homopolymer surfaces of P4VP and PS. (a) Fluorescence signal obtained from 0.5 μg/ml DTAF-antiIgG molecules adsorbed on the cylinder precursor PS-b-P4VP template. (b) Fluorescence signal from IgG/DTAF-antiIgG proteins interacting on the original PS-b-P4VP micellar template. A layer of 0.4 μg/ml IgG molecules was deposited onto the polymeric surface and a solution of 4 μg/ml DTAF-antiIgG was subsequently added to the sample, allowing protein-protein interaction. Before the fluorescence measurement, the sample was rinsed with an ample amount of PBS to remove any unbound proteins after the reaction. When 0.5 μg/ml of DTAF-antiIgG was identically applied on the two homopolymer surfaces of (c) P4VP and (d) PS, the strong adsorption preference of the protein to PS over P4VP was confirmed from the much higher surface coverage of the proteins on PS homopolymer.
In summary, we have determined the adsorption and assembly characteristics of anti-IgG on various PS-b-P4VP templates, whose unique and intricate nanoscale structures are produced by time-dependent annealing of the polymeric surfaces under a selective solvent vapor. Strong adsorption preference of the proteins to certain segment in the diblock copolymer templates leads to an easily predictable, controllable, and rich set of nanoscale protein arrays through self-assembly. In our approach, a rich set of intricate nanoscale morphologies of protein arrays is successfully produced via self-assembly of proteins on chemically treated diblock copolymer surfaces, without the use of clean room-based fabrication tools. These morphologies cannot be easily accessed by the well-known microphase separation of block copolymers or any other lithographic means. Therefore, our new method can offer a high degree of flexibility and versatility for the use of block copolymer-based protein arrays in biodetection. We also demonstrate that polymer compositions of hard-to-probe, sub-domain features within individual nanostructures can be readily elucidated by investigating the corresponding protein adsorption behavior. Due to the lack of available techniques simultaneously providing site-specific and high vertical and lateral resolution, correlating sub-nanometer scale topological details within individual nanostructures to their surface chemical compositions cannot be readily. In our approach, the specific arrangement of polymeric chains and the chemical identities associated with each nanostructure can be easily determined by utilizing the high discriminating power of the proteins during surface adsorption. Immediate applications of these protein nanoarrays will most likely involve the use of currently available microsample handling and conventional detection apparatuses. In this case, a known number of protein molecules in a group of nanoscale micelles will serve as an independently addressable, quantitative microscale unit in a given protein array. However, ideal applications of our protein arrays should address each micelle as an independent nanoscale assay unit that can be further utilized for protein detection apparatuses with nanoscale detection resolution. Together with the ease of fabrication in producing polymeric templates with well-defined nanoscale structures via self-assembly, the diverse arrays of proteins created using our approach can promote a large degree of versatility in creating high-density, high-payload, protein arrays. These advantages can be extremely beneficial to both miniaturized and low cost fundamental biology research as well as biomedical detection, especially in the areas of high-throughput protein sensing and screening.
Acknowledgments
The author acknowledges support on this work by the National Institutes of Health, National Research Service Award (1R01DK088016-01) from the National Institute of Diabetes And Digestive And Kidney Diseases and by the National Science Foundation under Grant Number CHE-REU0851581 from the Division of Chemistry under the Directorate for Mathematical and Physical Sciences.
References
- 1.Bang J, Jeong U, Ryu DY, Russell TP, Hawker CJ. Block copolymer nanolithography: translation of molecular level control to nanoscale patterns. Advanced Materials. 2009;21:4769–92. doi: 10.1002/adma.200803302. [DOI] [PubMed] [Google Scholar]
- 2.Bates FS, Fredrickson GH. Block copolymer thermodynamics - Theory and experiment. Annu Rev Phys Chem. 1990;41:525–57. doi: 10.1146/annurev.pc.41.100190.002521. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Wang Z, Gong Y, Huang H, He T. Solvent-induced novel morphologies in diblock copolymer blend thin films. J Phys Chem B. 2006;110:1647–55. doi: 10.1021/jp054260k. [DOI] [PubMed] [Google Scholar]
- 4.Chiu JJ, Kim BJ, Yi G-R, Bang J, Kramer EJ, Pine DJ. Distribution of nanoparticles in lamellar domains of block copolymers. Macromolecules. 2007;40:3361–5. [Google Scholar]
- 5.Choucair A, Eisenberg A. Control of amphiphilic block copolymer morphologies using solution conditions. Eur Phys J E. 2003;10:37–44. doi: 10.1140/epje/e2003-00002-5. [DOI] [PubMed] [Google Scholar]
- 6.Darling SB. Directing the self-assembly of block copolymers. Prog Polym Sci. 2007;32:1152–204. [Google Scholar]
- 7.Darling SB. Block copolymers for photovoltaics. Energy Environ Sci. 2009;2:1266–73. [Google Scholar]
- 8.Ekins R, Chu F. Immunoassay and other ligand assays: present status and future trends. J Int Fed Clin Chem. 1997;9:100–9. [PubMed] [Google Scholar]
- 9.Emili AQ, Cagney G. Large-scale functional analysis using peptide or protein arrays. Nat Biotechnol. 2000;18:393–7. doi: 10.1038/74442. [DOI] [PubMed] [Google Scholar]
- 10.Fredrickson GH, Bates FS. Dynamics of block copolymers: theory and experiment. Annu Rev Mater Sci. 1996;26:501–50. [Google Scholar]
- 11.Fukunaga K, Elbs H, Magerle R, Krausch G. Large-scale alignment of ABC block copolymer microdomains via solvent vapor treatment. Macromolecules. 2000;33:947–53. [Google Scholar]
- 12.Guo R, Huang H, Chen Y, Gong Y, Du B, He T. Effect of the nature of annealing solvent on the morphology of diblock copolymer blend thin films. Macromolecules. 2008;41:890–900. [Google Scholar]
- 13.Guo R, Huang H, Du B, He T. Solvent-induced morphology of the binary mixture of diblock copolymer in thin film: The block length and composition dependence of morphology. J Phys Chem B. 2009;113:2712–24. doi: 10.1021/jp808551j. [DOI] [PubMed] [Google Scholar]
- 14.Hahm J. Functional polymers in protein detection platforms: Optical, electrochemical, electrical, mass-sensitive and magnetic sensors. Sensors. 2011;11:3327–55. doi: 10.3390/s110303327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahm J. Polymeric surface-mediated protein assembly: Macroscopic and nanoscale approaches for biosensor applications. J Biomed Nanotech. 2011;7:731–42. doi: 10.1166/jbn.2011.1341. [DOI] [PubMed] [Google Scholar]
- 16.Kim BJ, Bang J, Hawker CJ, Kramer EJ. Effect of areal chain density on the location of polymer-modified gold nanoparticles in a block copolymer template. Macromolecules. 2006;39:4108–14. [Google Scholar]
- 17.Kim TH, Huh J, Hwang J, Kim H, Kim SH, Sohn B, Park C. Ordered arrays of PS-b-P4VP micelles by fusion and fission process upon solvent annealing. Macromolecules. 2009;42:6688–97. [Google Scholar]
- 18.Kumar N, Hahm J. Nanoscale protein patterning using self-assembled diblock copolymers. Langmuir. 2005;21:6652–5. doi: 10.1021/la050331v. [DOI] [PubMed] [Google Scholar]
- 19.Kumar N, Parajuli O, Dorfman A, Kipp D, Hahm J. Activity study of self-assembled proteins on nanoscale diblock copolymer templates. Langmuir. 2007;23:7416–22. doi: 10.1021/la063563i. [DOI] [PubMed] [Google Scholar]
- 20.Kumar N, Parajuli O, Gupta A, Hahm J. Elucidation of protein adsorption behavior on polymeric surfaces: Towards high density, high payload, protein templates. Langmuir. 2008;24:2688–94. doi: 10.1021/la7022456. [DOI] [PubMed] [Google Scholar]
- 21.Kumar N, Parajuli O, Hahm J. Two-dimensionally self-arranged protein nanoarrays on diblock copolymer templates. J Phys Chem B. 2007;111:4581–7. doi: 10.1021/jp068509p. [DOI] [PubMed] [Google Scholar]
- 22.Lau KHA, Bang J, Kim DH, Knoll W. Self-assembly of protein nanoarrays on block copolymer templates. Adv Funct Mater. 2008;18:3148–57. [Google Scholar]
- 23.Lee K-B, Park S-J, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295:1702–5. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Peng J, Wen Y, Kim DH, Knoll W. Morphology change of asymmetric diblock copolymer micellar films during solvent annealing. Polymer. 2007;48:2434–43. [Google Scholar]
- 25.Li Y, Huang H, He T, Gong Y. The effect of the preferential affinity of the solvent on the microstructure of solution-cast block copolymer thin films. J Phys Chem B. 2010;114:1264–70. doi: 10.1021/jp908852u. [DOI] [PubMed] [Google Scholar]
- 26.Lodge TP. Block copolymers: Past successes and future challenges. Macromol Chem Phys. 2003;204:265–73. [Google Scholar]
- 27.MacBeath G. Protein microarrays and proteomics. Nat Genetics. 2002;32:526–32. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 28.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–3. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 29.Niu S, Saraf RF. Stability of order in solvent-annealed block copolymer thin films. Macromolecules. 2003;36:2428–40. [Google Scholar]
- 30.Parajuli O, Gupta A, Kumar N, Hahm J. Evaluation of enzymatic activity on nanoscale PS-b-PMMA diblock copolymer domains. J Phys Chem B. 2007;111:14022–7. doi: 10.1021/jp075909j. [DOI] [PubMed] [Google Scholar]
- 31.Park C, Yoon J, Thomas EL. Enabling nanotechnology with self assembled block copolymer patterns. Polymer. 2003;44:6725–60. [Google Scholar]
- 32.Park S, Kim B, Yavuzcetin O, Tuominen MT, Russell TP. Ordering of PS-b-P4VP on patterned silicon surfaces. ACS Nano. 2008;2:1363–70. doi: 10.1021/nn800073f. [DOI] [PubMed] [Google Scholar]
- 33.Peng J, Kim DH, Knoll W, Xuan Y, Li B, Han Y. Morphologies in solvent-annealed thin films of symmetric diblock copolymer. J Chem Phys. 2006;125:064702. doi: 10.1063/1.2219446. [DOI] [PubMed] [Google Scholar]
- 34.Peng J, Xuan Y, Wang H, Li B, Han Y. Solvent vapor induced dewetting in diblock copolymer thin films. Polymer. 2005;46:5767–72. [Google Scholar]
- 35.Roach P, Farrar D, Perry CC. Interpretation of protein adsorption: Surface-induced donformational dhanges. JACS. 2005;127:8168–73. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- 36.Segalman R. Patterning with block copolymer thin films. Mater Sci Eng R. 2005;48:191–226. [Google Scholar]
- 37.Silverton EW, Navia MA, Davies DR. Three-dimensional structure of an intact human immunoglobulin. PNAS. 1977;74:5140–4. doi: 10.1073/pnas.74.11.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song S, Milchak M, Zhou H, Lee T, Hanscom M, Hahm J-i. Elucidation of novel nanostructures by time-lapse monitoring of polystyrene-block-polyvinylpyridine under chemical treatment. Langmuir. 2012 doi: 10.1021/la301585k. ASAP. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Hu Z, Chen Y, Gong Y, Huang H, He T. Rhythmic growth-induced concentric ring-banded structures in poly(e-caprolactone) solution-casting films obtained at the slow solvent evaporation rate. Macromolecules. 2007;40:4381–5. [Google Scholar]
- 40.Xuan Y, Peng J, Cui L, Wang H, Li B, Han Y. Morphology development of ultrathin symmetric diblock copolymer film via solvent vapor treatment. Macromolecules. 2004;37:7301–7. [Google Scholar]
- 41.Zhang L, Yu K, Eisenberg A. Ion-induced morphological changes in “crew-cut” aggregates of amphiphilic block copolymers. Science. 1996;272:1777–9. doi: 10.1126/science.272.5269.1777. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, Jiang S, Ji X, An L, Jiang B. Study of the time evolution of the surface morphology of thin asymmetric diblock copolymer films under solvent vapor. Polymer. 2005;46:6513–21. [Google Scholar]